Antinociceptive and Cytotoxic Activity of Opioid Peptides with Hydrazone and Hydrazide Moieties at the C-Terminus
Abstract
1. Introduction
2. Results and Discussion
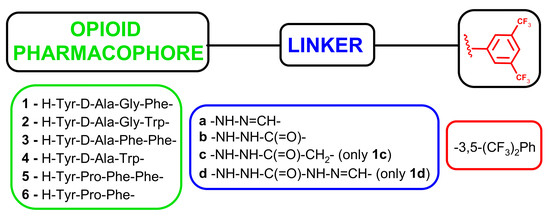
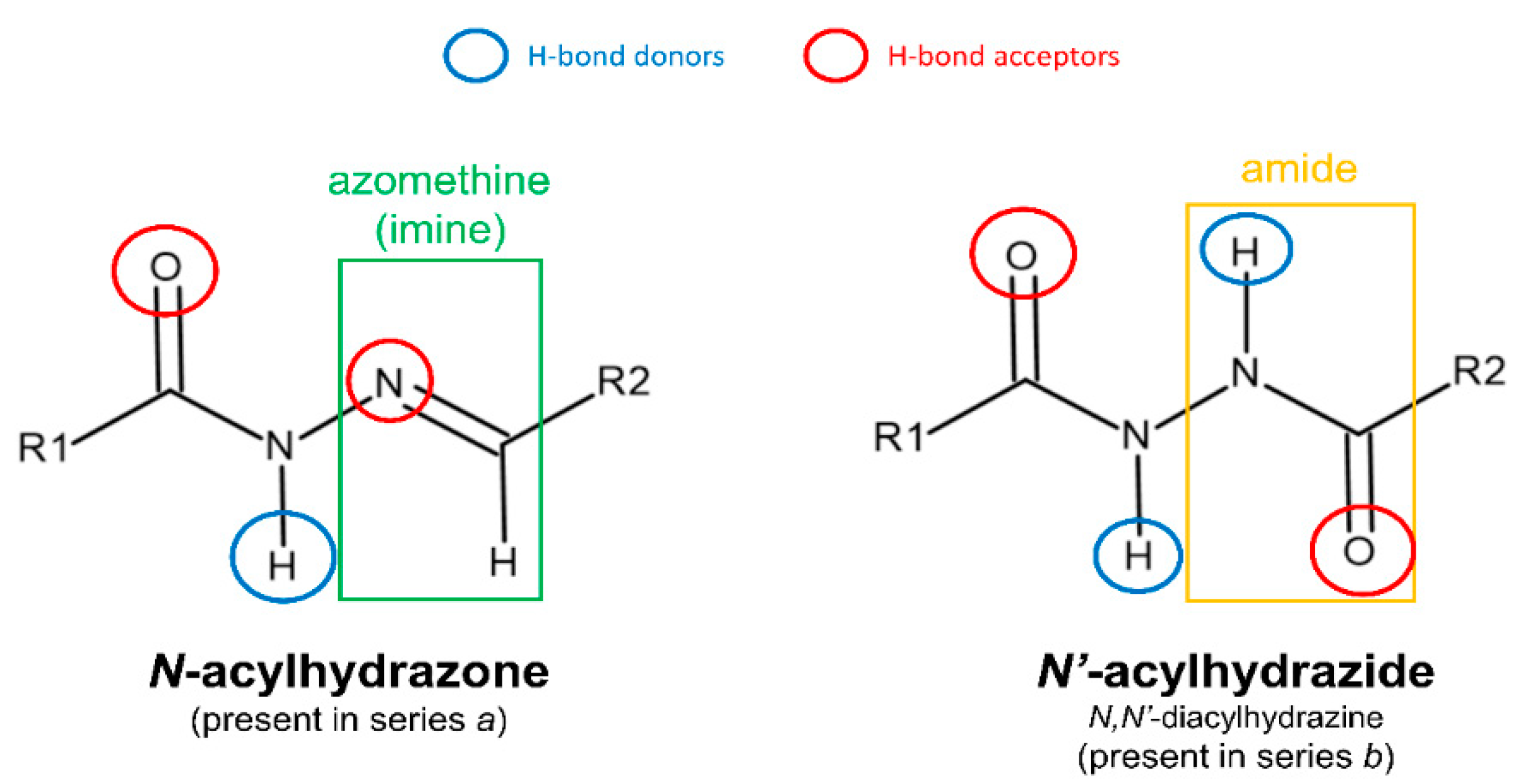
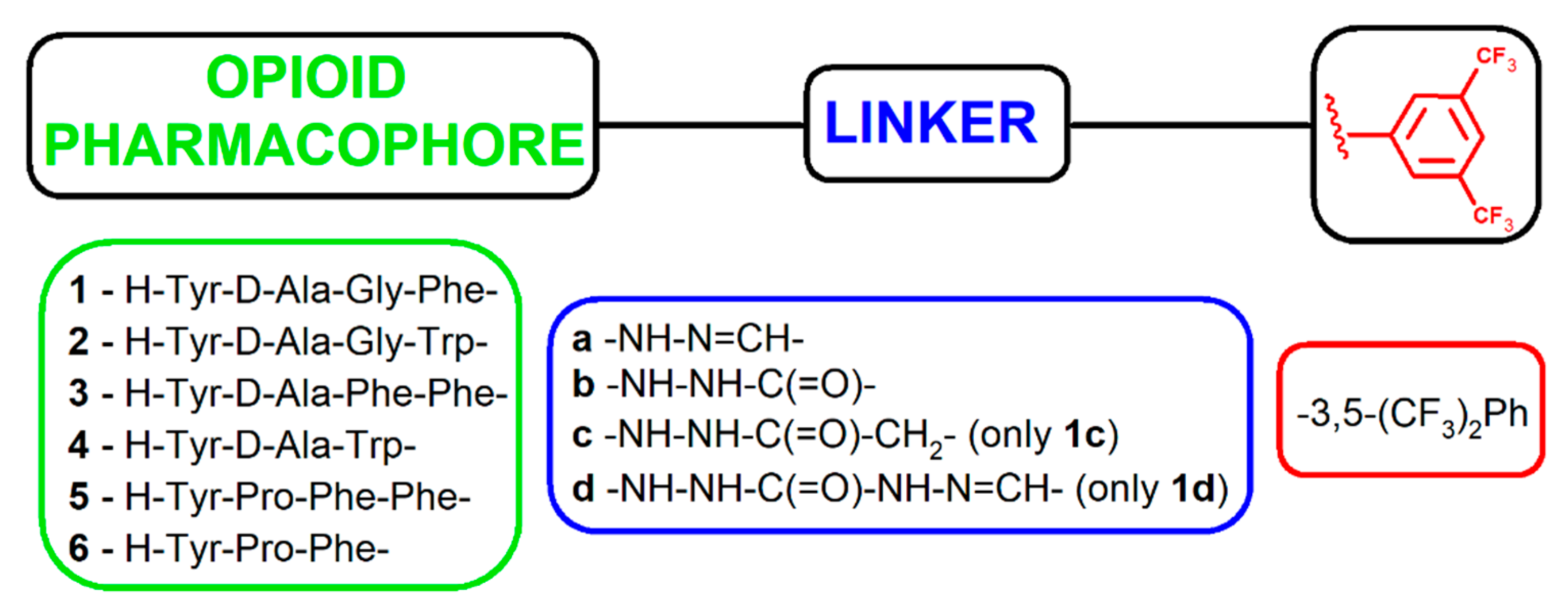
2.1. Chemistry
2.2. Binding Affinity
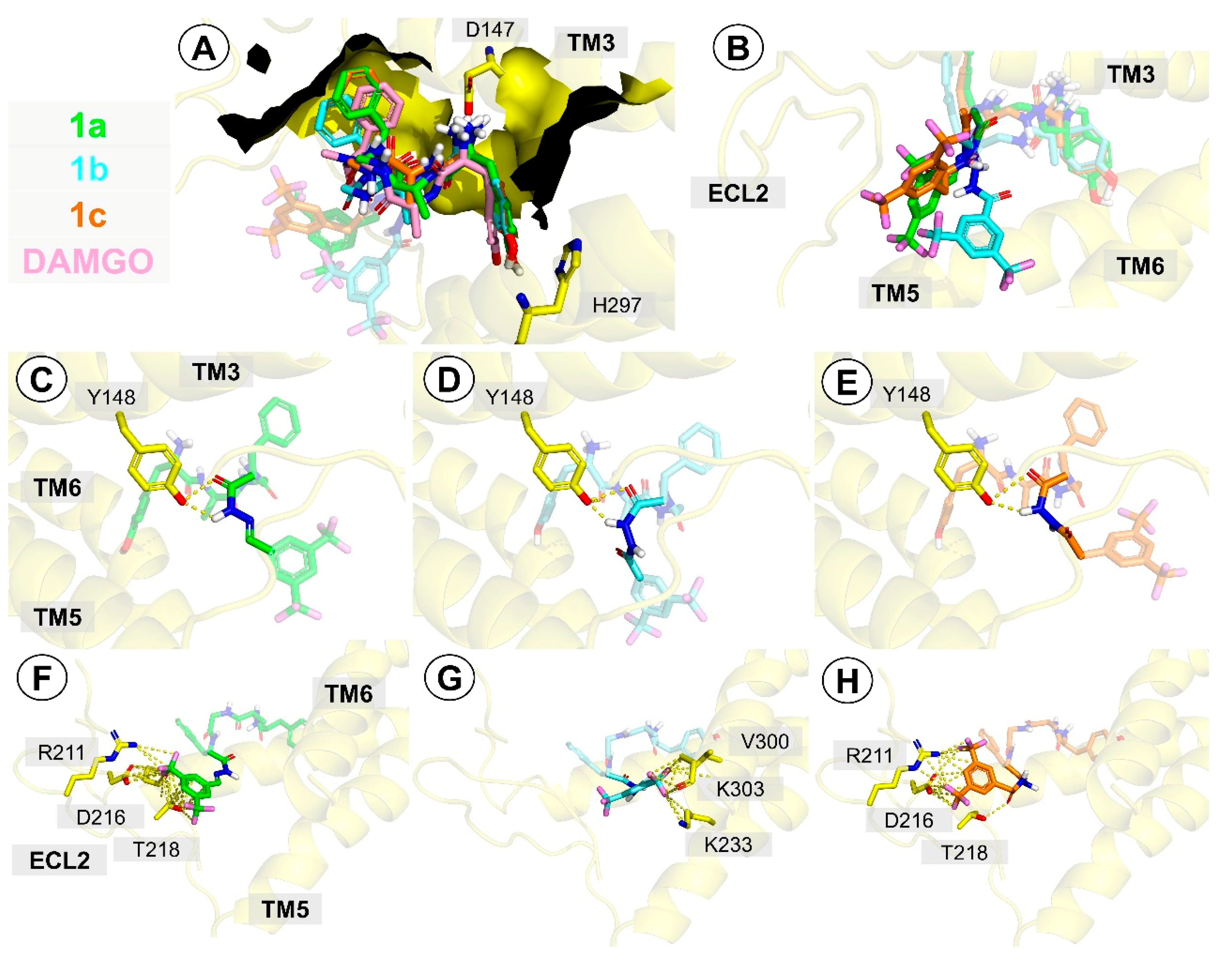
2.3. Molecular Docking
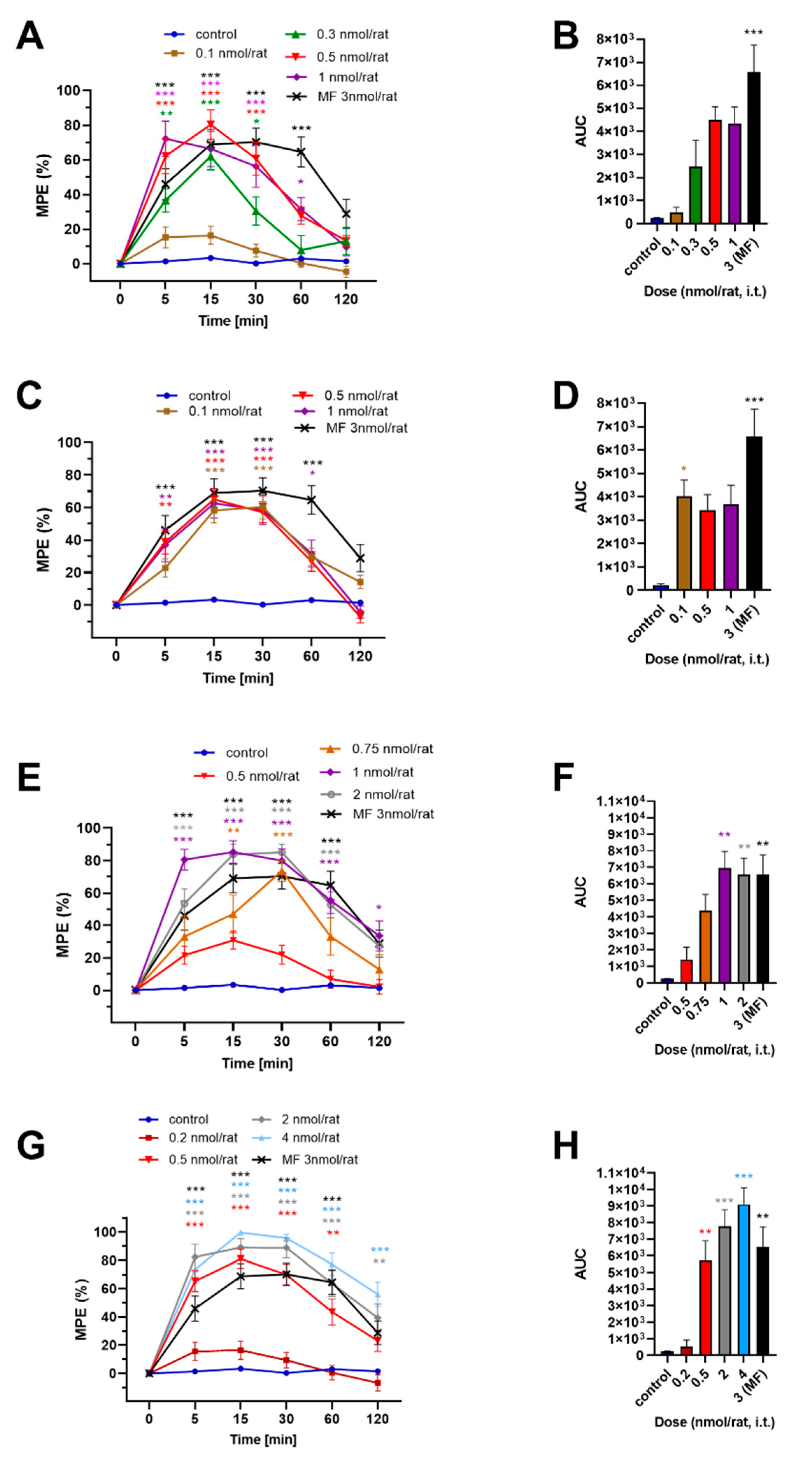
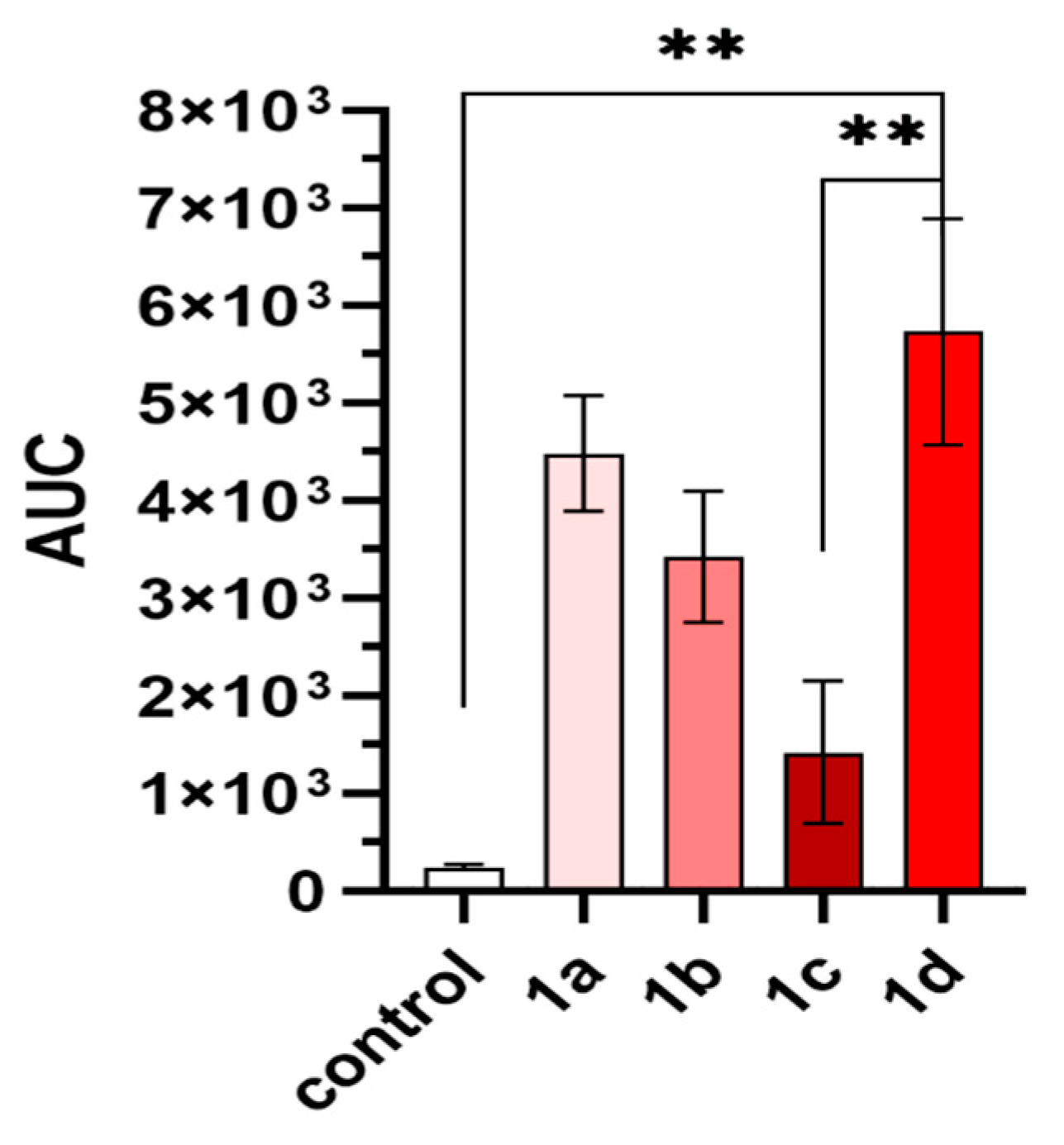
2.4. In Vivo Analgesic Activity
2.5. Cytotoxicity
3. Materials and Methods
3.1. Chemistry
3.2. Synthesis
3.2.1. General Method for Synthesis of Intermediate Hydrazides
3.2.2. General Method for Synthesis of N-acylhydrazone-Type Analogues (2a, 3a, 4a, 5a, 6a)
3.2.3. General Method for Synthesis of Hydrazide Type of Analogues (1b, 1c, 1d, 2b, 3b, 4b)
3.3. Binding Affinity Determinations
3.4. Molecular Docking
3.5. In Vivo Analgesic Activity after Intrathecal Administration
3.6. Evaluation of the Compounds’ Influence on Melanoma Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Duarte, C.D.; Barreiro, E.J.; Fraga, C.A.M. Privileged structures: A useful concept for the rational design of new lead drug candidates. Mini Rev. Med. Chem. 2007, 7, 1108–1119. [Google Scholar] [CrossRef]
- DeSimone, R.; Currie, K.; Mitchell, S.; Darrow, J.; Pippin, D. Privileged Structures: Applications in Drug Discovery. Comb. Chem. High Throughput Screen. 2004, 7, 473–493. [Google Scholar] [CrossRef] [PubMed]
- Thota, S.; Rodrigues, D.A.; de Sena MurteiraPinheiro, P.; Lima, L.M.; Fraga, C.A.M.; Barreiro, E.J. N-Acylhydrazones as drugs. Bioorg. Med. Chem. Lett. 2018, 28, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Fraga, C.; Barreiro, E. Medicinal Chemistry of N-Acylhydrazones: New Lead-Compounds of Analgesic, Antiinflammatory and Antithrombotic Drugs. Curr. Med. Chem. 2006, 13, 167–198. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł.; Biernasiuk, A. New Hydrazides and Hydrazide-Hydrazones of 2,3-Dihalogen Substituted Propionic Acids: Synthesis and in vitro Antimicrobial Activity Evaluation. Chem. Biodivers. 2017, 14, e1700075. [Google Scholar] [CrossRef]
