Biological Activities of Selected Plants and Detection of Bioactive Compounds from Ardisia elliptica Using UHPLC-Q-Exactive Orbitrap Mass Spectrometry
Abstract
1. Introduction
2. Results and Discussion
2.1. Total Phenolic Content (TPC) of the Selected Plant Extracts
2.2. DPPH and NO Free Radical Scavenging Activity of the Selected Plant Extracts
2.3. Anti-α-Glucosidase Activity of the Selected Plant Extracts
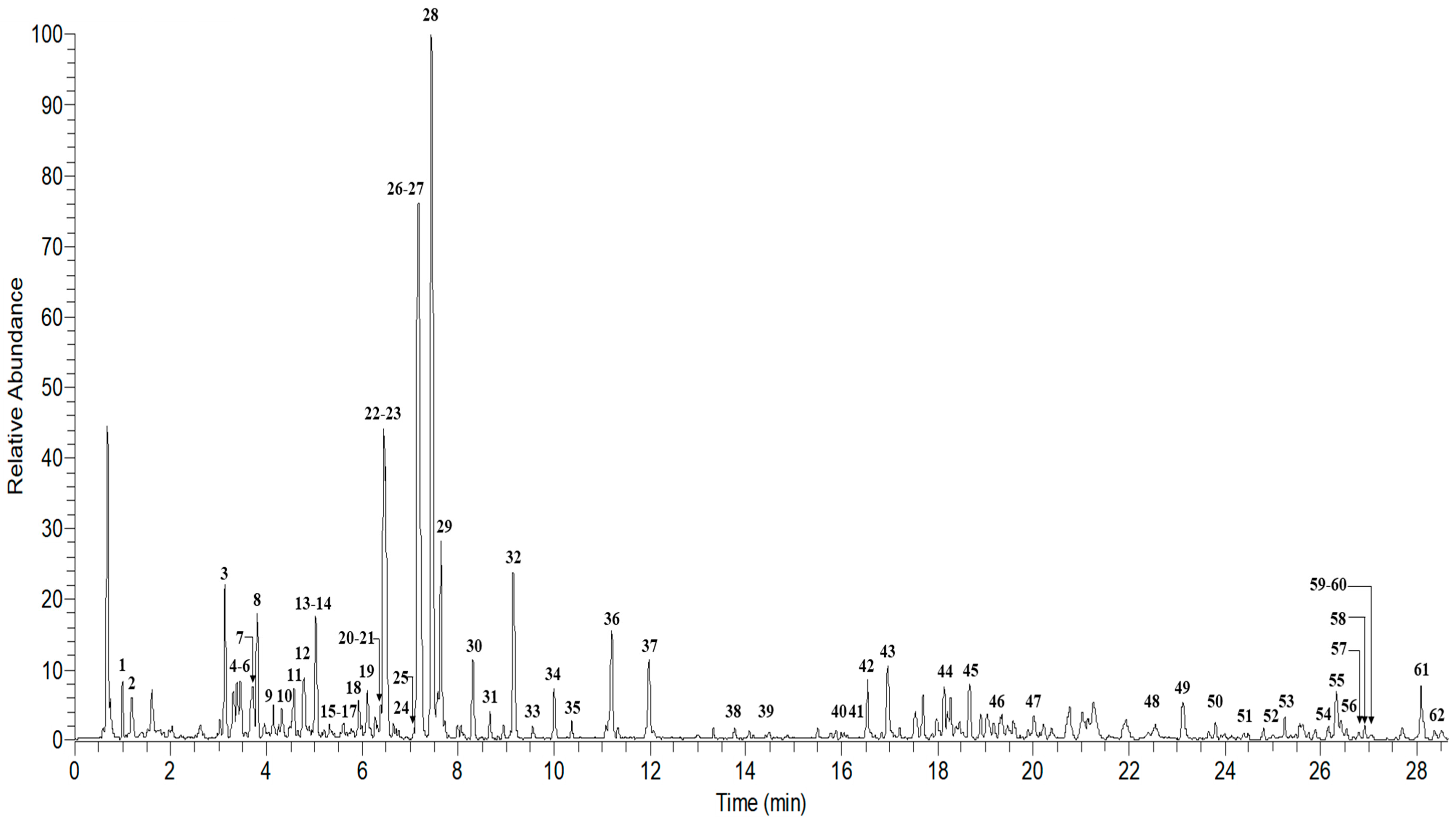
2.4. UHPLC–MS/MS Analysis of Ardisia Elliptica
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plant Collection and Sample Preparation
3.3. Sample Extraction
3.4. Total Phenolic Content (TPC) Determination
3.5. DPPH Free Radical Scavenging Assay
3.6. Nitric Oxide (NO) Scavenging Assay
3.7. Anti-α-Glucosidase Assay
3.8. UHPLC–MS/MS Analysis
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- IPGRI. Neglected and Underutilized Plant Species: Strategic Action Plan of the International Plant Genetic Resources Institute; International Plant Genetic Resources Institute: Rome, Italy, 2002; pp. 1–30. [Google Scholar]
- Feisul, M.I.; Azmi, S. National diabetes registry report. Minist. Health Malays. 2013, 1, 2009–2012. [Google Scholar]
- Trigwell, S.M.; Radford, P.M.; Page, S.R. Islet glutamic acid decarboxylase modified by reactive oxygen species is recognized by antibodies from patients with type 1 diabetes mellitus. Clin. Exp. Immunol. 2001, 126, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Bouayed, J.; Bohn, T. Exogenous antioxidants-Double-edged swords in cellular redox state: Health beneficial effects at physiologic doses versus deleterious effects at high doses. Oxid. Med. Cell Longev. 2010, 3, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Nam, K.A.; Kurihara, H.; Kim, S.M. Potent α-glucosidase inhibitors purified from the red alga Grateloupia elliptica. Phytochemistry 2008, 69, 2820–2825. [Google Scholar] [CrossRef] [PubMed]
- Triggle, C.R.; Ding, H. Cardiovascular impact of drugs used in the treatment of diabetes. Ther. Adv. Chronic Dis. 2014, 5, 245–268. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Narwal, S.; Kumar, V.; Prakash, O. α-glucosidase inhibitors from plants: A natural approach to treat diabetes. Pharmacogn. Rev. 2011, 5, 19–29. [Google Scholar] [CrossRef]
- Rahman, M.M.; Khan, F.E.; Das, R.; Hossain, M.A. Antioxidant activity and total phenolic content of some indigenous fruits of Bangladesh. Int. Food Res. J. 2016, 23, 2399–2404. [Google Scholar]
- Do, Q.D.; Angkawijaya, A.E.; Tran-Nguyen, P.L.; Huynh, L.H.; Soetaredjo, F.E.; Ismadji, S.; Ju, Y.H. Effect of extraction solvent on total phenol content, total flavonoid content, and antioxidant activity of Limnophila aromatica. J. Food Drug Anal. 2014, 22, 296–302. [Google Scholar] [CrossRef]
- Mariod, A.A.; Abdelwahab, S.I.; Elkheir, S.; Ahmed, J.M.; Fauzi, P.N.M.; Chuen, C.S. Antioxidant activity of different parts from Annona squamosa, and Catunaregam nilotica methanolic extract. Acta Sci. Pol. Technol. Aliment. 2012, 11, 249–257. [Google Scholar]
