Atomic Details of Carbon-Based Nanomolecules Interacting with Proteins
Abstract
1. Introduction
- Fullerene and nanotubes;
- Cryptophane and macrocages;
- Supramolecular host-guest molecules: cyclodextrins, calixarenes, cucurbiturils, and molecular tweezers.
2. All-Carbon Nanomolecules Interacting with Proteins
3. Carbon-Based Nanomolecules Interacting with Proteins
3.1. Molecular Cages
3.2. Calixarene Molecules
3.3. Cyclodextrins
3.4. Cucurbituril Molecules
3.5. Molecular Tweezers
4. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Kroto, H.W.; Heath, J.R.; O’Brien, S.C.; Curl, R.F.; Smalley, R.E. C60: Buckminsterfullerene. Nature 1985, 318, 162–163. [Google Scholar] [CrossRef]
- Iijima, S. Helical microtubules of graphitic carbon. Nature 1991, 354, 56–58. [Google Scholar] [CrossRef]
- Iijima, S.; Ichihashi, T. Single-shell carbon nanotubes of 1-nm diameter. Nature 1993, 363, 603–605. [Google Scholar] [CrossRef]
- Ragab, T.; Basaran, C. Joule heating in single-walled carbon nanotubes. J. Appl. Phys. 2009, 106, e063705. [Google Scholar] [CrossRef]
- Cai, L.; Wang, C. Carbon nanotube flexible and stretchable electronics. Nanoscale Res. Lett. 2015, 10, 320. [Google Scholar] [CrossRef]
- Park, S.; Vosguerichian, M.; Bao, Z. A review of fabrication and applications of carbon nanotube film-based flexible electronics. Nanoscale 2013, 5, 1727–1752. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew. Chem. Int. Ed. 2001, 40, 4128–4158. [Google Scholar] [CrossRef]
- Stankovich, S.; Dikin, D.A.; Dommett, G.H.B.; Kohlhaas, K.M.; Zimney, E.J.; Stach, E.A.; Piner, R.D.; Nguyen, S.B.T.; Ruoff, R.S. Graphene-based composite materials. Nature 2006, 442, 282–286. [Google Scholar] [CrossRef]
- Singh, V.; Joung, D.; Zhai, L.; Das, S.; Khondaker, S.I.; Seal, S. Graphene based materials: Past, present and future. Prog. Mater. Sci. 2011, 56, 1178–1271. [Google Scholar] [CrossRef]
- Reina, G.; González-Domínguez, J.M.; Criado, A.; Vázquez, E.; Bianco, A.; Prato, M. Promises, facts and challenges for graphene in biomedical applications. Chem. Soc. Rev. 2017, 46, 4400–4416. [Google Scholar] [CrossRef]
- Convertino, D.; Luin, S.; Marchetti, L.; Coletti, C. Peripheral neuron survival and outgrowth on graphene. Front. Neurosci. 2018, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.; Nilsson, K.; Sundahl, M.; Westman, G.; Wennerström, O. C60 embedded in γ-cyclodextrin: A water-soluble fullerene. J. Chem. Soc. Chem. Commun. 1992, 1992, 604–606. [Google Scholar]
- Wharton, T.; Kini, V.U.; Mortis, R.A.; Wilson, L.J. New non-ionic, highly water-soluble derivatives of C60 designed for biological compatibility. Tetrahedron Lett. 2001, 42, 5133–5134. [Google Scholar] [CrossRef]
- Zakharian, T.Y.; Christianson, D.W. Design and synthesis of C60-benzenesulfonamide conjugates. Tetrahedron Lett. 2010, 51, 3599–3696. [Google Scholar] [CrossRef]
- Pumera, M.; Sofer, Z. Towards stoichiometric analogues of graphene: Graphane, fluorographene, graphol, graphene acid and others. Chem. Soc. Rev. 2017, 46, 4450–4463. [Google Scholar] [CrossRef]
- Aaron, J.A.; Chambers, J.M.; Jude, K.M.; Di Costanzo, L.; Dmochowski, I.J.; Christianson, D.W. Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II. J. Am. Chem. Soc. 2008, 130, 6942–6943. [Google Scholar] [CrossRef]
- Calvaresi, M.; Zerbetto, F. The devil and holy water: Protein and carbon nanotube hybrids. Acc. Chem. Res. 2013, 46, 2454–2463. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; del Rodriguez-Torres, M.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef]
- Morgan, H.P.; McNae, I.W.; Hsin, K.Y.; Michels, P.A.M.; Fothergill-Gilmore, L.A.; Walkinshaw, M.D. An improved strategy for the crystallization of Leishmania mexicana pyruvate kinase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 215–218. [Google Scholar] [CrossRef]
- Goel, M.; Jain, D.; Kaur, K.J.; Kenoth, R.; Maiya, B.G.; Swamy, M.J.; Salunke, D.M. Functional equality in the absence of structural similarity. J. Biol. Chem. 2001, 276, 39277–39281. [Google Scholar] [CrossRef]
- Warren, A.J.; Crawshaw, A.D.; Trincao, J.; Aller, P.; Alcock, S.; Nistea, I.; Salgado, P.S.; Evans, G. In vacuo X-ray data collection from graphene-wrapped protein crystals. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.H.; DeCamp, D.L.; Kenyon, G.L.; Sijbesma, R.P.; Srdanov, G.; Wudl, F. Inhibition of the HIV-1 protease by fullerene derivatives: Model building studies and experimental verification. J. Am. Chem. Soc. 1993, 52, 2090–2100. [Google Scholar] [CrossRef]
- Pastorin, G.; Marchesan, S.; Hoebeke, J.; Da Ros, T.; Ehret-Sabatier, L.; Briand, J.-P.; Prato, M.; Bianco, A. Design and activity of cationic fullerene derivatives as inhibitors of acetylcholinesterase. Org. Biomol. Chem. 2006, 4, 2556–2562. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Shan, Y.; Guan, S.; Zhang, H.; Wang, S.; Han, W. Structural basis of fullerene derivatives as novel potent inhibitors of protein tyrosine phosphatase 1B: Insight into the inhibitory mechanism through molecular modeling studies. J. Chem. Inf. Model. 2016, 56, 2024–2034. [Google Scholar] [CrossRef]
