Evaluation of the Anticancer Activities of Novel Transition Metal Complexes with Berenil and Nitroimidazole
Abstract
1. Introduction
2. Results
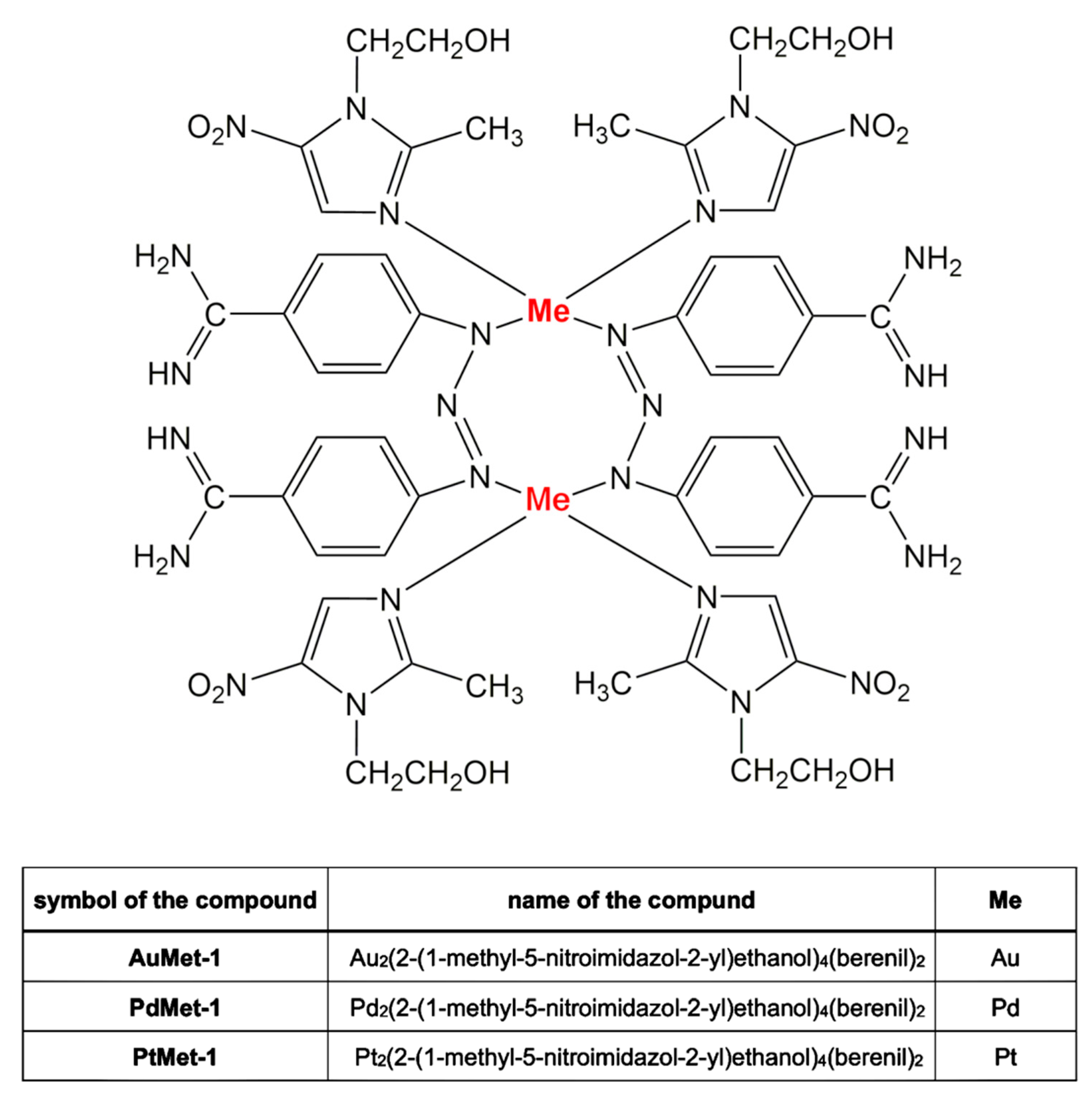
2.1. Chemistry Section
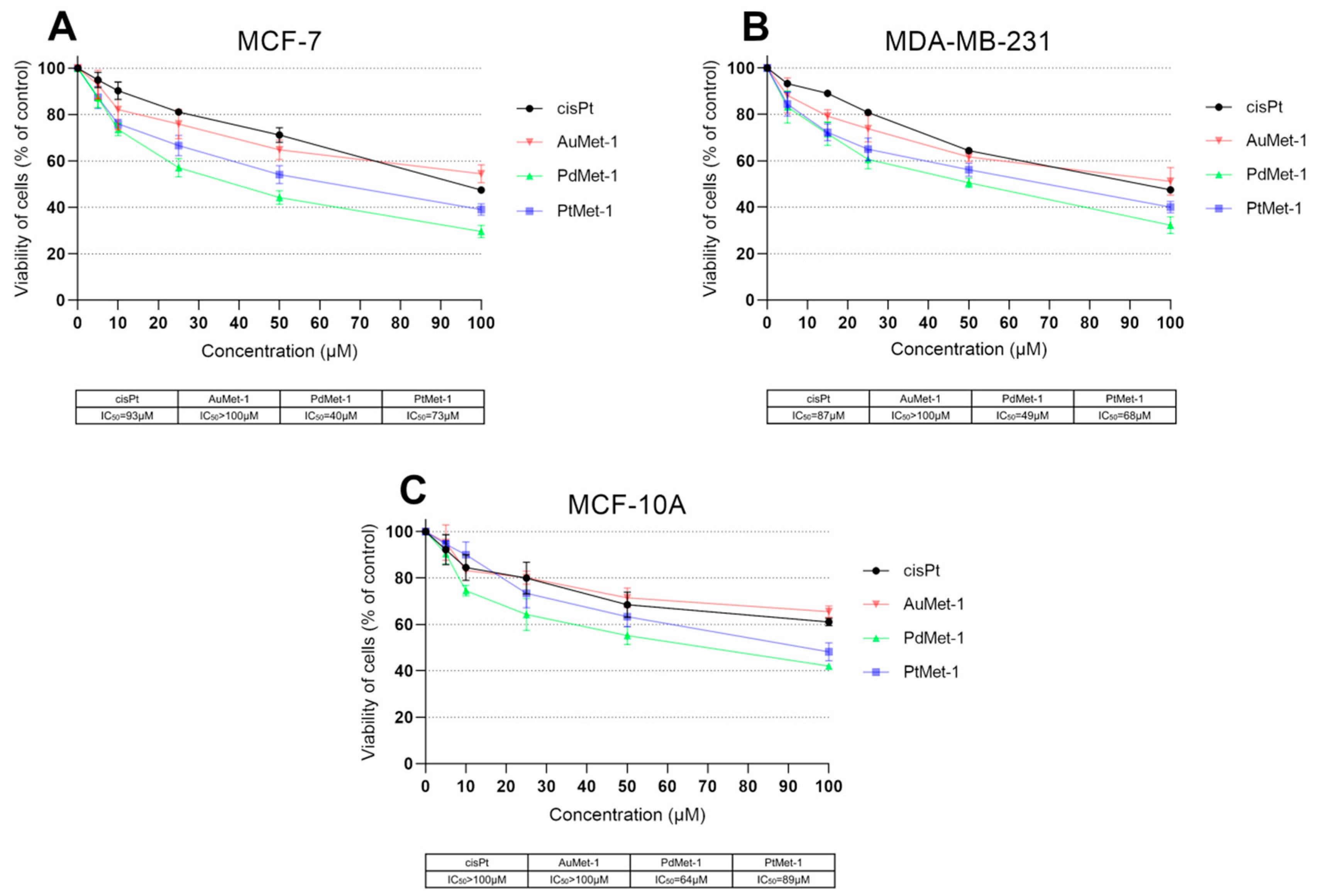
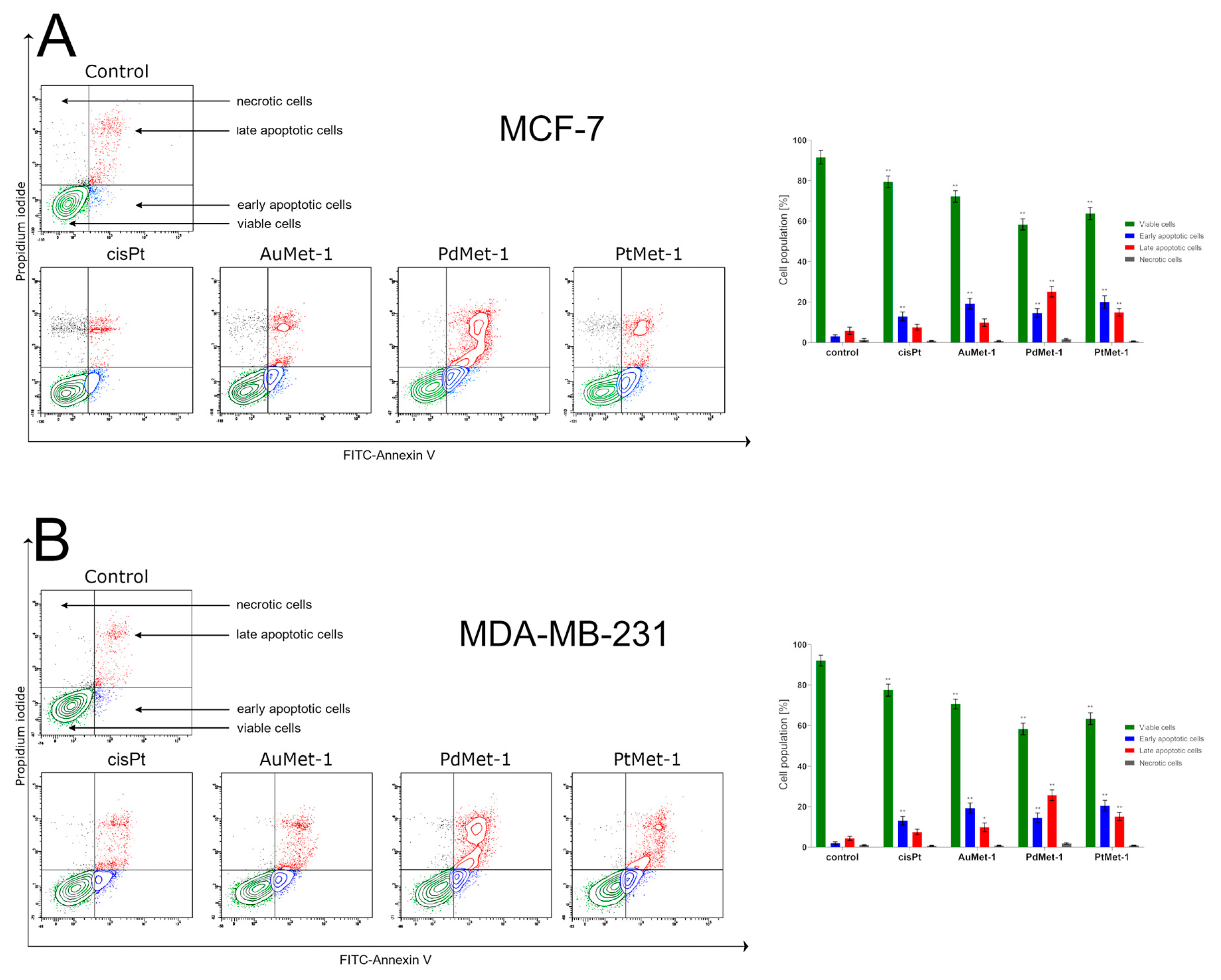
2.2. Biological Activity of Novel Series of Transition Metal Complexes with Nitroimidazole and Berenil (AuMet-1, PdMet-1, PtMet-1)
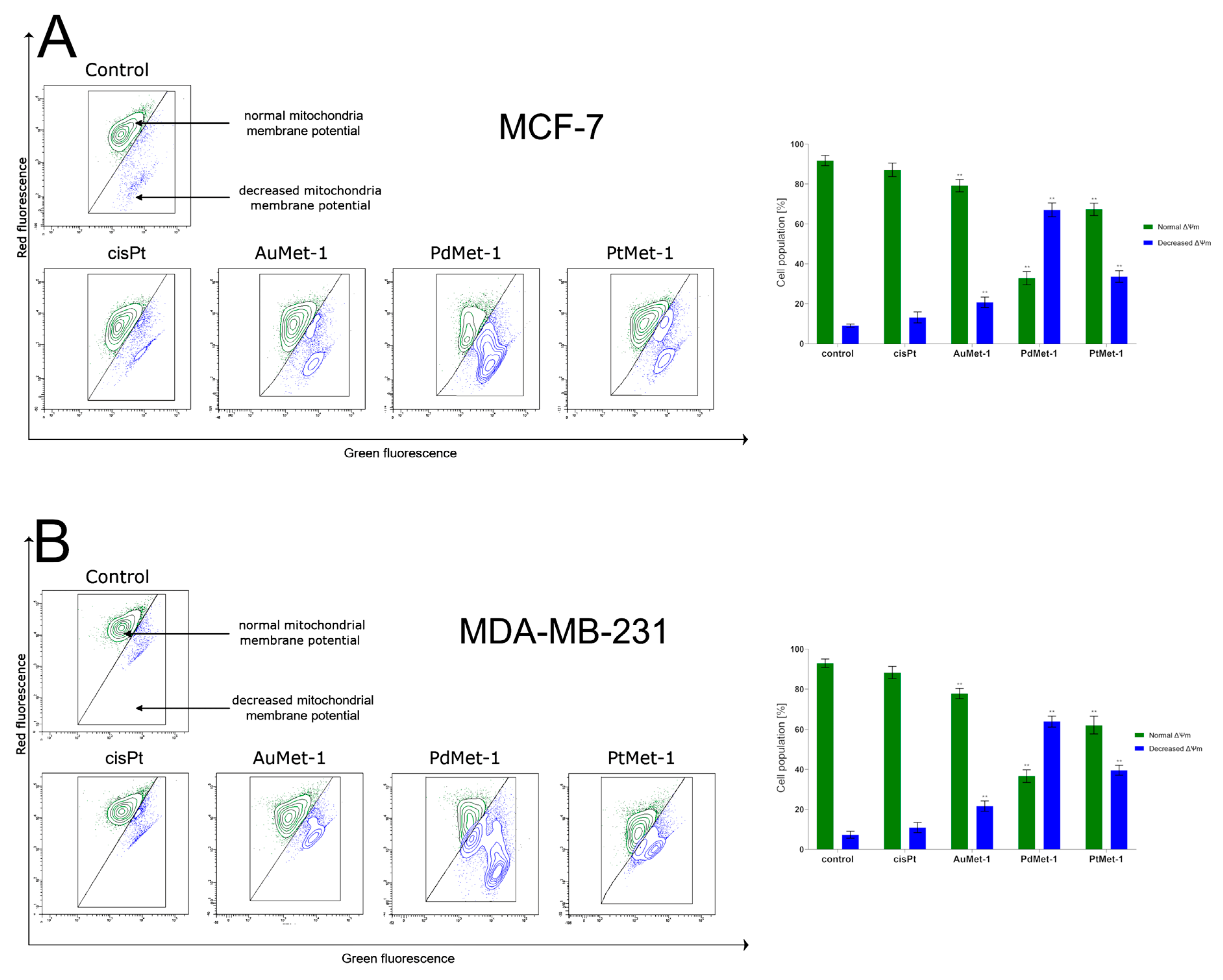
2.3. Novel Series of Transition Metal Compounds with Nitroimidazole and Berenil Moiety (AuMet-1, PdMet-1, PtMet-1) Induce Apoptosis by Decreasing Mitochondrial Membrane Potential
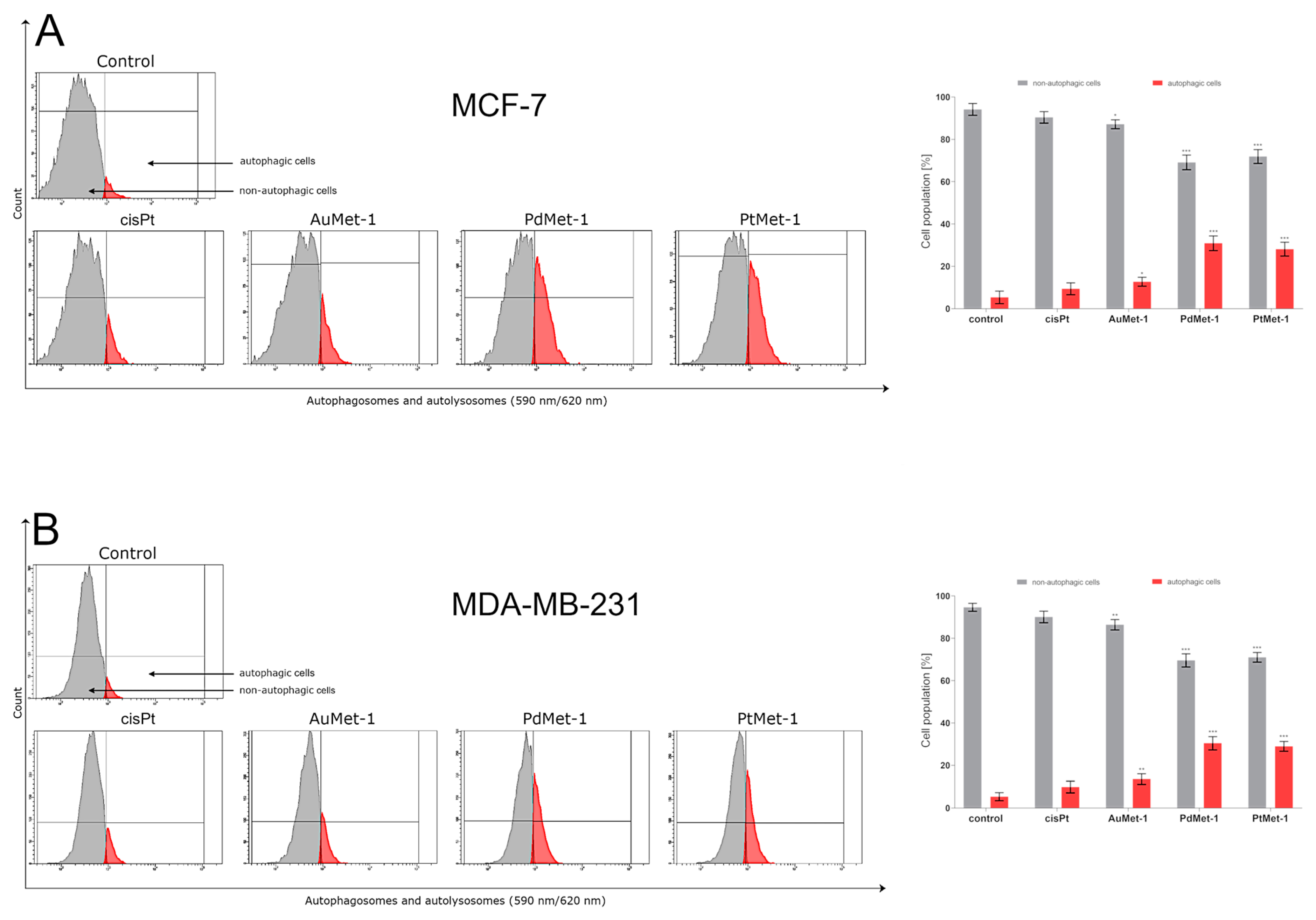
2.4. AuMet-1, PdMet-1 and PtMet-1 Induce Autophagy
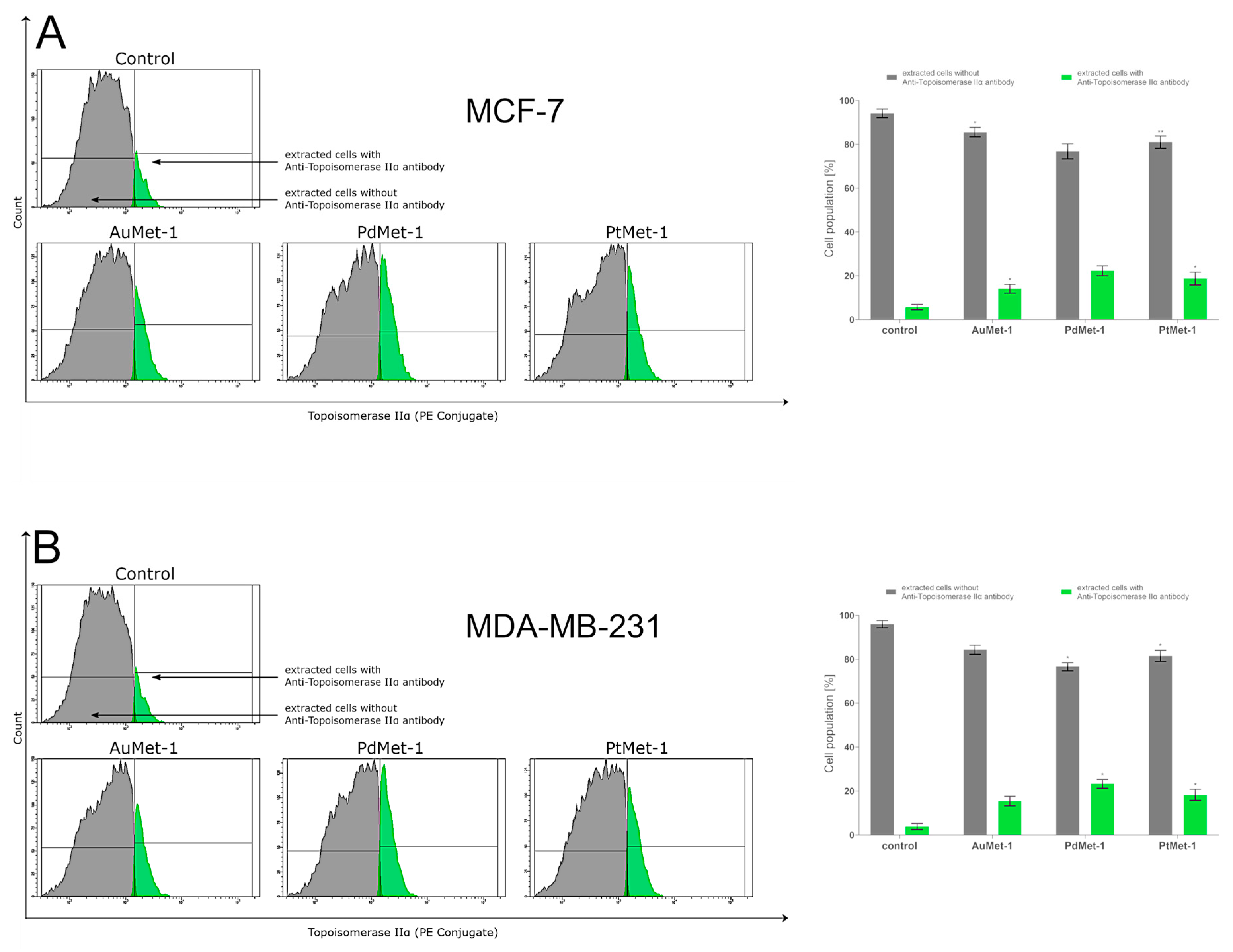
2.5. AuMet-1, PdMet-1, and PtMet-1 Increase Topoisomerase IIα Activity
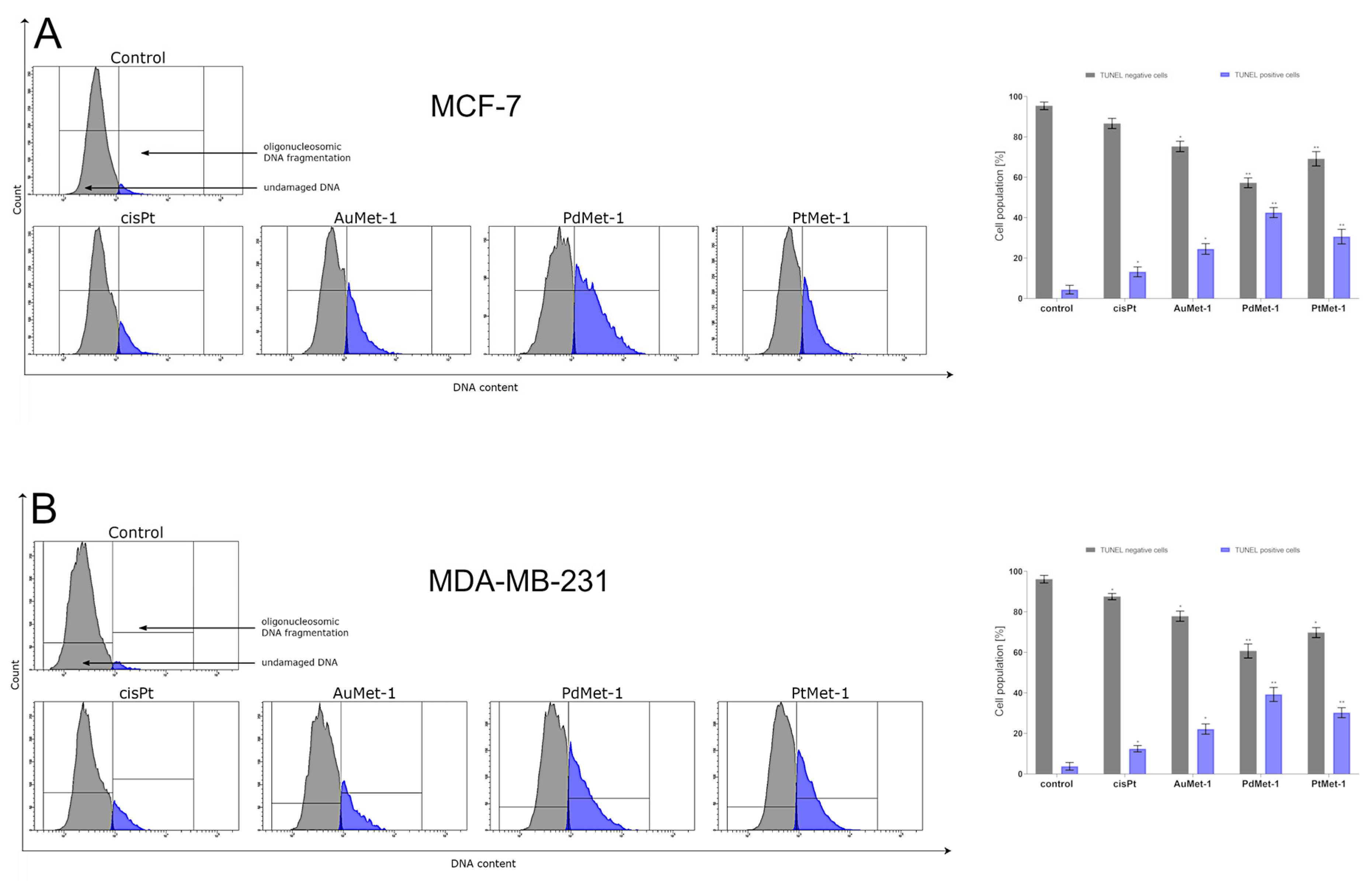
2.6. AuMet-1, PdMet-1 and PtMet-1 Promote DNA Fragmentation
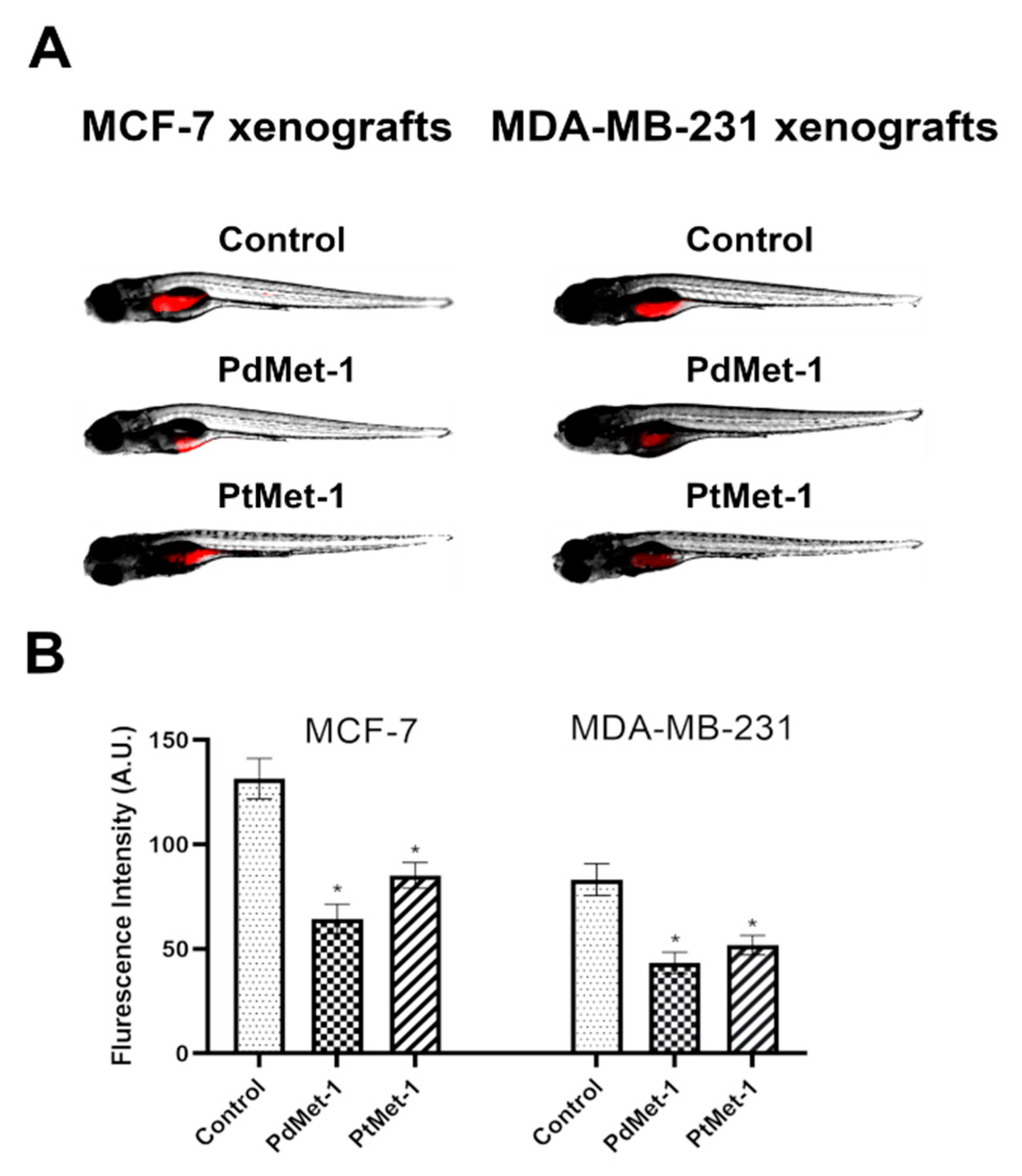
2.7. PdMet-1 and PtMet-1 Inhibit Tumor Development in Zebrafish Xenografts
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Physical Measurements
4.3. Chemistry
4.3.1. Preparation of [Au2(2-(1-methyl-5-nitroimidazol-2-yl)ethanol)4(berenil)2]·4HCl·2H2O (AuMet-1)