- Rabeta, M.S.; Nur Faraniza, R. Total phenolic content and ferric reducing antioxidant power of the leaves and fruits of Garcinia atrovirdis and Cynometra cauliflora. Int. Food Res. J. 2013, 20, 1691–1696. [Google Scholar]
- Zolkeflee, N.K.Z.; Isamail, N.A.; Maulidiani, M.; Abdul Hamid, N.A.; Ramli, N.S.; Azlan, A.; Abas, F. Metabolite variations and antioxidant activity of Muntingia calabura leaves in response to different drying methods and ethanol ratios elucidated by NMR-based metabolomics. Phytochem. Anal. 2020, in press, 1–15. [Google Scholar] [CrossRef]
- Al-Abd, N.M.; Nor, Z.M.; Mansor, M.; Zajmi, A.; Hasan, M.S.; Azhar, F.; Kassim, M. Phytochemical constituents, antioxidant and antibacterial activities of methanolic extract of Ardisia elliptica. Asian Pac. J. Trop. Biomed. 2017, 7, 569–576. [Google Scholar] [CrossRef]
- Rahardhian, M.R.R.; Murti, B.T.; Wigati, D.; Suharsanti, R.; Putri, C.N. Solvent concentration effect on total flavonoid and total phenolic contents of Averrhoa bilimbi leaf extract. Pharmaciana 2019, 9, 137–144. [Google Scholar] [CrossRef]
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Kitts, D. Antioxidant property of coffee components: Assessment of methods that define mechanism of action. Molecules 2014, 19, 19180–19208. [Google Scholar] [CrossRef]
- Sumanont, Y.; Murakami, Y.; Tohda, M.; Vajragupta, O.; Matsumoto, K.; Watanabe, H. Evaluation of the nitric oxide radical scavenging activity of manganese complexes of curcumin and its derivative. Biol. Pharm. Bull. 2004, 27, 170–173. [Google Scholar] [CrossRef]
- Ado, M.A.; Abas, F.; Ismail, I.S.; Ghazali, H.M.; Shaari, K. Chemical profile and antiacetylcholinesterase, antityrosinase, antioxidant and α-glucosidase inhibitory activity of Cynometra cauliflora L. leaves. J. Sci. Food Agric. 2014, 95, 635–642. [Google Scholar] [CrossRef]
- Lee, S.Y.; Mediani, A.; Ismail, I.S.; Maulidiani; Abas, F. Antioxidants and α-glucosidase inhibitors from Neptunia oleracea fractions using 1H NMR-based metabolomics approach and UHPLC-MS/MS analysis. BMC Complem. Altern. Med. 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Gavillán-Suárez, J.; Aguilar-Perez, A.; Rivera-Ortiz, N.; Rodríguez-Tirado, K.; Figueroa-Cuilan, W.; Morales-Santiago, L.; Martínez-Montemayor, G.; Cubano, L.A.; Martínez-Montemayor, M.M. Chemical profile and in vivo hypoglycemic effects of Syzygium jambos, Costus speciosus and Tapeinochilos ananassae plant extracts used as diabetes adjuvants in Puerto Rico. BMC Complem. Altern. Med. 2015, 15, 1–15. [Google Scholar] [CrossRef]
- Abdul-Hamid, N.A.; Maulidiani, M.; Mediani, A.; Yahya, U.I.I.; Ismail, I.S.; Tham, C.L.; Shadid, K.; Abas, F. Physicochemical characteristics, nutritional composition, and phytochemical profiles of nine Algerian date palm fruit (Phoenix dactylifera L.) varieties. J. Food Biochem. 2018, 42, e12663. [Google Scholar] [CrossRef]