- Kang, S.; Zhou, G.; Yang, P.; Liu, Y.; Sun, B.; Huynh, T.; Meng, H.; Zhao, L.; Xing, G.; Chen, C.; et al. Molecular mechanism of pancreatic tumor metastasis inhibition by Gd@C82(OH)22 and its implication for de novo design of nanomedicine. Proc. Natl. Acad. Sci. USA 2012, 109, 15431–15436. [Google Scholar] [CrossRef]
- Kang, S.; Araya-Secchi, R.; Wang, D.; Wang, B.; Huynh, T.; Zhou, R. Dual inhibitory pathways of metallofullerenol Gd@C82(OH)22 on matrix metalloproteinase-2: Molecular insight into drug-like nanomedicine. Sci. Rep. 2014, 4, 4775. [Google Scholar] [CrossRef]
- Basu-Dutt, S.; Minus, M.L.; Jain, R.; Nepal, D.; Kumar, S. Chemistry of carbon nanotubes for everyone. J. Chem. Educ. 2012, 89, 221–229. [Google Scholar] [CrossRef]
- Marchesan, S.; Prato, M. Under the lens: Carbon nanotube and protein interaction at the nanoscale. Chem. Commun. 2015, 51, 4347–4359. [Google Scholar] [CrossRef]
- Tasis, D.; Tagmatarchis, N.; Bianco, A.; Prato, M. Chemistry of carbon nanotubes. Chem. Rev. 2006, 3, 1105–1136. [Google Scholar] [CrossRef]
- Maggini, M.; Scorrano, G.; Prato, M. Addition of azomethine ylides to C60: Synthesis, characterization, and functionalization of fullerene pyrrolidines. J. Am. Chem. Soc. 1993, 115, 9798–9799. [Google Scholar] [CrossRef]
- Yang, J.; Alemany, L.B.; Driver, J.; Hartgerink, J.D.; Barron, A.R. Fullerene-derivatized amino acids: Synthesis, characterization, antioxidant properties, and solid-phase peptide synthesis. Chem.-A Eur. J. 2007, 13, 2530–2545. [Google Scholar] [CrossRef] [PubMed]
- Pochkaeva, E.I.; Podolsky, N.E.; Zakusilo, D.N.; Petrov, A.V.; Charykov, N.A.; Vlasov, T.D.; Penkova, A.V.; Vasina, L.V.; Murin, I.V.; Sharoyko, V.V.; et al. Fullerene derivatives with amino acids, peptides and proteins: From synthesis to biomedical application. Prog. Solid State Chem. 2020, 57, 100255. [Google Scholar] [CrossRef]
- Gao, J.; Wang, Y.; Folta, K.M.; Krishna, V.; Bai, W.; Indeglia, P.; Georgieva, A.; Nakamura, H.; Koopman, B.; Moudgil, B. Polyhydroxy fullerenes (fullerols or fullerenols): Beneficial effects on growth and lifespan in diverse biological models. PLoS ONE 2011, 6, e0019976. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Cruz, C.A.; Kang, S.; Zhou, R. Large scale molecular simulations of nanotoxicity. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 329–343. [Google Scholar] [CrossRef]
- Hernández-Cancel, G.; Suazo-Dávila, D.; Ojeda-Cruzado, A.J.; García-Torres, D.; Cabrera, C.R.; Griebenow, K. Graphene oxide as a protein matrix: Influence on protein biophysical properties. J. Nanobiotechnol. 2015, 13, 70. [Google Scholar] [CrossRef]
- Rahman, M.; Laurent, S.; Tawil, N.; Yahia, L.; Mahmoudi, M. Nanoparticle and protein corona. In Protein-Nanoparticle Interactions (The Bio-Nano Interface); Springer: London, UK, 2013; Volume 15, pp. 21–44. [Google Scholar]
- Othman, Z.; Cillero Pastor, B.; van Rijt, S.; Habibovic, P. Understanding interactions between biomaterials and biological systems using proteomics. Biomaterials 2018, 167, 191–204. [Google Scholar] [CrossRef]
- Baase, W.A.; Liu, L.; Tronrud, D.E.; Matthews, B.W. Lessons from the lysozyme of phage T4. Protein Sci. 2010, 19, 631–641. [Google Scholar] [CrossRef]
- Vaitheeswaran, S.; Garcia, A.E. Protein stability at a carbon nanotube interface. J. Chem. Phys. 2011, 134, 125101. [Google Scholar] [CrossRef]
- Asuri, P.; Bale, S.S.; Karajanagi, S.S.; Kane, R.S. The protein-nanomaterial interface. Curr. Opin. Biotechnol. 2006, 17, 562–568. [Google Scholar] [CrossRef]
- Trozzi, F.; Marforio, T.D.; Bottoni, A.; Zerbetto, F.; Calvaresi, M. Engineering the fullerene-protein interface by computational design: The sum is more than its parts. Isr. J. Chem. 2017, 57, 547–552. [Google Scholar] [CrossRef]
- Dieckmann, G.R.; Dalton, A.B.; Johnson, P.A.; Razal, J.; Chen, J.; Giordano, G.M.; Muñoz, E.; Musselman, I.H.; Baughman, R.H.; Draper, R.K. Controlled assembly of carbon nanotubes by designed amphiphilic peptide helices. J. Am. Chem. Soc. 2003, 125, 1770–1777. [Google Scholar] [CrossRef]
- Xie, H.; Ortiz-Acevedo, A.; Zorbas, V.; Baughman, R.H.; Draper, R.K.; Musselman, I.H.; Dalton, A.B.; Dieckmann, G.R. Peptide cross-linking modulated stability and assembly of peptide-wrapped single-walled carbon nanotubes. J. Mater. Chem. 2005, 15, 1734–1741. [Google Scholar] [CrossRef]
- Burley, S.K.; Petsko, G.A. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science 1985, 229, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Bignucolo, O.; Leung, H.T.A.; Grzesiek, S.; Berneìche, S. Backbone hydration determines the folding signature of amino acid residues. J. Am. Chem. Soc. 2015, 137, 4300–4303. [Google Scholar] [CrossRef] [PubMed]
- Ferreira De Freitas, R.; Schapira, M. A systematic analysis of atomic protein-ligand interactions in the PDB. Medchemcomm 2017, 8, 1970–1981. [Google Scholar] [CrossRef]