4.3.2. Preparation of [Pd2(2-(1-methyl-5-nitroimidazol-2-yl)ethanol)4(berenil)2]·4HCl·2H2O (PdMet-1)
4.3.3. Preparation of [Pt2(2-(1-methyl-5-nitroimidazol-2-yl)ethanol)4(berenil)2]·4HCl·2H2O (PtMet-1)
4.4. Biological Activity
4.4.1. Cell Lines and Cell Culture
4.4.2. Cell Viability Assay
4.4.3. Flow Cytometry Assessment of Annexin V Binding
4.4.4. Analysis of Mitochondrial Membrane Potential
4.4.5. Measuring the Number of Autophagosomes and Autolysosomes by Autophagy Assay, Red
4.4.6. Antibody Topoisomerase IIα Detection
4.4.7. DNA Fragmentation Assay
4.4.8. Zebrafish Handling, Establishment of Xenograft
4.4.9. Microscope Imaging
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Riddell, I.A.; Lippard, S.J. Cisplatin and Oxaliplatin: Our Current Understanding of Their Actions. Met. Ions Life Sci. 2018, 18, 1–42. [Google Scholar] [CrossRef]
- Yeo, C.I.; Ooi, K.K.; Tiekink, E.R.T. Gold-Based Medicine: A Paradigm Shift in Anti-Cancer Therapy? Molecules 2018, 23, 1410. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Lok, C.N.; Wan, P.K.; Zhang, Z.F.; Fung, S.K.; Che, C.M. Anticancer metal-N-heterocyclic carbene complexes of gold, platinum and palladium. Curr. Opin. Chem. Biol. 2018, 43, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Deo, K.M.; Pages, B.J.; Ang, D.L.; Gordon, C.P.; Aldrich-Wright, J.R. Transition Metal Intercalators as Anticancer Agents-Recent Advances. Int. J. Mol. Sci. 2016, 17, 1818. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; AlAjmi, M.F.; Rehman, M.T.; Khan, A.A.; Shaikh, P.A.; Khan, R.A. Evaluation of Transition Metal Complexes of Benzimidazole-Derived Scaffold as Promising Anticancer Chemotherapeutics. Molecules 2018, 23, 1232. [Google Scholar] [CrossRef]
- Zhao, H.; Tao, L.; Zhang, F.; Zhang, Y.; Liu, Y.; Xu, H.; Diao, G.; Ni, L. Transition metal substituted sandwich-type polyoxometalates with a strong metal-C (imidazole) bond as anticancer agents. Chem. Commun. (Camb) 2019, 55, 1096–1099. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, J.; Rasheed, S.; Zhou, C. Design, synthesis, and biological evaluation of novel benzimidazole derivatives and their interaction with calf thymus DNA and synergistic effects with clinical drugs. Sci. China Chem. 2014, 57, 807–822. [Google Scholar] [CrossRef]
- Islami-Moghaddam, M.; Mansouri-Torshizi, H.; Divsalar, A.; Saboury, A.A. Synthesis, characterization, cytotoxic and DNA binding studies of diimine Platinum (II) and Palladium (II) complexes of short hydrocarbon chain ethyldithiocarbamate ligand. JICS 2009, 6, 552–569. [Google Scholar] [CrossRef]