- Hvattum, E.; Ekeberg, D. Study of the collision-induced radical cleavage of flavonoid glycosides using negative electrospray ionization tandem quadrupole mass spectrometry. J. Mass Spectrom. 2003, 38, 43–49. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Zhang, X.; Kirisawa, M.; Uzawa, J.; Sumatra, M. New flavones from Artocarpus communis forst. Chem. Pharm. Bull. 1990, 38, 1787–1789. [Google Scholar] [CrossRef][Green Version]
- Hatano, T.; Kusuda, M.; Hori, M.; Shiota, S.; Tsuchiya, T.; Yoshida, T. Theasinensin A, a tea polyphenol formed from (−)-epigallocatechin gallate, suppresses antibiotic resistance of methicillin-resistant Staphylococcus aureus. Planta Med. 2003, 69, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.Y.; Seok, J.K.; Suh, H.J.; Choi, Y.H.; Hong, S.S.; Kim, D.S.; Boo, Y.C. Anti-melanogenic effects of luteolin 7-sulfate isolated from Phyllospadix iwatensis Makino. Brit. J. Dermatol. 2016, 175, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Teles, Y.C.F.; Souza, M.S.R. Sulphated flavonoids: Biosynthesis, structures, and biological activities. Molecules 2018, 23, 480. [Google Scholar] [CrossRef]
- Cuyckens, F.; Ma, Y.L.; Pocsfalvi, G.; Claeysi, M. Tandem mass spectral strategies for the structural characterization of flavonoid glycosides. Analusis 2000, 28, 888–895. [Google Scholar] [CrossRef]
- Chang, X.; Li, W.; Jia, Z.; Satou, T.; Fushiya, S.; Koike, K. Biologically active triterpenoid saponins from Ardisia japonica. J. Nat. Prod. 2007, 70, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Hisham, A.; Jayakumar, G.; Fujimoto, Y.; Hara, N. 1β,15α-dihydroxyfriedelan-3-one, a triterpene from Salacia beddomei. Phytochemistry 1996, 43, 843–845. [Google Scholar] [CrossRef]
- Naumoska, K.; Vovk, I. Analysis of triterpenoids and phytosterols in vegetables by thin-layer chromatography coupled to tandem mass spectrometry. J. Chromatogr. A 2015, 1381, 229–238. [Google Scholar] [CrossRef]
- Fukuyama, Y.; Kiriyama, Y.; Okino, J.; Kodama, M.; Iwaki, H.; Hosozawa, S.; Matsui, K. Naturally occuring 5-lipoxygenase inhibitor. II. Structures and syntheses of ardisianones A and B, and maesanin, alkenyl-1,4-benzoquinones from the rhizome of Ardisia japonica. Chem. Pharm. Bull. 1993, 41, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Joy, B.; Sivadasan, R.; Emilia, A.T.; John, M.; Sobhan, P.K.; Seervi, M.; Santhoshkumar, T.R. Lysosomal destabilization and Cathepsin B contributes for cytochrome c release and caspase activation in embelin-induced apoptosis. Mol. Carcinogen. 2010, 49, 324–336. [Google Scholar] [CrossRef]
- Yang, L.; Khoo-beattie, C.; Goh, K.; Chng, B.; Yoganathan, K.; Lai, Y.; Butler, M.S. Ardisiaquinones from Ardisia teysmanniana. Phytochemistry 2001, 58, 1235–1238. [Google Scholar] [CrossRef]
- Ogawa, H.; Natori, S. Hydroxybenzoquinones from Myrsinaceae plants. The structures of 2-hydroxy-5-methoxy-3-pentadecenylbenzoquinone and ardisiaquinones A, B and C from Ardisia spp. Chem. Pharm. Bull. 1968, 16, 1709–1720. [Google Scholar] [CrossRef]