- Wang, S.; Humphreys, E.S.; Chung, S.Y.; Delduco, D.F.; Lustig, S.R.; Wang, H.; Parker, K.N.; Rizzo, N.W.; Subramoney, S.; Chiang, Y.M.; et al. Peptide with selective activity for carbon nanotube. Nat. Mater. 2003, 2, 196–200. [Google Scholar]
- Ge, C.; Du, J.; Zhao, L.; Wang, L.; Liu, Y.; Li, D.; Yang, Y.; Zhou, R.; Zhao, Y.; Chai, Z.; et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. USA 2011, 108, 16968–16973. [Google Scholar] [CrossRef]
- Johnson, R.R.; Johnson, A.T.C.; Klein, M.L. Probing the structure of DNA-carbon nanotube hybrids with molecular dynamics. Nano Lett. 2008, 8, 69–75. [Google Scholar] [CrossRef]
- Calvaresi, M.; Zerbetto, F. Fullerene sorting proteins. Nanoscale 2011, 3, 2873–2881. [Google Scholar] [CrossRef]
- Di Giosia, M.; Valle, F.; Cantelli, A.; Bottoni, A.; Zerbetto, F.; Fasoli, E.; Calvaresi, M. High-throughput virtual screening to rationally design protein-carbon nanotube interactions. Identification and preparation of stable water dispersions of protein-carbon nanotube hybrids and efficient design of new functional materials. Carbon 2019, 147, 112–119. [Google Scholar] [CrossRef]
- Fjodorova, N.; Novič, M.K.; Venko, A.; Rasulev, A. A comprehensive cheminformatics analysis of structural features affecting the binding activity of fullerene derivatives. Nanomater. 2020, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Mikami, B.; Hehre, E.J.; Sato, M.; Katsube, Y.; Hirose, M.; Morita, Y.; Sacchettini, J.C. The 2.0-A resolution structure of soybean beta-amylase complexed with alpha-cyclodextrin. Biochemistry 1993, 32, 6836–6845. [Google Scholar] [CrossRef] [PubMed]
- Alex, J.M.; Brancatelli, G.; Volpi, S.; Bonaccorso, C.; Casnati, A.; Geremia, S.; Crowley, P.B. Probing the determinants of porosity in protein frameworks: Co-crystals of cytochrome: C and an octa-anionic calix[4]arene. Org. Biomol. Chem. 2020, 18, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Brancatelli, G.; Dalcanale, E.; Pinalli, R.; Geremia, S. Probing the structural determinants of amino acid recognition: X-ray studies of crystalline ditopic host-guest complexes of the positively charged amino acids, ARg, Lys, and His with a cavitand molecule. Molecules 2018, 23, 3368. [Google Scholar] [CrossRef] [PubMed]
- Pinalli, R.; Brancatelli, G.; Pedrini, A.; Menozzi, D.; Hernández, D.; Ballester, P.; Geremia, S.; Dalcanale, E. The origin of selectivity in the complexation of N-methyl amino acids by tetraphosphonate cavitands. J. Am. Chem. Soc. 2016, 138, 8569–8580. [Google Scholar] [CrossRef]
- Westbrook, J.D.; Shao, C.; Feng, Z.; Zhuravleva, M.; Velankar, S.; Young, J. The chemical component dictionary: Complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the Protein Data Bank. Bioinformatics 2015, 31, 1274–1278. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Zardecki, C.; Di Costanzo, L.; Duarte, J.M.; Hudson, B.P.; Persikova, I.; Segura, J.; Shao, C.; Voigt, M.; Westbrook, J.D.; et al. RCSB Protein Data Bank: Enabling biomedical research and drug discovery. Protein Sci. 2020, 29, 52–65. [Google Scholar] [CrossRef]
- Sobolev, V.; Sorokine, A.; Prilusky, J.; Abola, E.E.; Edelman, M. Automated analysis of interatomic contacts in proteins. Bioinformatics 1999, 15, 327–332. [Google Scholar] [CrossRef]
- Chen, R.J.; Zhang, Y.; Wang, D.; Dai, H. Noncovalent sidewall functionalization of single-walled carbon nanotubes for protein immobilization. J. Am. Chem. Soc. 2001, 123, 3838–3839. [Google Scholar] [CrossRef]
- Chen, B.X.; Wilson, S.R.; Das, M.; Coughlin, D.J.; Erlanger, B.F. Antigenicity of fullerenes: Antibodies specific for fullerenes and their characteristics. Proc. Natl. Acad. Sci. USA 1998, 95, 10809–10813. [Google Scholar] [CrossRef]
- Hendrickson, O.; Fedyunina, N.; Zherdev, A.; Solopova, O.; Sveshnikov, P.; Dzantiev, B. Production of monoclonal antibodies against fullerene C 60 and development of a fullerene enzyme immunoassay. Analyst 2012, 137, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Braden, B.C.; Goldbaum, F.A.; Chen, B.X.; Kirschner, A.N.; Wilson, S.R.; Erlanger, B.F. X-ray crystal structure of an anti-Buckminsterfullerene antibody Fab fragment: Biomolecular recognition of C60. Proc. Natl. Acad. Sci. USA 2000, 97, 12193–12197. [Google Scholar] [CrossRef] [PubMed]
- Osipov, E.M.; Hendrickson, O.D.; Tikhonova, T.V.; Zherdev, A.V.; Solopova, O.N.; Sveshnikov, P.G.; Dzantiev, B.B.; Popov, V.O. Structure of the anti-C60 fullerene antibody fab fragment: Structural determinants of fullerene binding. Acta Naturae 2019, 11, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Erlanger, B.F.; Chen, B.X.; Zhu, M.; Brus, L. Binding of an anti-fullerene IgG monoclonal antibody to single wall carbon nanotubes. Nano Lett. 2001, 9, 465–467. [Google Scholar] [CrossRef]