- Mansouri-Torshizi, H.; Eslami-Moghadam, M.; Divsalar, A.; Saboury, A.A. DNA-Binding Studies of Some Potential Antitumor 2,2′-bipyridine Pt(II)/Pd(II) Complexes of piperidinedithiocarbamate. Their Synthesis, Spectroscopy and Cytotoxicity. Acta Chim. Slovenica 2011, 58, 811–822. [Google Scholar]
- Ali, I.; Lone, M.N.; Aboul-Enein, H.Y. Imidazoles as potential anticancer agents. MedChemComm 2017, 8, 1742–1773. [Google Scholar] [CrossRef] [PubMed]
- Czarnomysy, R.; Bielawska, A.; Bielawski, K. Effect of 2nd and 3rd generation PAMAM dendrimers on proliferation, differentiation, and pro-inflammatory cytokines in human keratinocytes and fibroblasts. Int. J. Nanomed. 2019, 14, 7123–7139. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Bialik, S.; Dasari, S.K.; Kimchi, A. Autophagy-dependent cell death-where, how and why a cell eats itself to death. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Deng, S.; Metzger, A.; Gödtel-Armbrust, U.; Porter, A.C.; Wojnowski, L. Topoisomerase IIα-dependent and -independent apoptotic effects of dexrazoxane and doxorubicin. Mol. Cancer Ther. 2009, 8, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Wilson, D.M., 3rd. DNA Damage and Associated DNA Repair Defects in Disease and Premature Aging. Am. J. Hum. Genet. 