- Popiołek, Ł. Hydrazide–hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2017, 26, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, R.; Reddy, K.T.; Sujitha, P.; Kumar, C.G.; Raju, R.R. Synthesis, antiproliferative and apoptosis induction potential activities of novel bis(indolyl)hydrazide-hydrazone derivatives. Bioorg. Med. Chem. 2019, 27, 1043–1055. [Google Scholar] [CrossRef]
- Settypalli, T.; Chunduri, V.R.; Maddineni, A.K.; Begari, N.; Allagadda, R.; Kotha, P.; Chippada, A.R. Design, synthesis, in silico docking studies and biological evaluation of novel quinoxaline-hydrazide hydrazone-1,2,3-triazole hybrids as α-glucosidase inhibitors and antioxidants. NewJ. Chem. 2019, 43, 15435–15452. [Google Scholar] [CrossRef]
- Blanco, F.; Egan, B.; Caboni, L.; Elguero, J.; O’Brien, J.; McCabe, T.; Fayne, D.; Meegan, M.J.; Lloyd, D.G. Study of E/Z Isomerization in a Series of Novel Non-ligand Binding Pocket Androgen Receptor Antagonists. J. Chem. Inf. Model. 2012, 52, 2387–2397. [Google Scholar] [CrossRef]
- Rodrigues, D.A.; Guerra, F.S.; Sagrillo, F.S.; Sena, M.; Pinheiro, P.; Alves, M.A.; Thota, S.; Chaves, L.S.; Sant’Anna, C.M.R.; Fernandes, P.D.; et al. Design, Synthesis, and Pharmacological Evaluation of First-in-Class Multitarget N -Acylhydrazone Derivatives as Selective HDAC6/8 and PI3Kα Inhibitors. ChemMedChem 2020, 15, 539–551. [Google Scholar] [CrossRef]
- Reynolds, C.H.; Hormann, R.E. Theoretical Study of the Structure and Rotational Flexibility of Diacylhydrazines: Implications for the Structure of Nonsteroidal Ecdysone Agonists and Azapeptides. J. Am. Chem. Soc. 1996, 118, 9395–9401. [Google Scholar] [CrossRef]
- Lipkowski, A.W.; Tam, S.W.; Portoghese, P.S. Peptides as receptor selectivity modulators of opiate pharmacophores. J. Med. Chem. 1986, 29, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, T.; Kasten, B.B.; Bučar, D.-K.; MacGillivray, L.R.; Berkman, C.E.; Benny, P.D. The hydrazide/hydrazone click reaction as a biomolecule labeling strategy for M(CO)3 (M = Re, 99mTc) radiopharmaceuticals. Chem. Commun. 2011, 47, 12846–12848. [Google Scholar] [CrossRef] [PubMed]