- Grigoryan, G.; Kim, Y.H.; Acharya, R.; Axelrod, K.; Jain, R.M.; Willis, L.; Drndic, M.; Kikkawa, J.M.; DeGrado, W.F. Computational design of virus-like protein assemblies on carbon nanotube surfaces. Science 2011, 332, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Ko, D.K.; Kim, Y.T.; Kim, N.H.; Paul, J.; Zhang, S.Q.; Murray, C.B.; Acharya, R.; Degrado, W.F.; Kim, Y.H.; et al. Protein-directed self-assembly of a fullerene crystal. Nat. Commun. 2016, 7, 11429. [Google Scholar] [CrossRef]
- Lehn, J.-M. From supramolecular chemistry towards constitutional dynamic chemistry and adaptive chemistry. Chem. Soc. Rev. 2007, 36, 151–160. [Google Scholar] [CrossRef]
- Pedersen, C.J. The discovery of crown ethers (noble lecture). Angew. Chem. Int. Ed. Engl. 1988, 27, 1021–1027. [Google Scholar] [CrossRef]
- Lehn, J.-M. Supramolecular Chemistry. Concepts and Perspectives; VCH: Weinheim, Germany, 1995; p. 195. [Google Scholar]
- Jones, S.; Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef]
- Milroy, L.G.; Grossmann, T.N.; Hennig, S.; Brunsveld, L.; Ottmann, C. Modulators of protein-protein interactions. Chem. Rev. 2014, 114, 4695–4748. [Google Scholar] [CrossRef]
- Van Dun, S.; Ottmann, C.; Milroy, L.G.; Brunsveld, L. Supramolecular chemistry targeting proteins. J. Am. Chem. Soc. 2017, 139, 13960–13968. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Langlois d’Estaintot, B.; Granier, T.; Mackereth, C.D.; Fischer, L.; Huc, I. Structure elucidation of helical aromatic foldamer–protein complexes with large contact surface areas. Chem.-A Eur. J. 2019, 25, 11042–11047. [Google Scholar] [CrossRef] [PubMed]
- Spence, M.M.; Rubin, S.M.; Dimitrov, I.E.; Ruiz, E.J.; Wemmer, D.E.; Pines, A.; Yao, S.Q.; Tian, F.; Schultz, P.G. Functionalized xenon as a biosensor. Proc. Natl. Acad. Sci. USA 2001, 98, 10654–10657. [Google Scholar] [CrossRef] [PubMed]
- Chattaraj, R.; Hwang, M.; Zemerov, S.D.; Dmochowski, I.J.; Hammer, D.A.; Lee, D.; Sehgal, C.M. Ultrasound responsive noble gas microbubbles for applications in image-guided gas delivery. Adv. Healthc. Mater. 2020, 9, e1901721. [Google Scholar] [CrossRef] [PubMed]
- Oros, A.M.; Shah, N.J. Hyperpolarized xenon in NMR and MRI. Phys. Med. Biol. 2004, 49, 105–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Collet, A.; Dutasta, J.-P.; Lozach, B.; Canceill, J. Cyclotriveratrylenes and cryptophanes: Their synthesis and applications to host-guest chemistry and to the design of new materials. Supramol. Chem. I 1993, 165, 103–129. [Google Scholar]
- Brotin, T.; Dutasta, J.P. Cryptophanes and their complexes-present and future. Chem. Rev. 2009, 109, 88–130. [Google Scholar] [CrossRef]
- Gutsche, C.D. Calixarenes: An Introduction; RCS Publishing: Marshfield, MO, USA, 2008. [Google Scholar]
- Reinhoudt, D.N. Introduction and history. In Calixarenes and Beyond; Neri, P., Sessler, J.L., Wang, M.-X., Eds.; Springer: London, UK, 2016; pp. 1–11. [Google Scholar]
- Guérineau, V.; Rollet, M.; Viel, S.; Lepoittevin, B.; Costa, L.; Saint-Aguet, P.; Laurent, R.; Roger, P.; Gigmes, D.; Martini, C.; et al. The synthesis and characterization of giant Calixarenes. Nat. Commun. 2019, 10, 113. [Google Scholar] [CrossRef]
- Sliwa, W.; Girek, T. Calixarene complexes with metal ions. J. Incl. Phenom. Macrocycl. Chem. 2010, 73, 7768–7771. [Google Scholar] [CrossRef]
- Arena, G.; Contino, A.; Gulino, F.G.; Magri, A.; Sciotto, D.; Ungaro, R. Complexation of small neutral organic molecules by water soluble calix[4]arenes. Tetrahedron Lett. 2000, 41, 9327–9330. [Google Scholar] [CrossRef]
- Selkti, M.; Coleman, A.W.; Nicolis, I.; Douteau-Guével, N.; Villain, F.; Tomas, A.; De Rango, C. The first example of a substrate spanning the calix[4]arene bilayer: The solid state complex of p-sulfonatocalix[4]arene with L-lysine. Chem. Commun. 2000, 2000, 161–162. [Google Scholar] [CrossRef]
- Corbellini, F.; Di Costanzo, L.; Crego-Calama, M.; Geremia, S.; Reinhoudt, D.N. Guest encapsulation in a water-soluble molecular capsule based on ionic interactions. J. Am. Chem. Soc. 2003, 125, 9946–9947. [Google Scholar] [CrossRef]
- Durso, A.; Brancatelli, G.; Hickey, N.; Farnetti, E.; De Zorzi, R.; Bonaccorso, C.; Purrello, R.; Geremia, S. Interactions of a water-soluble calix[4]arene with spermine: Solution and solid-state characterisation. Supramol. Chem. 2016, 28, 499–505. [Google Scholar] [CrossRef]
- Brancatelli, G.; Gattuso, G.; Geremia, S.; Manganaro, N.; Notti, A.; Pappalardo, S.; Parisi, M.F.; Pisagatti, I. Encapsulation of biogenic polyamines by carboxylcalix[5]arenes: When solid-state design beats recognition in solution. CrystEngComm 2016, 18, 5012–5016. [Google Scholar] [CrossRef]
- Ribeiro, R.C.; Sandrini, F.; Figueiredo, B.; Zambetti, G.P.; Michalkiewicz, E.; Lafferty, A.R.; DeLacerda, L.; Rabin, M.; Cadwell, C.; Sampaio, G.; et al. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc. Natl. Acad. Sci. USA 2001, 98, 9930–9935. [Google Scholar] [CrossRef] [PubMed]