2019, 105, 237–257. [Google Scholar] [CrossRef]
- Franich, A.A.; Živković, M.D.; Ilić-Tomić, T.; Đorđević, I.S.; Nikodinović-Runić, J.; Pavić, A.; Janjić, G.V.; Rajković, S. New minor groove covering DNA binding mode of dinuclear Pt(II) complexes with various pyridine-linked bridging ligands and dual anticancer-antiangiogenic activitie. J. Biol. Inorg. Chem. 2020, 25, 395–409. [Google Scholar] [CrossRef]
- Ruiz, M.C.; Resasco, A.; Di Virgilio, A.L.; Ayala, M.; Cavaco, I.; Cabrera, S.; Aleman, J.; León, I.E. In vitro and in vivo anticancer effects of two quinoline-platinum(II) complexes on human osteosarcoma models. Cancer Chemother. Pharmacol. 2019, 83, 681–692. [Google Scholar] [CrossRef]
- Kumar, S.; Saha, S.T.; Gu, L.; Palma, G.; Perumal, S.; Singh-Pillay, A.; Singh, P.; Anand, A.; Kaur, M.; Kumar, V. 1H-1,2,3-Triazole Tethered Nitroimidazole-Isatin Conjugates: Synthesis, Docking, and Anti-Proliferative Evaluation against Breast Cancer. ACS Omega 2018, 3, 12106–12113. [Google Scholar] [CrossRef]