- Tymecka, D.; Misicka, A. Solution Phase Peptide Synthesis: The Case of Biphalin. In Peptide Synthesis; Hussein, W., Skwarczynski, M., Toth, I., Eds.; Humana: New York, NY, USA, 2020; pp. 1–11. ISBN 978-1-0716-0226-3. [Google Scholar]
- Lipkowski, A.W.; Konecka, A.M.; Sroczynska, I.; Przewlocki, R.; Stala, L.; Tam, S.W. Bivalent opioid peptide analogues with reduced distances between pharmacophores. Life Sci. 1987, 40, 2283–2288. [Google Scholar] [CrossRef]
- Feliciani, F.; Pinnen, F.; Stefanucci, A.; Costante, R.; Cacciatore, I.; Lucente, G.; Mollica, A. Structure-activity relationships of biphalin analogs and their biological evaluation on opioid receptors. Mini Rev. Med. Chem. 2013, 13, 11–33. [Google Scholar] [CrossRef]
- Misicka, A.; Lipkowski, A.W.; Horvath, R.; Davis, P.; Porreca, F.; Yamamura, H.I.; Hruby, V.J. Structure-activity relationship of biphalin. The synthesis and biological activities of new analogues with modifications in positions 3 and 4. Life Sci. 1997, 60, 1263–1269. [Google Scholar] [CrossRef]
- Cowell, S.M.; Sun Lee, Y. Biphalin: The Foundation of Bivalent Ligands. Curr. Med. Chem. 2016, 23, 3267–3284. [Google Scholar]
- Mollica, A.; Costante, R.; Stefanucci, A.; Pinnen, F.; Lucente, G.; Fidanza, S.; Pieretti, S. Antinociceptive profile of potent opioid peptide AM94, a fluorinated analogue of biphalin with non-hydrazine linker. J. Pept. Sci. 2013, 19, 233–239. [Google Scholar] [CrossRef]
- Garbuz, O.; Gulea, A.; Dyniewicz, J.; Zablocka, B.; Lipkowski, A.W. The non-opioid receptor, antioxidant properties of morphine and the opioid peptide analog biphalin. Peptides 2015, 63, 1–3. [Google Scholar] [CrossRef]
- Mollica, A.; Carotenuto, A.; Novellino, E.; Limatola, A.; Costante, R.; Pinnen, F.; Stefanucci, A.; Pieretti, S.; Borsodi, A.; Samavati, R.; et al. Novel Cyclic Biphalin Analogue with Improved Antinociceptive Properties. ACS Med. Chem. Lett. 2014, 5, 1032–1036. [Google Scholar] [CrossRef]