- Gordo, S.; Martos, V.; Santos, E.; Menéndez, M.; Bo, C.; Giralt, E.; De Mendoza, J. Stability and structural recovery of the tetramerization domain of p53-R337H mutant induced by a designed templating ligand. Proc. Natl. Acad. Sci. USA 2008, 105, 16426–16431. [Google Scholar] [CrossRef]
- De March, M.; Demitri, N.; De Zorzi, R.; Casini, A.; Gabbiani, C.; Guerri, A.; Messori, L.; Geremia, S. Nitrate as a probe of cytochrome c surface: Crystallographic identification of crucial “hot spots” for protein-protein recognition. J. Inorg. Biochem. 2014, 135, 58–67. [Google Scholar] [CrossRef]
- McGovern, R.E.; Fernandes, H.; Khan, A.R.; Power, N.P.; Crowley, P.B. Protein camouflage in cytochrome c-calixarene complexes. Nat. Chem. 2012, 4, 527–533. [Google Scholar] [CrossRef]
- McGovern, R.E.; McCarthy, A.A.; Crowley, P.B. Protein assembly mediated by sulfonatocalix[4]arene. Chem. Commun. 2014, 50, 10412–10415. [Google Scholar] [CrossRef]
- McGovern, R.E.; Snarr, B.D.; Lyons, J.A.; McFarlane, J.; Whiting, A.L.; Paci, I.; Hof, F.; Crowley, P.B. Structural study of a small molecule receptor bound to dimethyllysine in lysozyme. Chem. Sci. 2015, 6, 442–449. [Google Scholar] [CrossRef]
- Mummidivarapu, V.V.S.; Rennie, M.L.; Doolan, A.M.; Crowley, P.B. Noncovalent PEGylation via sulfonatocalix[4]arene-a crystallographic proof. Bioconjug. Chem. 2018, 29, 3999–4003. [Google Scholar] [CrossRef] [PubMed]
- Doolan, A.M.; Rennie, M.L.; Crowley, P.B. Protein recognition by functionalized sulfonatocalix[4]arenes. Chem.-A Eur. J. 2018, 24, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Engilberge, S.; Rennie, M.L.; Crowley, P.B. Calixarene capture of partially unfolded cytochrome c. FEBS Lett. 2019, 593, 2112–2117. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.L.; Fox, G.C.; Pérez, J.; Crowley, P.B. Auto-regulated protein assembly on a supramolecular scaffold. Angew. Chemie-Int. Ed. 2018, 20, 1011–1017. [Google Scholar]
- Engilberge, S.; Rennie, M.L.; Dumont, E.; Crowley, P.B. Tuning protein frameworks via auxiliary supramolecular interactions. ACS Nano 2019, 13, 10343–10350. [Google Scholar] [CrossRef] [PubMed]
- De Zorzi, R.; Guidolin, N.; Randaccio, L.; Purrello, R.; Geremia, S. Nanoporous crystals of calixarene/porphyrin supramolecular complex functionalized by diffusion and coordination of metal ions. J. Am. Chem. Soc. 2009, 131, 2487–2489. [Google Scholar] [CrossRef]
- De Zorzi, R.; Guidolin, N.; Randaccio, L.; Geremia, S. A bifunctionalized porous material containing discrete assemblies of copper-porphyrins and calixarenes metallated by ion diffusion. CrystEngComm 2010, 12, 4056–4058. [Google Scholar] [CrossRef]
- Giuliani, M.; Morbioli, I.; Sansone, F.; Casnati, A. Moulding calixarenes for biomacromolecule targeting. Chem. Commun. 2015, 52, 14140–14159. [Google Scholar] [CrossRef]
- Di Costanzo, L.; Geremia, S.; Randaccio, L.; Purrello, R.; Lauceri, R.; Sciotto, D.; Gulino, F.G.; Pavone, V. Calixarene-porphyrin supramolecular complexes: pH-tuning of the complex stoichiometry. Angew. Chem. Int. Ed. 2001, 40, 4245–4247. [Google Scholar] [CrossRef]
- Brancatelli, G.; De Zorzi, R.; Hickey, N.; Siega, P.; Zingone, G.; Geremia, S. New multicomponent porous architecture of self-assembled porphyrins/calixarenes driven by nickel ions. Cryst. Growth Des. 2012, 12, 5111–5117. [Google Scholar] [CrossRef]
- Alex, J.M.; Rennie, M.L.; Engilberge, S.; Lehoczki, G.; Dorottya, H.; Fizil, Á.; Batta, G.; Crowley, P.B. Calixarene-mediated assembly of a small antifungal protein. IUCrJ 2019, 6, 2238–2247. [Google Scholar] [CrossRef] [PubMed]
- Alex, J.M.; Rennie, M.L.; Volpi, S.; Sansone, F.; Casnati, A.; Crowley, P.B. Phosphonated calixarene as a “molecular glue” for protein crystallization. Cryst. Growth Des. 2018, 18, 2467–2473. [Google Scholar] [CrossRef]
- Rennie, M.L.; Doolan, A.M.; Raston, C.L.; Crowley, P.B. Protein dimerization on a phosphonated calix[6]arene disc. Angew. Chem.-Int. Ed. 2017, 56, 5517–5521. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.C.; Mijts, B.N.; Swaminathan, K.; Patel, B.K.C.; Divne, C. Crystal structure of the polyextremophilic α-amylase amyb from halothermothrix orenii: Details of a productive enzyme-substrate complex and an N domain with a role in binding raw starch. J. Mol. Biol. 2008, 378, 852–870. [Google Scholar] [CrossRef]
- Homburg, C.; Bommer, M.; Wuttge, S.; Hobe, C.; Beck, S.; Dobbek, H.; Deutscher, J.; Licht, A.; Schneider, E. Inducer exclusion in Firmicutes: Insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr. Mol. Microbiol. 2017, 105, 25–45. [Google Scholar] [CrossRef]
- Buedenbender, S.; Schulz, G.E. Structural base for enzymatic cyclodextrin hydrolysis. J. Mol. Biol. 2009, 385, 606–617. [Google Scholar] [CrossRef]
- Feng, L.; Fawaz, R.; Hovde, S.; Sheng, F.; Nosrati, M.; Geiger, J.H. Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 7641–7647. [Google Scholar] [CrossRef]
- Freeman, W.A.; Mock, W.L.; Shih, N.Y. Cucurbituril. J. Am. Chem. Soc. 1981, 103, 7367–7368. [Google Scholar] [CrossRef]