- Ndonsta, B.L.; Tatsimo, J.S.N.; Csupor, D.; Forgo, P.; Berkecz, R.; Berenyi, A.; Tene, M.; Molnar, J.; Zupko, I.; Hohmann, J.; et al. Alkylbenzoquinones with antiproliferative effect against human cancer cell lines from stem of Ardisia kivuensis. Phytochem. Lett. 2011, 4, 227–230. [Google Scholar] [CrossRef]
- Sumino, M.; Sekine, T.; Ruangrungsi, N.; Ikegami, F. Ardisiphenols A-C, novel antioxidants from the fruits of Ardisia colorata. Chem. Pharm. Bull. 2001, 49, 1664–1665. [Google Scholar] [CrossRef]
- Sun, W.Y.; Zong, Q.; Gu, R.L.; Pan, B.C. Synthesis of ardisinol II. Synthesis 1998, 11, 1619–1622. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, Y.; Wu, F. Ardisinones A-E, novel diarylundecanones from Ardisia arborescens. J. Nat. Prod. 2004, 67, 1617–1619. [Google Scholar] [CrossRef]
- Ndontsa, B.L.; Dongmo, F.L.M.; Tala, M.F.; Wabo, K.; Zeng, G.; Tan, N.; Tane, P. A new cytotoxic alkenylresorcinol from Embelia schimperi. Rec. Nat. Prod. 2013, 8, 37–40. [Google Scholar]
- David Horgen, F.; Guinaudeau, H.; Pezzuto, J.M.; Soejarto, D.D.; Farnsworth, N.R. Isolation and structure elucidation of ardisenone: A new, cytotoxic alkenylphenol from Ardisia iwahigensis. J. Nat. Prod. 1997, 60, 533–535. [Google Scholar] [CrossRef]
- Taamalli, A.; Arráez-Román, D.; Abaza, L.; Iswaldi, I.; Fernández-Gutiérrez, A.; Zarrouk, M.; Segura-Carretero, A. LC-MS-based metabolite profiling of methanolic extracts from the medicinal and aromatic species Mentha pulegium and Origanum majorana. Phytochem. Analysis 2015, 26, 320–330. [Google Scholar] [CrossRef]
- Mackeen, M.M.; Mooi, L.Y.; Amran, M.; Mat, N.; Lajis, N.H.; Ali, A.M. Noncytotoxic and antitumour-promoting activities of garcinia acid esters from Garcinia atroviridis Griff. ex T. Anders (Guttiferae). Evid. Based Compl. Alt. 2012, 2012, 1–5. [Google Scholar] [CrossRef]
- Scazzocchio, F.; Cometa, M.F.; Tomassini, L.; Palmery, M. Antibacterial activity of Hydrastis canadensis extract and its major isolated alkaloids. Planta Med. 2001, 67, 561–564. [Google Scholar] [CrossRef]
- Li, Y.Q.; Zhou, F.C.; Gao, F.; Bian, J.S.; Shan, F. Comparative evaluation of quercetin, isoquercetin and rutin as inhibitors of α-Glucosidase. J. Agric. Food Chem. 2009, 57, 11463–11468. [Google Scholar] [CrossRef]
- Atoui, A.K.; Mansouri, A.; Boskou, G.; Kefalas, P. Tea and herbal infusions: Their antioxidant activity and phenolic profile. Food Chem. 2005, 89, 27–36. [Google Scholar] [CrossRef]
- Mediani, A.; Abas, F.; Tan, C.; Khatib, A. Effects of different drying methods and storage time on free radical scavenging activity and total phenolic content of Cosmos Caudatus. Antioxidants 2014, 3, 358–370. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, J.; Shen, J.; Silva, A.; Dennis, D.A.; Barrow, C.J. A simple 96-well microplate method for estimation of total polyphenol content in seaweeds. J. Appl. Phycol. 2006, 18, 445–450. [Google Scholar] [CrossRef]