- Mollica, A.; Davis, P.; Ma, S.-W.; Porreca, F.; Lai, J.; Hruby, V.J. Synthesis and biological activity of the first cyclic biphalin analogues. Bioorg. Med. Chem. Lett. 2006, 16, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Frączak, O.; Lasota, A.; Tymecka, D.; Kosson, P.; Muchowska, A.; Misicka, A.; Olma, A. Synthesis, binding affinities and metabolic stability of dimeric dermorphin analogs modified with β3-homo-amino acids. J. Pept. Sci. 2016, 22, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Lipkowski, A.W.; Misicka, A.; Kosson, D.; Kosson, P.; Lachwa-From, M.; Brodzik-Bienkowska, A.; Hruby, V.J. Biological properties of a new fluorescent biphalin fragment analogue. Life Sci. 2002, 70, 893–897. [Google Scholar] [CrossRef]
- Lukowiak, M.; Kosson, P.; Hennink, W.E.; Lipkowski, A.W. Synthesis and pharmacological properties of a new fluorescent opioid peptide analog. Pharm. Rep. 2009, 61, 727–731. [Google Scholar] [CrossRef]
- Lee, Y.S.; Agnes, R.S.; Badghisi, H.; Davis, P.; Ma, S.; Lai, J.; Porreca, F.; Hruby, V.J. Design and Synthesis of Novel Hydrazide-Linked Bifunctional Peptides as δ/μ Opioid Receptor Agonists and CCK-1/CCK-2 Receptor Antagonists. J. Med. Chem. 2006, 49, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Bonney, I.M.; Foran, S.E.; Marchand, J.E.; Lipkowski, A.W.; Carr, D.B. Spinal antinociceptive effects of AA501, a novel chimeric peptide with opioid receptor agonist and tachykinin receptor antagonist moieties. Eur. J. Pharm. 2004, 488, 91–99. [Google Scholar] [CrossRef]
- Matalinska, J.; Skurzak, H.; Markowicz, S.; Lesniak, A.; Sacharczuk, M.; Molnar, G.; Varga, E.; Lipkowski, A.W. Opioid agonist–tachykinin antagonist as a new analgesic with adjuvant anticancer properties. Folia Neuropathol. 2013, 2, 132–139. [Google Scholar] [CrossRef]
- Wtorek, K.; Adamska-Bartłomiejczyk, A.; Piekielna-Ciesielska, J.; Ferrari, F.; Ruzza, C.; Kluczyk, A.; Piasecka-Zelga, J.; Calo’, G.; Janecka, A. Synthesis and Pharmacological Evaluation of Hybrids Targeting Opioid and Neurokinin Receptors. Molecules 2019, 24, 4460. [Google Scholar] [CrossRef]