- Lee, J.W.; Samal, S.; Selvapalam, N.; Kim, H.J.; Kim, K. Cucurbituril homologues and derivatives: New opportunities in supramolecular chemistry. Acc. Chem. Res. 2003, 36, 621–636. [Google Scholar] [CrossRef]
- Kim, J.; Jung, I.S.; Kim, S.Y.; Lee, E.; Kang, J.K.; Sakamoto, S.; Yamaguchi, K.; Kim, K. New cucurbituril homologues: Syntheses, isolation, characterization, and X-ray crystal structures of cucurbit[n]uril (n = 5, 7, and 8). J. Am. Chem. Soc. 2000, 122, 540–541. [Google Scholar] [CrossRef]
- Barrow, S.J.; Kasera, S.; Rowland, M.J.; Del Barrio, J.; Scherman, O.A. Cucurbituril-based molecular recognition. Chem. Rev. 2015, 115, 12320–12406. [Google Scholar] [CrossRef] [PubMed]
- Chinai, J.M.; Taylor, A.B.; Ryno, L.M.; Hargreaves, N.D.; Morris, C.A.; Hart, P.J.; Urbach, A.R. Molecular recognition of insulin by a synthetic receptor. J. Am. Chem. Soc. 2011, 133, 8810–8813. [Google Scholar] [CrossRef] [PubMed]
- De Vink, P.J.; Briels, J.M.; Schrader, T.; Milroy, L.G.; Brunsveld, L.; Ottmann, C. A binary bivalent supramolecular assembly platform based on cucurbit[8]uril and dimeric adapter protein 14-3-3. Angew. Chem. Int. Ed. 2017, 56, 8998–9002. [Google Scholar] [CrossRef] [PubMed]
- Guagnini, F.; Antonik, P.M.; Rennie, M.L.; O’Byrne, P.; Khan, A.R.; Pinalli, R.; Dalcanale, E.; Crowley, P.B. Cucurbit[7]uril-dimethyllysine recognition in a model protein. Angew. Chem.-Int. Ed. 2018, 57, 7126–7130. [Google Scholar] [CrossRef]
- Klärner, F.G.; Kahlert, B. Molecular tweezers and clips as synthetic receptors. Molecular recognition and dynamics in receptor-substrate complexes. Acc. Chem. Res. 2003, 36, 919–932. [Google Scholar] [CrossRef]
- Fokkens, M.; Schrader, T.; Klärner, F.G. A molecular tweezer for lysine and arginine. J. Am. Chem. Soc. 2005, 127, 14415–14421. [Google Scholar] [CrossRef]
- Sinha, S.; Lopes, D.H.J.; Du, Z.; Pang, E.S.; Shanmugam, A.; Lomakin, A.; Talbiersky, P.; Tennstaedt, A.; McDaniel, K.; Bakshi, R.; et al. Lysine-specific molecular tweezers are broad-spectrum inhibitors of assembly and toxicity of amyloid proteins. J. Am. Chem. Soc. 2011, 92, 947–956. [Google Scholar] [CrossRef]
- Tainer, J.A.; Getzoff, E.D.; Beem, K.M.; Richardson, J.S.; Richardson, D.C. Determination and analysis of the 2 Å structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982, 160, 181–217. [Google Scholar] [CrossRef]
- Malik, R.; Meng, H.; Wongkongkathep, P.; Corrales, C.I.; Sepanj, N.; Atlasi, R.S.; Klärner, F.G.; Schrader, T.; Spencer, M.J.; Loo, J.A.; et al. The molecular tweezer CLR01 inhibits aberrant superoxide dismutase 1 (SOD1) self-assembly in vitro and in the G93A-SOD1 mouse model of ALS. J. Biol. Chem. 2019, 294, 3501–3513. [Google Scholar] [CrossRef]
- Baccarini, M. Second nature: Biological functions of the Raf-1 “kinase”. FEBS Lett. 2005, 39, 1007–1012. [Google Scholar] [CrossRef]
- Pandit, B.; Sarkozy, A.; Pennacchio, L.A.; Carta, C.; Oishi, K.; Martinelli, S.; Pogna, E.A.; Schackwitz, W.; Ustaszewska, A.; Landstrom, A.; et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007, 39, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Molzan, M.; Schumacher, B.; Ottmann, C.; Baljuls, A.; Polzien, L.; Weyand, M.; Thiel, P.; Rose, R.; Rose, M.; Kuhenne, P.; et al. Impaired binding of 14-3-3 to C-RAF in noonan syndrome suggests new approaches in diseases with increased ras signaling. Mol. Cell. Biol. 2010, 30, 4698–4711. [Google Scholar] [CrossRef] [PubMed]
- Bier, D.; Rose, R.; Bravo-Rodriguez, K.; Bartel, M.; Ramirez-Anguita, J.M.; Dutt, S.; Wilch, C.; Klärner, F.G.; Sanchez-Garcia, E.; Schrader, T.; et al. Molecular tweezers modulate 14-3-3 protein-protein interactions. Nat. Chem. 2013, 5, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Bier, D.; Mittal, S.; Bravo-Rodriguez, K.; Sowislok, A.; Guillory, X.; Briels, J.; Heid, C.; Bartel, M.; Wettig, B.; Brunsveld, L.; et al. The molecular tweezer CLR01 stabilizes a disordered protein-protein interface. J. Am. Chem. Soc. 2017, 139, 16256–16263. [Google Scholar] [CrossRef] [PubMed]
- Bustos, D.M.; Iglesias, A.A. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins Struct. Funct. Genet. 2006, 63, 35–42. [Google Scholar] [CrossRef]
- Bhattarai, A.; Emerson, I.A. Computational investigations on the dynamic binding effect of molecular tweezer CLR01 toward intrinsically disordered HIV-1 Nef. Biotechnol. Appl. Biochem. 2020. [Google Scholar] [CrossRef]
- Yan, X.; Sedykh, A.; Wang, W.; Yan, B.; Zhu, H. Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nat. Commun. 2020, 11, 2519. [Google Scholar] [CrossRef]
- Shi, J.; Votruba, A.R.; Farokhzad, O.C.; Langer, R. Nanotechnology in drug delivery and tissue engineering: From discovery to applications. Nano Lett. 2010, 10, 3223–3230. [Google Scholar] [CrossRef]
- Malik, S.A.; Mohanta, Z.; Srivastava, C.; Atreya, H.S. Modulation of protein-graphene oxide interactions with varying degrees of oxidation. Nanoscale Adv. 2020, 2, 1904–1912. [Google Scholar] [CrossRef]