- Wan, C.; Yuan, T.; Cirello, A.L.; Seeram, N.P. Antioxidant and α-glucosidase inhibitory phenolics isolated from highbush blueberry flowers. Food Chem. 2012, 135, 1929–1937. [Google Scholar] [CrossRef]
- Tsai, P.; Tsai, T.; Yu, C.; Ho, S. Comparison of NO-scavenging and NO-suppressing activities of different herbal teas with those of green tea. Food Chem. 2007, 103, 181–187. [Google Scholar] [CrossRef]
- Abd Ghafar, S.Z.; Mediani, A.; Maulidiani; Ramli, N.Z.; Abas, F. Antioxidant, α-glucosidase, and nitric oxide inhibitory activities of Phyllanthus acidus and LC–MS/MS profile of the active extract. Food Bioscience 2018, 25, 134–140. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
| Leaves Extracts/Standard | Total Phenolic Content (mg GAE/g Sample) | IC50 Value (µg/mL) | ||
|---|---|---|---|---|
| DPPH Free Radical Scavenging Activity | NO Free Radical Scavenging Activity | Anti-α-Glucosidase Activity | ||
| Leucaena leucocephala | 175.75 ± 3.48 d | 8.67 ± 0.29 d | 65.57 ± 4.57 b | 6.62 ± 0.19 c |
| Muntingia calabura | 172.32 ± 3.39 d | 4.67 ± 0.21 c | 59.40 ± 3.39 b | 0.51 ± 0.01 a |
| Spondias dulcis | 50.90 ± 0.69 f | 14.22 ± 0.82 e | 301.66 ± 23.06 f | 45.52 ± 2.18 e |
| Annona squamosa | 199.62 ± 7.40 c | 5.00 ± 0.20 c | 109.02 ± 3.18 c | 3.59 ± 0.18 b |
| Ardisia elliptica | 253.10 ± 1.19 b | 2.17 ± 0.08 a | 49.43 ± 0.18 b | 0.29 ± 0.01 a |
| Cynometra cauliflora | 344.17 ± 10.80 a | 2.88 ± 0.05 ab | 118.62 ± 3.44 cd | 0.90 ± 0.02 a |
| Ficus auriculata | 167.15 ± 2.04 d | 5.06 ± 0.35 c | 169.65 ± 1.53 e | 0.36 ± 0.02 a |
| Averrhoa bilimbi | 97.50 ± 3.46 e | 16.80 ± 0.04 f | 134.33 ± 2.46 d | 26.91 ± 0.58 d |
| Quercetin | - | 3.55 ± 0.28 b | 15.85 ± 0.58 a | 6.62 ± 0.03 c |
| Gallic acid | - | - | 15.41 ± 0.63 a | - |
| Peak No. | Retention Time, Min | Exact Mass | Deprotonated Molecule [M − H]− (m/z) | Delta | MS/MS Fragment Ions | Tentative Identification | Molecular Formula |
|---|---|---|---|---|---|---|---|
| 1 | 1.00 | 192.0197 | 191.0184 | 0.0013 | 173.0080, 129.0180, 111.0074, 87.0073 | Citric acid | C6H8O7 |
| 2 | 1.19 | 332.0671 | 331.0662 | 0.0009 | 312.1069, 271.0454, 241.0342, 211.0238, 169.0130 | Monogalloylglucose | C13H16O10 |
| 3 | 3.14 | 452.4087 | 451.3394 | 0.0693 | 302.5710, 289.0708, 210.2854, 151.2638 | (+)-Catechin 6-C-glucoside | C21H24O11 |
| 4 | 3.31 | 282.0817 | 281.0331 | 0.0486 | 239.1548, 219.1387, 207.1384, 201.1277, 165.0904 | 5,7-Dimethoxyflavone | C17H14O4 |
| 5 | 3.40 | 282.0817 | 281.0329 | 0.0488 | 239.1548, 219.1387, 207.1384, 201.1277, 165.0904 | 5,7-Dimethoxyflavone isomer | C17H14O4 |
| 6 | 3.45 | 282.0817 | 281.0328 | 0.0489 | 239.1548, 219.1387, 207.1384, 201.1277, 165.0904 | 5,7-Dimethoxyflavone isomer | C17H14O4 |
| 7 | 3.70 | 762.1571 | 761.1343 | 0.0228 | 610.1257, 301.0352, 169.0131 | Quercetin 3-O-(2″-O-galloyl)-rutinoside | C34H34O20 |