- Guillemyn, K.; Kleczkowska, P.; Lesniak, A.; Dyniewicz, J.; Van der Poorten, O.; Van den Eynde, I.; Keresztes, A.; Varga, E.; Lai, J.; Porreca, F.; et al. Synthesis and biological evaluation of compact, conformationally constrained bifunctional opioid agonist-neurokinin-1 antagonist peptidomimetics. Eur. J. Med. Chem. 2015, 92, 64–77. [Google Scholar] [CrossRef]
- Betti, C.; Starnowska, J.; Mika, J.; Dyniewicz, J.; Frankiewicz, L.; Novoa, A.; Bochynska, M.; Keresztes, A.; Kosson, P.; Makuch, W.; et al. Dual Alleviation of Acute and Neuropathic Pain by Fused Opioid Agonist-Neurokinin 1 Antagonist Peptidomimetics. ACS Med. Chem. Lett. 2015, 6, 1209–1214. [Google Scholar] [CrossRef]
- Guillemyn, K.; Starnowska, J.; Lagard, C.; Dyniewicz, J.; Rojewska, E.; Mika, J.; Chung, N.N.; Utard, V.; Kosson, P.; Lipkowski, A.W.; et al. Bifunctional Peptide-Based Opioid Agonist–Nociceptin Antagonist Ligands for Dual Treatment of Acute and Neuropathic Pain. J. Med. Chem. 2016, 59, 3777–3792. [Google Scholar] [CrossRef] [PubMed]
- Dyniewicz, J.; Lipiński, P.F.J.; Kosson, P.; Leśniak, A.; Bochyńska-Czyż, M.; Muchowska, A.; Tourwé, D.; Ballet, S.; Misicka, A.; Lipkowski, A.W. Hydrazone Linker as a Useful Tool for Preparing Chimeric Peptide/Nonpeptide Bifunctional Compounds. ACS Med. Chem. Lett. 2017, 8, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Advanced Chemistry Development Inc. ACD/ChemSketch version. Available online: https://www.acdlabs.com/resources/freeware/chemsketch/ (accessed on 2 July 2020).
- Lee, Y.S.; Petrov, R.; Park, C.K.; Ma, S.; Davis, P.; Lai, J.; Porreca, F.; Vardanyan, R.; Hruby, V.J. Development of Novel Enkephalin Analogues that Have Enhanced Opioid Activities at Both μ and δ Opioid Receptors. J. Med. Chem. 2007, 50, 5528–5532. [Google Scholar] [CrossRef]
- Lipkowski, A.W.; Misicka, A.; Davis, P.; Stropova, D.; Janders, J.; Lachwa, M.; Porreca, F.; Yamamura, H.I.; Hruby, V.J. Biological activity of fragments and analogues of the potent dimeric opioid peptide, biphalin. Bioorg. Med. Chem. Lett. 1999, 9, 2763–2766. [Google Scholar] [CrossRef]
- McGregor, W.H.; Stein, L.; Belluzzi, J.D. Potent analgesic activity of the enkephalin-like tetrapeptide H-Tyr-D-Ala-Gly-Phe-NH2. Life Sci. 1978, 23, 1371–1376. [Google Scholar] [CrossRef]