- Hu, Q.D.; Tang, G.P.; Chu, P.K. Cyclodextrin-based host-guest supramolecular nanoparticles for delivery: From design to applications. Acc. Chem. Res. 2014, 47, 2017–2025. [Google Scholar] [CrossRef]
- Vitali, M.; Rigamonti, V.; Natalello, A.; Colzani, B.; Avvakumova, S.; Brocca, S.; Santambrogio, C.; Narkiewicz, J.; Legname, G.; Colombo, M.; et al. Conformational properties of intrinsically disordered proteins bound to the surface of silica nanoparticles. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Arai, R. Design and construction of self-assembling supramolecular protein complexes using artificial and fusion proteins as nanoscale building blocks. Curr. Opin. Biotechnol. 2017, 46, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Seisdedos, H.; Empereur-Mot, C.; Elad, N.; Levy, E.D. Proteins evolve on the edge of supramolecular self-assembly. Nature 2017, 548, 244–247. [Google Scholar] [CrossRef] [PubMed]




| Carbon-Based Nano-Molecule Name | Cif Code of Bound Nanomolecule | Proteins Interacting with Nanomolecules, (Identifiers of PDB Entry and Number of Bound Nanomolecules) | Main Interacting Protein Residues. Identifiers of PDB Entry, Nanomolecule, Protein Chain(s), Residue(s) or Atom(s), and Total Atomic Contacts | Buried Surface (Å2) Versus Nanomolecule Surface Area (Å2) | Protein and Nanomolecule Functions |
|---|---|---|---|---|---|
| Fullerene | - | Fab antibody fragment on an antifullerene antibody (1EMT: unbound) | 1EMT: CDR region contains aromatic residues: Y36(L), W47(H), Y91(L), F96(L); residues N35(L), Q89(L) | n/a | Antibody binding fullerene. |
| Fullerene | - | Fab antibody FabC 60 on an antifullerene antibody (6H3H: unbound) | 6H3H: CDR region contains aromatic residues: Y50(H), Y101(H), Y34(L), W93(L), W98(L); residue D100 (L) | n/a | Antibody binding fullerene. |
| Fullerene, buckminsterfullerene | 60C | Fullerene organizing protein (C60SOL-COP-3) (5ET3: 1, 5HKN: 1, 5HKR: 1) | 5ET3: 101 A Y9/A6/E2/S5 5HKN: 101 B Y9/A6/E2/S5 5HKR: 101 B Y9/A6/E2/S5 | 388/547 381/542 316/545 | Designed peptide binding fullerene. Crystal of COP protein in complex with fullerene does conduct electricity |
| Single wall nanotube (SWNT) | - | De novo designed helical assembly (Hexcoil-Ala) (3S0R: unbound) | 3S0R: Ala-rich HexCoil-Ala | n/a | Designed virus-like protein assembling on a carbon nanotube |
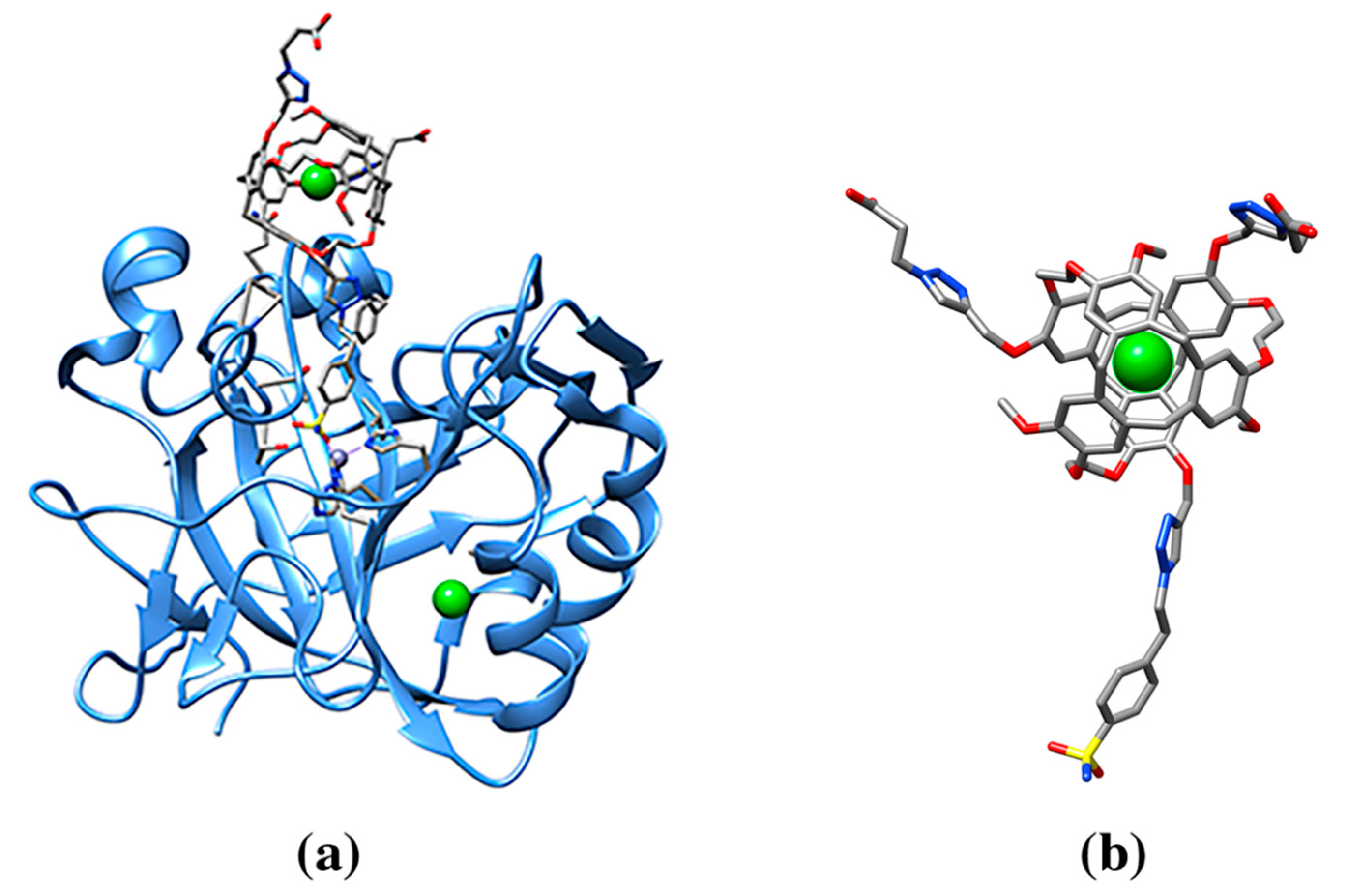
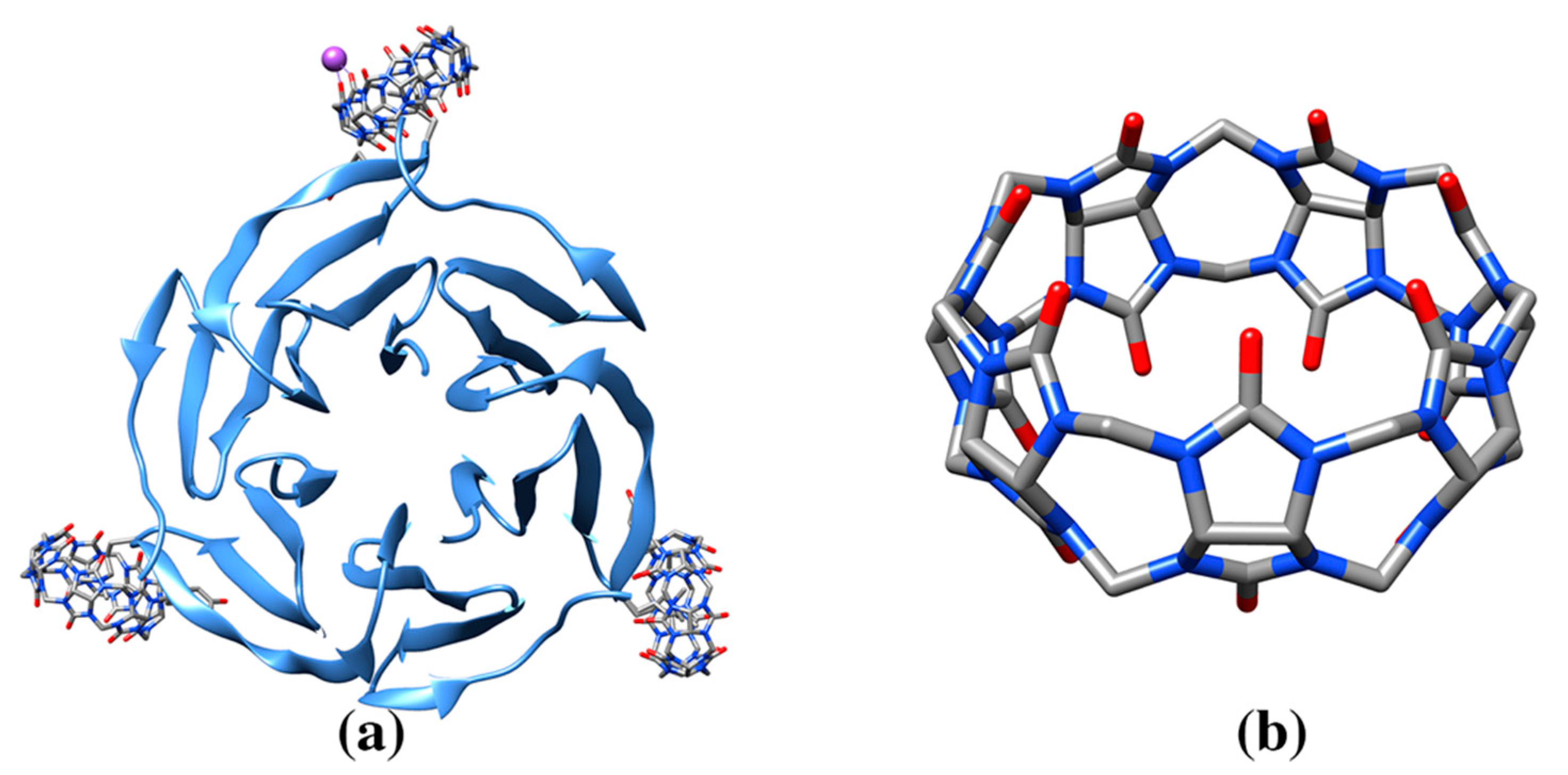
| 129Xe-Cryptophane biosensor (racemic mixture) | 0CR, 1CR | Human Carbonic Anhydrase II (3CYU: 1) | 3CYU: 263A Q136/ZN/XE (158) | 993/1515 | Enzyme lyase. Nanomolecule functions as an inhibitor of the enzyme and a xenon biosensor |
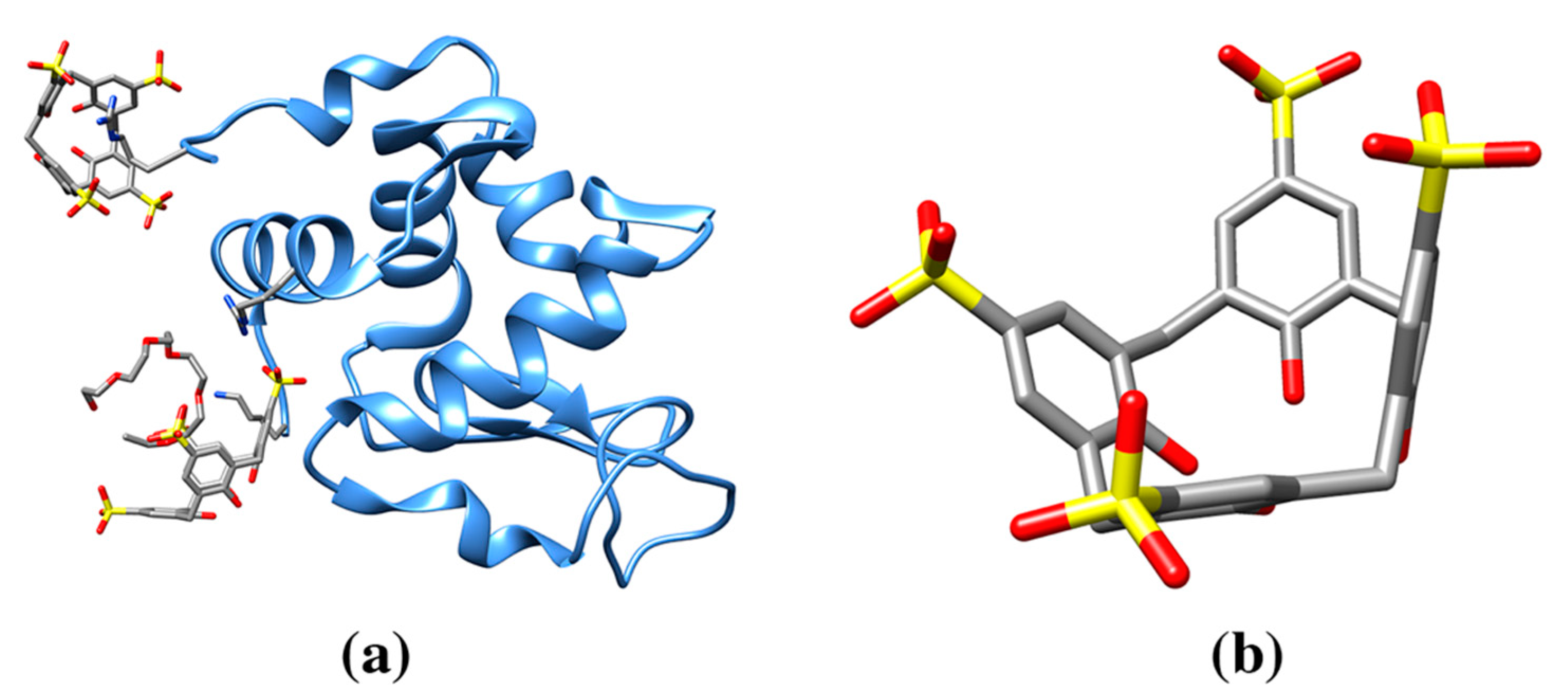
| Sulfonatocalix[4]arene (sclx4) | T3Y | Cytochrome c (3TYI: 3, 4YE1: 3, 4N0K: 3) | 3TYI: 105A K89 (83), 105B K4 (63), 106B K22 (60) | 472/815, 610/826, 622/808 | Electron carrier protein. Calixarene “camouflages” protein surface charges or promote quaternary structure formation. |
| 4YE1: 202A K89 (79), 202B K4 (71), 203B K89/K5 (48) | 486/814, 589/840, 601/823 | ||||
| 4N0K: 202A K89/K87 (84), 202B K4 (64), 203B K22 (60) | 482/815, 627/830, 625/814 | ||||
| Lysozyme C (4PRQ: 5, 4PRU: 4, 4N0J: 4) | 4PRQ: 201A R128 (67), 202A P6G (163), 201B R128 (68), 202B P6G (171), 203B PG4 (112) | 598/826, 174/830, 593/825, 167/833, 426/835 | Antibacterial protein | ||
| 4PRU: 201A MeK116 (70), 202A R14 (103), 201B MeK116/R112 (127), 202B R14 (97) | 533/794, 459/815, 316/800, 483/805 | ||||
| 4N0J: 201A MeK116 (73), 202A R14 (85), 201B MeK116/R112 (102), 202B R14 (86) | 534/789, 506/812, 426/797, 488/800 | ||||
| PAF (6HA4: 1) | 6HA4: 202A K30 (70) | 545/824 | Antifungal protein | ||
| Lectin protein (6GL5: 6) | 6GL5: 101A MeS1/T3Y (97), 101B MeS1 (73), 102B PEG (92), 101C MeK83 (57), 102C MeS1//T3Y (82), 103C S57 (80) | 481/806, 558/813, 524/828, 642/812, 502/807, 487/831 | Fucose-binding lectin protein.Calixarene is recognized by methylated serine | ||
| PEGylated sulfonatocalix[4]arene | B4T | Cytochrome c (6EGY: 4) | 6EGY: 203A Y97/K100/K4 (101), 201B K73 (47), 203B B4T/K86 (135), 204B R13/B4T (63) | 430/824, 636/840, 368/922, 133/419 | Calixarene derivative as a probe to increase protein half-life. |
| Di-PEGylated sulfonatocalix[4]arene | B4X | Cytochrome c (6EGZ: 4) | 6EGZ: 202A R13/B4X (51), 202B K27/B4X (78), 203 B K86/K87/B4X (137), 203B K72/B4X (73) | 60/275, 123/463, 377/901, 655/889 | |
| Bromo-trisulfonatocalix[4]arene | 6VB | Cytochrome c (5LFT: 3) | 5LFT: 204A K86/K87 (58), 205A 6VB/K89 (91), 206A 6VB (79) | 559/776, 425/777, 501/776 | Calixarene used as programmable molecule to control specific protein assemblies, or as a probe for “hiding” specific genetic mutations. |
| Phenyl-trisulfonatocalix[4]arene | 6VJ | Cytochrome c (5KPF: 1) | 5KPF: 202B K4/K5/(71) | 621/854 | |
| p-methylphosphonatocalix[4]arene | 8TE | Cytochrome c (5NCV: 3) | 5NCV: 202A K54/D50 (65), 203A K87 (74), 202B K86 (74) | 682/941, 687/942, 651/932 | Water soluble calixarene is used to study the effect on protein quaternary structure. |