| 8 | 3.80 | 550.1402 | 549.1448 | 0.0046 | 429.1028, 369.0819, 339.0715, 309.0611, 269.0662 | Formononetin 7-O-(2″-p-hydroxybenzoylglucoside) | C29H26O11 |
| 9 | 4.14 | 463.1168 | 461.1610 * | 0.0442 | 314.0427, 300.1083, 287.0562, 255.0286 | Oxycoccicyanin/Peonidin-3-glucoside | C22H23O11 |
| 10 | 4.34 | 454.1555 | 453.1605 | 0.0050 | 386.9793, 367.0700, 301.0338, 284.0323, 176.0435 | KB-2/5,2″,4′,5′-Tetrahydroxy-3-(3-hydroxy-3-methylbutyl)-6″,6″-dimethylpyrano[2″,3″:7,8]flavone | C25H26O8 |
| 11 | 4.57 | 914.1469 | 913.1451 | 0.0018 | 761.1329, 743.1249, 609.1331, 591.1135, 575.0852, 453.0853 | Theasinensin A | C44H34O22 |
| 12 | 4.79 | 350.0024 | 349.0591 | 0.0567 | 269.6351, 241.0011, 227.0375, 152.0433 | Apigenin 7-sulfate | C15H10O8S |
| 13 | 4.95 | 290.0790 | 289.1802 | 0.1012 | 245.0812, 203.0703, 179.0335, 137.0230, 123.0437 | Catechin | C15H14O6 |
| 14 | 5.02 | 458.0776 | 457.0766 | 0.0010 | 331.0454, 305.0666, 287.0562, 269.0456, 193.0132, 169.0131 | (−)-epigallocatechin-3-gallate | C22H18O11 |
| 15 | 5.61 | 756.2040 | 755.2025 | 0.0015 | 489.1044, 301.0315, 300.0270, 271.0243, 255.0294, 178.9978 | Quercetin 3-O-(2,6-di-O-rhamnosylglucoside) | C33H40O20 |
| 16 | 5.71 | 480.0831 | 479.0816 | 0.0015 | 316.0219, 287.0189, 271.0241, 178.9979, 151.0025 | Myricetin-3-O-glucoside | C21H20O13 |
| 17 | 5.76 | 626.1410 | 625.1396 | 0.0014 | 478.0751, 317.0288, 316.0212, 271.0243, 178.9976 | Myricetin-3-O-rutinoside | C27H30O17 |
| 18 | 5.93 | 596.1305 | 595.1293 | 0.0012 | 463.0802, 301.0349, 300.0271, 283.0230, 271.0244, 178.9975 | Quercetin 3-lathyroside | C26H28O16 |
| 19 | 6.11 | 450.0726 | 449.0713 | 0.0013 | 316.0220, 287.0198, 271.0241, 178.9975 | Myricetin-3-O-arabinoside | C20H18O12 |
| 20 | 6.32 | 610.1461 | 609.1451 | 0.0010 | 463.0797, 447.0925, 301.0349, 300.0247 | Quercetin 3-O-α-l-rhamnoside-7-O-β-d-glucoside | C27H30O16 |
| 21 | 6.38 | 610.1461 | 609.1450 | 0.0011 | 301.0339, 300.0269, 271.0244, 178.9973, 151.0026 | Quercetin-3-O-rutinoside (Rutin) | C27H30O16 |
| 22 | 6.45 | 464.0882 | 463.0869 | 0.0013 | 316.0218, 287.0193, 178.9977, 151.0026 | Myricetin-3-O-rhamnoside | C21H20O12 |
| 23 | 6.47 | 381.9922 | 380.9911 | 0.0011 | 301.0349, 300.0247, 283.0245, 271.0238, 257.0445, 229.0496, 193.0133 | Quercetin 3-O-sulfate | C15H10O10S |
| 24 | 6.66 | 464.0882 | 463.0875 | 0.0007 | 301.0339, 300.0270, 271.0244, 255.0293, 178.9976, 151.0024 | Quercetin-3-O-glucoside | C21H20O12 |
| 25 | 6.95 | 434.0776 | 433.0768 | 0.0008 | 301.0346, 300.0271, 271.0243, 255.0291, 178.9982, 151.0023 | Quercetin-3-O-arabinoside | C20H18O11 |
| 26 | 7.18 | 286.0405 | 285.0395 | 0.0012 | 257.0450, 213.0545, 187.0391, 163.0021 | Kaempferol | C15H10O6 |
| 27 | 7.19 | 365.9973 | 364.9961 | 0.0010 | 285.0400, 267.0294, 255.0291, 241.0501, 229.0500, 213.0548, 178.4121, 133.0279, 105.6355 | Luteolin 7-sulfate | C15H10O9S |
| 28 | 7.44 | 448.0933 | 447.0917 | 0.0016 | 301.0339, 300.0271, 271.0243, 255.0291, 178.9975, 151.0024 | Quercetin-3-O-rhamnoside | C21H20O11 |