- Tymecka, D.; Lipiński, P.F.J.; Kosson, P.; Misicka, A. β2-Homo-Amino Acid Scan of µ-Selective Opioid Tetrapeptide TAPP. Molecules 2020, 25, 2461. [Google Scholar] [CrossRef] [PubMed]
- Laskowska, A.K.; Puszko, A.K.; Sosnowski, P.; Różycki, K.; Kosson, P.; Matalińska, J.; Durlik, M.; Misicka, A. Opioid Tripeptides Hybridized with trans -1-Cinnamylpiperazine as Proliferation Inhibitors of Pancreatic Cancer Cells in Two- and Three-Dimensional in vitro Models. Chem. Med. Chem. 2017, 12, 1637–1644. [Google Scholar] [CrossRef]
- Fichna, J.; Do-Rego, J.-C.; Kosson, P.; Costentin, J.; Janecka, A. Characterization of antinociceptive activity of novel endomorphin-2 and morphiceptin analogs modified in the third position. Biochem. Pharm. 2005, 69, 179–185. [Google Scholar] [CrossRef]
- Okada, Y.; Fukumizu, A.; Takahashi, M.; Shimizu, Y.; Tsuda, Y.; Yokoi, T.; Bryant, S.D.; Lazarus, L.H. Synthesis of Stereoisomeric Analogues of Endomorphin-2, H-Tyr-Pro-Phe-Phe-NH2, and Examination of Their Opioid Receptor Binding Activities and Solution Conformation. Biochem. Biophys. Res. Commun. 2000, 276, 7–11. [Google Scholar] [CrossRef]
- Lipiński, P.F.J.; Kosson, P.; Matalińska, J.; Roszkowski, P.; Czarnocki, Z.; Jarończyk, M.; Misicka, A.; Dobrowolski, J.; Sadlej, J. Fentanyl Family at the Mu-Opioid Receptor: Uniform Assessment of Binding and Computational Analysis. Molecules 2019, 24, 740. [Google Scholar] [CrossRef]
- Koehl, A.; Hu, H.; Maeda, S.; Zhang, Y.; Qu, Q.; Paggi, J.M.; Latorraca, N.R.; Hilger, D.; Dawson, R.; Matile, H.; et al. Structure of the µ-opioid receptor–Gi protein complex. Nature 2018, 558, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Manglik, A.; Venkatakrishnan, A.J.; Laeremans, T.; Feinberg, E.N.; Sanborn, A.L.; Kato, H.E.; Livingston, K.E.; Thorsen, T.S.; Kling, R.C.; et al. Structural insights into µ-opioid receptor activation. Nature 2015, 524, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Lipiński, P.F.J.; Jarończyk, M.; Dobrowolski, J.C.; Sadlej, J. Molecular dynamics of fentanyl bound to μ-opioid receptor. J. Mol. Model. 2019, 25, 144. [Google Scholar] [CrossRef] [PubMed]