| Sulfonatocalix[6]arene | FWQ | Cytochrome c (6RGI: 1) | 6RGI: 202A R13/K73 (133) | 668/1183 | Increased calixarene size to study effect on protein crystallinity. Nanomolecule shows irregular conformation. |
| PAF, Antifungal protein (6HAH: 1) | 6HAH: K30/PEG (161) | 690/1184 | |||
| Phosphonato-calix[6]arene | 7AZ | Cytochrome c (5LYC: 2) | 5LYC: 202A K4/K100 (67), 202B K4/K11/K100 (95) | 900/1205, 835/1207 | Water soluble calixarene is used to study the effect on protein quaternary structure. |
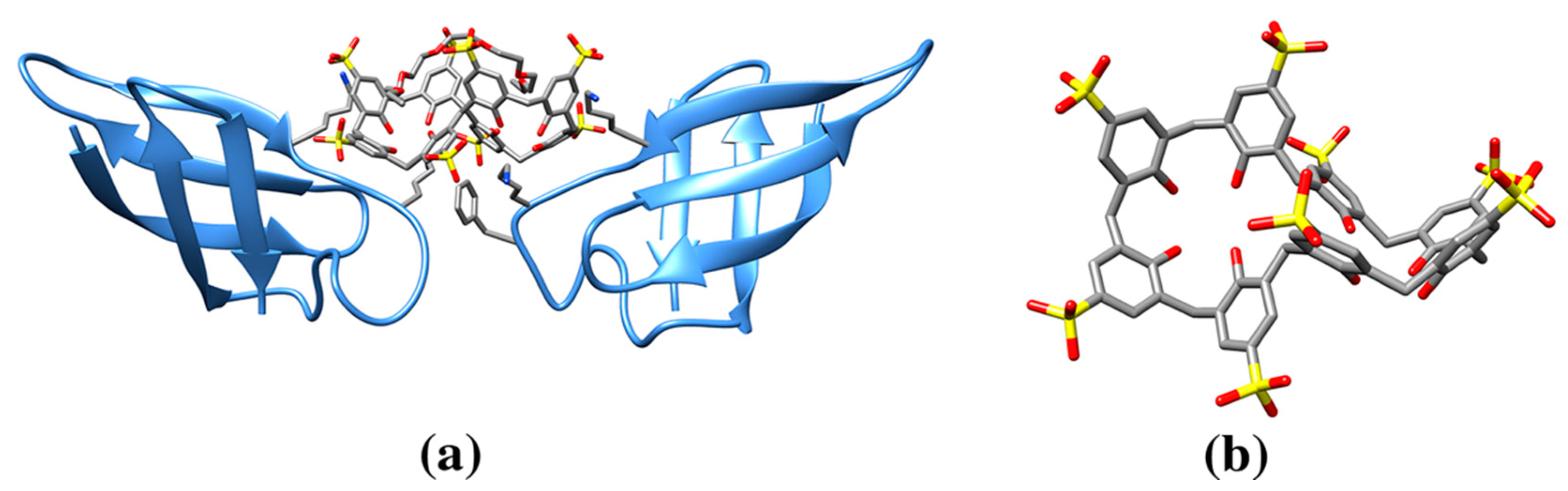
| p-sulfonatocalix[8]arene | EVB | Cytochrome c (6GD6: 1, 6GD7: 3, 6GD8: 3, 6GD9: 3, 6RSI: 3, 6RSL: 6, 6RSK: 6, 6RSJ: 6, 6GDA: 3) | 6GD6: K4/K100/A8 (147) | 1086/1538 | Larger, eight membered ring calixarene and effect on protein crystallinity. Effect of a mediator molecule (e.g., spermine) between protein and calixarene. |
| 6RSI: 202A K27/K11/Q16/L15 (120), 203A GOL/K100 (165), 204A K73/K86 (159) | 1158/1626, 928/1492, 1038/1665 | ||||
| 6RSL: 202A K27/K11/Q16/L15 (131), 203A SPM/K100/K4 (190), 204A SPM/EVB/K86/K87 (223), 202B K27/K11/Q16 (125), 203B SPM/K100/K4(205), 204B SPM/EVB/K73 (223) | 1163/1625, 819/1505, 753/1561, 1154/1627, 805/1510, 693/1559 | ||||
| PAF, Antifungal protein (6HAJ: 1) | 6HAJ: PEG/K27/K30/F31(256) | 720/1510 | |||
| Octaanionic calix[4]arene | LVT | Yeast cytochrome c (6SUY: 2) | 6SUY: 202A R13/K92/K87 (83), 202B K87/K89/E88/D90 (59) | 664/942, 783/954 | Octa-anionic charged calixarene and effect on protein quaternary structure. Larger assembly is formed |
| LVQ | Horse cytochrome c (6SUV: 8) | 6SUV: 203A K86/K87/K88/T89/E90 (175), 203B K86/K87/K88/T89/E90 (161), 203C K86/K87/K88/T89/E90 (122), 203D K86/K87/K88/T89/E90 (120), 203E K86/K87/K88/T89/E90 (181), 203F K86/K87/K88/T89/E90 (172), 203G K86/K87/K88/T89/E90 (123), 203H K86/K87/K88/T89/E90 (123) | 346/1031, 346/1009, 551/1017, 558/1028, 337/1001, 334/1021, 550/999, 534,984 | ||
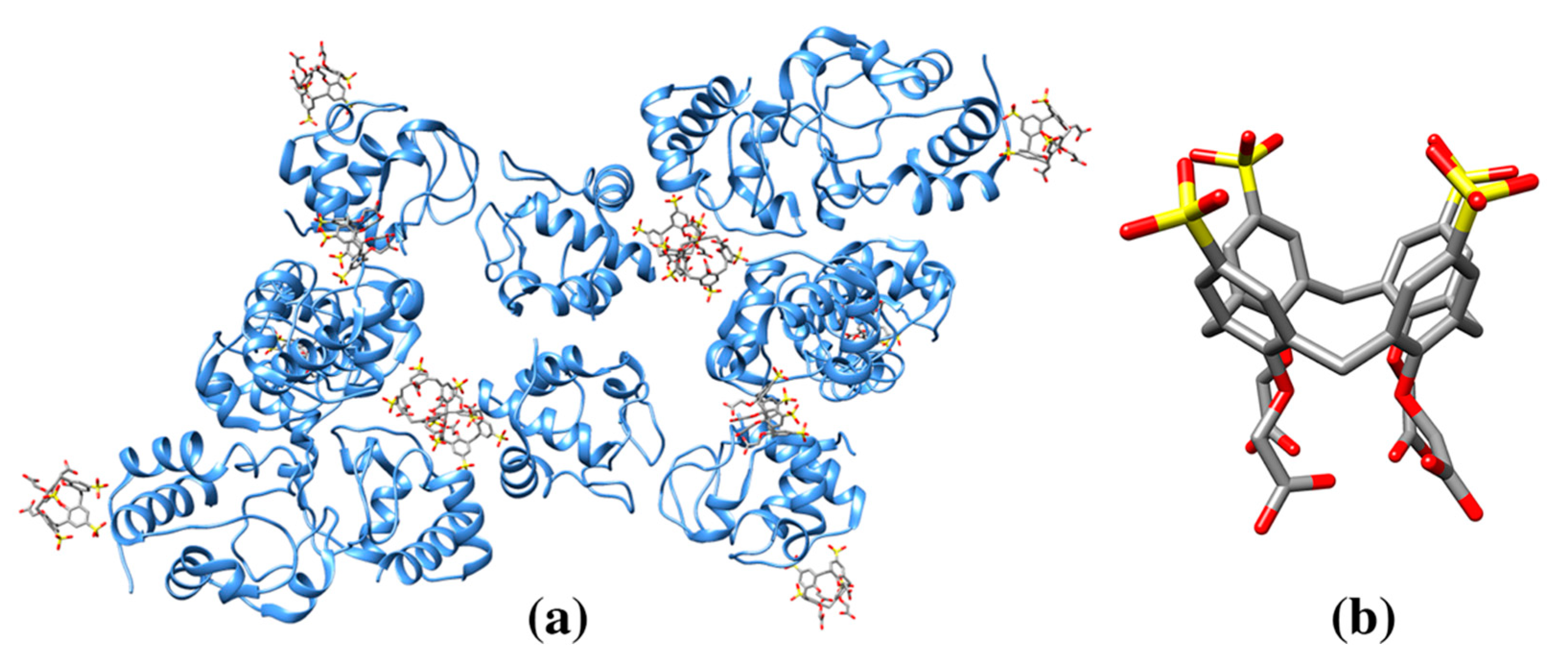
| α-cyclodextrin | ACX | Thermostable alpha-amylase (3BCD: 1) | 3BCD: 901A M176/W260/W287 (88) | 649/991 | Hydrolytic enzyme for starch degradation |
| β-cyclodextrin | BCD | Cytochrome P450 2R1 (3CZH: 1) | 3CZH: 603A F240/P239 (120) | 724/1119 | Oxidoreductase |
| γ-cyclodextrin | RCD | Maltodextrin binding protein MalE1 (5MKA: 1) | 5MKA: 401A A58/N59/N46/W234/Y164/W(353) (186) | 560/1281 | Protein belongs to the ABC transporter complex involved in maltose/maltodextrin import. Protein binds maltose and maltodextrins. Protein functions to linearize cyclomaltodextrin. |
| 3EDK: 700A R464/D466/D418/W342/ D311/F274/Y178 (180) | 513/1263 | ||||
| Cyclomaltodextrinase (3EDK: 2) | 700B R464/D466/D418/W342/D311/F274/ Y178 (196) | 485/1273 | |||
| Molecular tweezer, CLR01 | 9SZ | 14-3-3 adapter protein (5OEH:1, 5OEG:1, 5M36:3, 5M37:4) | 5OEH: 301 A K214/Y213 (74) | 579/832 | Dimeric binding protein. Molecular tweezer is used as an inhibitor or a binding modulator. |
| 5M37: 301A N183/K138/9SZ301D (56), 301B K74/M78/E73 (121), 301C R208/N183 (138), 301D R208/9SZ301A (153) | 640/837, 504/830, 363/833, 277/831 | ||||
| Cucurbit[7]uril | QQ7 | Human insulin (3Q6E: 1) Lectin binding protein (6F7W: 3, 6F7X: 2, 6SU0: 12) | 3Q6E: F1/V2 (64) | 827/1031 | Hormone protein. Cucurbituril molecules are used as probe for engineering arrays of protein assembly. Methylated lysine residue is recognized |
| 6F7W: 101AMeK34 (71), 101B MeK34 (75), 101C MeK34 (74) | 744/998, 712/993, 748/998 | ||||
| 6F7X: 101AMeK34 (78), 103B MeK(34) (64) | 749/996, 760/992 | ||||
| 6SU0: 202A MeK34/Y37 (79), 102B MeK34/Y37 (83), 204C MeK34/Y37 (91), 201I MeK79B/Y82B (80), 201J MeK79C/W81C/Y82C (80), 201K MeK79A/W81A/Y82A (88), 201M MeK79D/W81D/Y82D (80), 202M MeK34F/Y37F (74), 201N MeK34D/Y37D (87), 202N MeK79E/W81E/Y82E (82), 201O MeK34E/Y37E (82), 202O MeK79F/Y82F (86) | 742/1005, 754/1011, 694/1007, 749/1007, 717/1004, 717/1002, 716/991, 738/992, 712/999, 740/1001, 736/988, 708/992 | ||||
| Cucurbit[8]uril | C8L | 14-3-3 adapter protein (5N10: 1) | 5N10: 601C: F581C/F581D (130) | 643/1165 | Dimeric binding protein. Larger size cucurbituril is used to study effect on protein assembly |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Costanzo, L.; Geremia, S. Atomic Details of Carbon-Based Nanomolecules Interacting with Proteins. Molecules 2020, 25, 3555. https://doi.org/10.3390/molecules25153555
Di Costanzo L, Geremia S. Atomic Details of Carbon-Based Nanomolecules Interacting with Proteins. Molecules. 2020; 25(15):3555. https://doi.org/10.3390/molecules25153555
Chicago/Turabian StyleDi Costanzo, Luigi, and Silvano Geremia. 2020. "Atomic Details of Carbon-Based Nanomolecules Interacting with Proteins" Molecules 25, no. 15: 3555. https://doi.org/10.3390/molecules25153555
APA StyleDi Costanzo, L., & Geremia, S. (2020). Atomic Details of Carbon-Based Nanomolecules Interacting with Proteins. Molecules, 25(15), 3555. https://doi.org/10.3390/molecules25153555