| 29 | 7.64 | 416.1035 | 415.1962 | 0.0927 | 252.1096, 238.9105, 177.2131, 123.0804 | 7,2′-Dihydroxyflavone 7-glucoside | C21H20O9 |
| 30 | 8.31 | 432.0984 | 431.0970 | 0.0014 | 285.0393, 284.0321, 255.0293, 227.0341 | Kaempferol-3-O-rhamnoside | C21H20O10 |
| 31 | 8.66 | 458.0776 | 457.0766 | 0.0010 | 331.0454, 305.0666, 287.0562, 269.0456, 193.0132, 169.0131 | Epigallocatechin-3-gallate isomer | C22H18O11 |
| 32 | 9.15 | 396.0079 | 395.0064 | 0.0015 | 315.0607, 272.0317, 259.0608, 151.0027 | Persicarin/Isorhamnetin 3-sulfate | C16H12O10S |
| 33 | 9.57 | 302.0354 | 301.0348 | 0.0006 | 273.0403, 178.9974, 151.0024, 121.0281 | Quercetin | C15H10O7 |
| 34 | 10.00 | 380.0129 | 379.0117 | 0.0012 | 299.0555, 284.0321, 257.0403, 243.0656, 228.0399, 211.0385, 162.5436, 151.0027, 110.0001 | Rhamnocitrin 3-O-sulfate | C16H12O9S |
| 35 | 10.37 | 542.0658 | 541.0644 | 0.0014 | 461.1082, 314.0426, 299.0188, 271.0243, 256.0363, 158.7938 | Isoscutellarein 4′-methyl ether 8-(2′-sulfatoglucoside) | C22H22O14S |
| 36 | 11.20 | 328.2177 | 327.2171 | 0.0006 | 229.1440, 211.1331, 171.1015 | Trihydroxy octadecadienoic acid | C18H32O5 |
| 37 | 11.98 | 328.2177 | 327.2171 | 0.0006 | 229.1440, 211.1331, 171.1015 | Trihydroxy octadecadienoic acid isomer | C18H32O5 |
| 38 | 13.78 | 376.2541 | 375.1778 | 0.0763 | 333.6364, 330.1770, 329.1730, 307.1919, 235.1334, 207.0993 | Ardisiphenol B | C23H36O4 |
| 39 | 14.49 | 290.2173 | 289.1803 | 0.0370 | 245.1902, 161.9148, 148.7701, 123.0794 | Ardisinol II | C19H30O2 |
| 40 | 16.00 | 884.5061 | 883.4165 | 0.0896 | 837.4141, 559.1864, 456.2514, 397.1332, 277.2172 | Ardisianoside D | C46H76O16 |
| 41 | 16.37 | 346.2497 | 345.1830 | 0.0667 | 306.9802, 292.2949, 192.5377 | Ardisianone A | C22H34O3 |
| 42 | 16.54 | 722.1410 | 721.3632 | 0.2222 | 675.3585, 569.1615, 415.1444, 400.9850, 305.0875, 277.2165 | Thonningianin B | C35H30O17 |
| 43 | 16.96 | 722.1410 | 721.3631 | 0.2221 | 675.3580, 569.1614, 415.1447, 400.9853, 305.0875, 277.2166 | Thonningianin B isomer | C35H30O17 |
| 44 | 18.14 | 294.1758 | 293.2114 | 0.0356 | 275.2011, 155.1072, 141.1270, 127.1115, 121. 1009 | Embelin | C17H26O4 |
| 45 | 18.67 | 560.0399 | 559.1269 | 0.0870 | 354.6981, 286.8783, 228.8837, 121.6282 | Gossypetin 8-glucoside-3-sulfate | C21H20O16S |
| 46 | 19.33 | 426.3789 | 425.2304 | 0.1485 | 271.0612, 245.0811, 203.0705, 177.0179, 151.0386 | Friedelan-3-one | C30H50O |
| 47 | 20.00 | 572.2621 | 571.2880 | 0.0259 | 530.8586, 487.1684, 391.2236, 255.2324, 241.0111, 223.0012 | Ardisiaquinone G | C31H40O10 |
| 48 | 22.76 | 378.2334 | 377.2329 | 0.0005 | 359.2229, 335.2515, 316.2326, 152.0106 | Cornudentanone | C22H34O5 |
| 49 | 23.12 | 324.0328 | 323.1162 | 0.0834 | 279.2323, 265.8335, 216.0093, 184.0194 | 6-Chlorocatechin | C15H13ClO6 |
| 50 | 23.80 | 360.0772 | 359.0612 | 0.0160 | 317.2494, 315.2661, 245.0694, 211.2592 | Acerosin | C18H16O8 |
| 51 | 24.48 | 386.2093 | 385.2741 | 0.0648 | 268.6892, 176.3903, 153.4738 | Ardisinone E | C23H30O5 |