- Kosson, D.; Bonney, I.; Carr, D.B.; Mayzner-Zawadzka, E.; Lipkowski, A.W. Antinociception after intrathecal biphalin application in rats: A reevaluation and novel, rapid method to confirm correct catheter tip position. Pharmacol. Rep. 2005, 57, 545–549. [Google Scholar] [PubMed]
- Muñoz, M.; Coveñas, R. Neurokinin-1 Receptor Antagonists as Anticancer Drugs. Lett. Drug Des. Discov. 2019, 16, 1110–1129. [Google Scholar] [CrossRef]
- Muñoz, M.; Coveñas, R.; Esteban, F.; Redondo, M. The substance P/NK-1 receptor system: NK-1 receptor antagonists as anti-cancer drugs. J. Biosci. 2015, 40, 441–463. [Google Scholar] [CrossRef]
- Muñoz, M.; Rosso, M. The NK-1 receptor antagonist aprepitant as a broad spectrum antitumor drug. Invest. New Drugs 2010, 28, 187–193. [Google Scholar] [CrossRef]
- Muñoz, M.; Bernabeu-Wittel, J.; Coveñas, R. NK-1 as a melanoma target. Expert Opin. Targets 2011, 15, 889–897. [Google Scholar] [CrossRef]
- Matalińska, J.; Lipiński, P.F.J.; Kotlarz, A.; Kosson, P.; Muchowska, A.; Dyniewicz, J. Evaluation of Receptor Affinity, Analgesic Activity and Cytotoxicity of a Hybrid Peptide, AWL3020. Int. J. Pept. Res. 2020. [Google Scholar] [CrossRef]
- McLaughlin, P.J.; Zagon, I.S. The opioid growth factor–opioid growth factor receptor axis: Homeostatic regulator of cell proliferation and its implications for health and disease. Biochem. Pharm. 2012, 84, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Lipkowski, A. A method of producing an N-blocked peptide. WO2010077155 A1, 8 July 2010. [Google Scholar]
- Tóth, G.; Lovas, S.; Ötvös, F. Tritium Labeling of Neuropeptides. In Neuropeptide Protocols; Humana Press: Totowa, NJ, USA; pp. 219–230.
- GraphPad Prism 8 version 8.3.1 San Diego, C.A. Available online: https://www.graphpad.com/ (accessed on 2 July 2020).
- Schrödinger LLC The PyMOL Molecular Graphics System The PyMOL Molecular Graphics System. Available online: https://www.acdlabs.com/resources/freeware/chemsketch/ (accessed on 2 July 2020).