- Ravera, M.; Gabano, E.; Sardi, M.; Ermondi, G.; Caron, G.; McGlinchey, M.J.; Müller-Bunz, H.; Monti, E.; Gariboldi, M.B.; Osella, D. Synthesis, characterization, structure, molecular modeling studies and biological activity of sterically crowded Pt(II) complexes containing bis(imidazole) ligands. J. Inorg. Biochem. 2011, 105, 400–409. [Google Scholar] [CrossRef]
- Rocha, C.; Silva, M.M.; Quinet, A.; Cabral-Neto, J.B.; Menck, C. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics (Sao Paulo) 2018, 73 (suppl 1), e478s. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, X.M.; Damu, G.L.; Geng, R.X.; Zhou, C.H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2014, 34, 340–437. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, K.I.; Bando, T.; Sugiyama, H. Anticancer activities of alkylating pyrrole-imidazole polyamides with specific sequence recognition. Anticancer Drugs 2010, 21, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.L.; Hsieh, C.M.; Chan, N.L.; Hiasa, H. Topoisomerases as anticancer targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef] [PubMed]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 2016, 8, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A Target for Anticancer Therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Duan, W.M.; Wu, M.Y.; Wang, W.J.; Liu, L.; Xu, M.D.; Zhu, J.; Li, D.M.; Gui, Q.; Lian, L.; et al. Participation of autophagy in the cytotoxicity against breast cancer cells by cisplatin. Oncol. Rep. 2015, 34, 359–367. [Google Scholar] [CrossRef][Green Version]
- Lin, X.; Han, L.; Weng, J.; Wang, K.; Chen, T. Rapamycin inhibits proliferation and induces autophagy in human neuroblastoma cells. Biosci. Rep. 2018, 38, BSR20181822. [Google Scholar] [CrossRef]
- Yu, H.C.; Lin, C.S.; Tai, W.T.; Liu, C.Y.; Shiau, C.W.; Chen, K.F. Nilotinib induces autophagy in hepatocellular carcinoma through AMPK activation. J. Biol. Chem. 2013, 288, 18249–18259. [Google Scholar] [CrossRef]
- Chen, K.; Shou, L.M.; Lin, F.; Duan, W.M.; Wu, M.Y.; Xie, X.; Xie, Y.F.; Li, W.; Tao, M. Artesunate induces G2/M cell cycle arrest through autophagy induction in breast cancer cells. Anticancer Drugs 2014, 25, 652–662. [Google Scholar] [CrossRef]
- Czarnomysy, R.; Surażyński, A.; Muszyńska, A.; Gornowicz, A.; Bielawska, A.; Bielawski, K. A novel series of pyrazole-platinum(II) complexes as potential anticancer agents that induce cell cycle arrest and apoptosis in breast cancer cells. J. Enzyme Inhib. Med. Chem. 2018, 33, 1006–1023. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czarnomysy, R.; Radomska, D.; Muszyńska, A.; Hermanowicz, J.M.; Prokop, I.; Bielawska, A.; Bielawski, K. Evaluation of the Anticancer Activities of Novel Transition Metal Complexes with Berenil and Nitroimidazole. Molecules 2020, 25, 2860. https://doi.org/10.3390/molecules25122860
Czarnomysy R, Radomska D, Muszyńska A, Hermanowicz JM, Prokop I, Bielawska A, Bielawski K. Evaluation of the Anticancer Activities of Novel Transition Metal Complexes with Berenil and Nitroimidazole. Molecules. 2020; 25(12):2860. https://doi.org/10.3390/molecules25122860
Chicago/Turabian StyleCzarnomysy, Robert, Dominika Radomska, Anna Muszyńska, Justyna Magdalena Hermanowicz, Izabela Prokop, Anna Bielawska, and Krzysztof Bielawski. 2020. "Evaluation of the Anticancer Activities of Novel Transition Metal Complexes with Berenil and Nitroimidazole" Molecules 25, no. 12: 2860. https://doi.org/10.3390/molecules25122860
APA StyleCzarnomysy, R., Radomska, D., Muszyńska, A., Hermanowicz, J. M., Prokop, I., Bielawska, A., & Bielawski, K. (2020). Evaluation of the Anticancer Activities of Novel Transition Metal Complexes with Berenil and Nitroimidazole. Molecules, 25(12), 2860. https://doi.org/10.3390/molecules25122860