| 52 | 25.02 | 318.2558 | 317.2477 | 0.0081 | 300.0236, 231.3264, 192.0054, 178.9974, 151.0025 | Bilobol | C21H34O2 |
| 53 | 25.24 | 502.1919 | 501.3211 | 0.1292 | 486.2979, 473.2810, 456.2825, 443.2783, 435.2527 | Cycloheterophyllin | C30H30O7 |
| 54 | 26.18 | 418.3083 | 417.2634 | 0.0449 | 401.6784, 375.2524, 335.2505, 308.5951, 193.0859 | Maesaquinone | C26H42O4 |
| 55 | 26.33 | 332.1551 | 331.2267 | 0.0716 | 313.2373, 287.2391, 254.8929, 225.6182, 213.1681 | Gibberellin A4 | C19H24O5 |
| 56 | 26.54 | 336.1163 | 335.1342 | 0.0179 | 332.2440, 317.2474, 305.2462, 279.2698, 230.0207 | Berberine | C20H18NO4 |
| 57 | 26.80 | 542.2879 | 541.3524 | 0.0645 | 526.3293, 511.3054, 493.2956, 359.2590, 183.6019 | Ardisiaquinone D | C31H42O8 |
| 58 | 26.93 | 426.3861 | 425.2300 | 0.1489 | 407.0769, 381.0991, 339.0862, 257.0452, 216.0410, 167.2586 | Alpha-amyrin | C30H50O |
| 59 | 27.05 | 528.2723 | 527.3369 | 0.0646 | 514.3290, 499.3061, 191.0708, 165.0543, 151.0386 | Ardisiaquinone A | C30H40O8 |
| 60 | 27.08 | 530.2952 | 529.3524 | 0.0572 | 514.3292, 499.3058, 481.2948, 453.2976, 347.2579, 225.7453 | Ardisiaquinone J | C30H42O8 |
| 61 | 28.09 | 334.1555 | 333.2426 | 0.0871 | 279.2678, 186.7317, 134.0364 | 1,5-Dibutyl methyl hydroxycitrate | C15H26O8 |
| 62 | 28.37 | 496.2824 | 495.2625 | 0.0199 | 316.1812, 278.8960, 205.1232, 181.0499, 169.0134 | Ardisenone | C30H40O6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, P.L.; Fauzi, N.A.; Mohamed Yunus, S.N.; Abdul Hamid, N.A.; Abd Ghafar, S.Z.; Azizan, A.; Zolkeflee, N.K.Z.; Abas, F. Biological Activities of Selected Plants and Detection of Bioactive Compounds from Ardisia elliptica Using UHPLC-Q-Exactive Orbitrap Mass Spectrometry. Molecules 2020, 25, 3067. https://doi.org/10.3390/molecules25133067
Wong PL, Fauzi NA, Mohamed Yunus SN, Abdul Hamid NA, Abd Ghafar SZ, Azizan A, Zolkeflee NKZ, Abas F. Biological Activities of Selected Plants and Detection of Bioactive Compounds from Ardisia elliptica Using UHPLC-Q-Exactive Orbitrap Mass Spectrometry. Molecules. 2020; 25(13):3067. https://doi.org/10.3390/molecules25133067
Chicago/Turabian StyleWong, Pei Lou, Nurul Azila Fauzi, Siti Norhamimah Mohamed Yunus, Nur Ashikin Abdul Hamid, Siti Zulaikha Abd Ghafar, Awanis Azizan, Nur Khaleeda Zulaikha Zolkeflee, and Faridah Abas. 2020. "Biological Activities of Selected Plants and Detection of Bioactive Compounds from Ardisia elliptica Using UHPLC-Q-Exactive Orbitrap Mass Spectrometry" Molecules 25, no. 13: 3067. https://doi.org/10.3390/molecules25133067
APA StyleWong, P. L., Fauzi, N. A., Mohamed Yunus, S. N., Abdul Hamid, N. A., Abd Ghafar, S. Z., Azizan, A., Zolkeflee, N. K. Z., & Abas, F. (2020). Biological Activities of Selected Plants and Detection of Bioactive Compounds from Ardisia elliptica Using UHPLC-Q-Exactive Orbitrap Mass Spectrometry. Molecules, 25(13), 3067. https://doi.org/10.3390/molecules25133067