- Yaksh, T.L.; Rudy, T.A. Chronic catheterization of the spinal subarachnoid space. Physiol. Behav. 1976, 17, 1031–1036. [Google Scholar] [CrossRef]
Sample Availability:
Samples of the compounds 1a–6a are available from the authors. |





| No | Compound | Formula | ESI-MS [M + H] + m/z | tR [min] | LogP | |
|---|---|---|---|---|---|---|
| Calcd | Obsvd | |||||
| 1a[33] | H-Tyr-D-Ala-Gly-Phe-NH-N=CH-3,5-(CF3)2Ph | C32H32F6N6O5 | 695.24 | 694.95 | 12.91 | 5.55 |
| 1b | H-Tyr-D-Ala-Gly-Phe-NH-NH-C(=O)-3,5-(CF3)2Ph | C32H32F6N6O6 | 711.24 | 710.85 | 11.82 | 4.5 |
| 1c | H-Tyr-D-Ala-Gly-Phe-NH-NH-C(=O)-CH2-3,5-(CF3)2Ph | C33H34F6N6O6 | 725.25 | 724.90 | 12.10 | 3.54 |
| 1d | H-Tyr-D-Ala-Gly-Phe-NH-NH-C(=O)-NH-N=CH-3,5-(CF3)2Ph | C33H34F6N8O6 | 753.26 | 752.85 | 12.53 | 4.19 |
| 2a | H-Tyr-D-Ala-Gly-Trp-NH-N=CH-3,5-(CF3)2Ph | C34H33F6N7O5 | 734.25 | 733.90 | 12.88 | 5.48 |
| 2b | H-Tyr-D-Ala-Gly-Trp-NH-NH-C(=O)-3,5-(CF3)2Ph | C34H33F6N7O6 | 750.25 | 749.95 | 12.03 | 4.43 |
| 3a | H-Tyr-D-Ala-Phe-Phe-NH-N=CH-3,5-(CF3)2Ph | C39H38F6N6O5 | 785.29 | 784.85 | 14.20 | 7.74 |
| 3b | H-Tyr-D-Ala-Phe-Phe-NH-NH-C(=O)-3,5-(CF3)2Ph | C39H38F6N6O6 | 801.28 | 800.85 | 13.40 | 6.69 |
| 4a | H-Tyr-D-Ala-Trp-NH-N=CH-3,5-(CF3)2Ph | C32H30F6N6O4 | 677.23 | 677.15 | 12.94 | 6.09 |
| 4b | H-Tyr-D-Ala-Trp-NH-NH-C(=O)-3,5-(CF3)2Ph | C32H30F6N6O5 | 693.23 | 692.85 | 12.25 | 5.04 |
| 5a | H-Tyr-Pro-Phe-Phe-NH-N=CH-3,5-(CF3)2Ph | C41H40F6N6O5 | 811.30 | 810.80 | 14.09 | 8.03 |
| 6a | H-Tyr-Pro-Phe-NH-N=CH-3,5-(CF3)2Ph | C32H31F6N5O4 | 664.24 | 664.16 | 13.25 | 6.03 |
| IC50 ± SEM 1 (nM) | |||
|---|---|---|---|
| Compound | MOR | DOR | NK1R |
| YaGF- (enkephalin-derivative) | |||
| 1a [33] | 12.73 ± 0.75 | 74.20 ± 0.40 | 4.24 µM ± 0.80 |
| 1b | 110.64 ± 12.66 | 413.45 ± 105.15 | >1000 |
| 1c | 5.25 ± 1.14 | 161.65 ± 8.75 | >1000 |
| 1d | 4.73 ± 0.02 | 44.04 ± 2.75 | >1000 |
| YaGW- (enkephalin-derivative) | |||
| 2a | 112.01 ± 16.89 | 914.4 ± 43.50 | >1000 |
| 2b | 93.465 ± 32.84 | 467.05 ± 36.95 | >1000 |
| YaFF- (TAPP) | |||
| 3a | 90.4 ± 9.91 | >1000 | 722.8 ± 58.2 |
| 3b | 391.05 ± 57.85 | >1000 | >1000 |
| YaW- | |||
| 4a | 118.11 ± 40.19 | 621.0 ± 65.20 | >1000 |
| 4b | >1000 | >1000 | >1000 |
| YPFF- (endomorphin-2) | |||
| 5a | 28.5 ± 9.41 | >1000 | >1000 |
| YPF- (truncated endomorphin-2) | |||
| 6a | 113.1 ± 7.00 | >1000 | >1000 |
| Cmpd | Compound | ED50 ± SEM (nmol/rat) 1 |
|---|---|---|
| 1a | H-Tyr-D-Ala-Gly-Phe-NH-N=CH-3,5-(CF3)2Ph | 0.18 ± 0.05 |
| 1b | H-Tyr-D-Ala-Gly-Phe-NH-NH-C(=O)-3,5-(CF3)2Ph | n./a 2 |
| 1c | H-Tyr-D-Ala-Gly-Phe-NH-NH-C(=O)-CH2-3,5-(CF3)2Ph | 0.56 ± 0.04 |
| 1d | H-Tyr-D-Ala-Gly-Phe-NH-NH-C(=O)-NH-N=CH-3,5-(CF3)2Ph | 0.32 ± 0.04 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyniewicz, J.; Lipiński, P.F.J.; Kosson, P.; Bochyńska-Czyż, M.; Matalińska, J.; Misicka, A. Antinociceptive and Cytotoxic Activity of Opioid Peptides with Hydrazone and Hydrazide Moieties at the C-Terminus. Molecules 2020, 25, 3429. https://doi.org/10.3390/molecules25153429
Dyniewicz J, Lipiński PFJ, Kosson P, Bochyńska-Czyż M, Matalińska J, Misicka A. Antinociceptive and Cytotoxic Activity of Opioid Peptides with Hydrazone and Hydrazide Moieties at the C-Terminus. Molecules. 2020; 25(15):3429. https://doi.org/10.3390/molecules25153429
Chicago/Turabian StyleDyniewicz, Jolanta, Piotr F. J. Lipiński, Piotr Kosson, Marta Bochyńska-Czyż, Joanna Matalińska, and Aleksandra Misicka. 2020. "Antinociceptive and Cytotoxic Activity of Opioid Peptides with Hydrazone and Hydrazide Moieties at the C-Terminus" Molecules 25, no. 15: 3429. https://doi.org/10.3390/molecules25153429
APA StyleDyniewicz, J., Lipiński, P. F. J., Kosson, P., Bochyńska-Czyż, M., Matalińska, J., & Misicka, A. (2020). Antinociceptive and Cytotoxic Activity of Opioid Peptides with Hydrazone and Hydrazide Moieties at the C-Terminus. Molecules, 25(15), 3429. https://doi.org/10.3390/molecules25153429