Quinoxaline Derivatives as Antiviral Agents: A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Background Definition
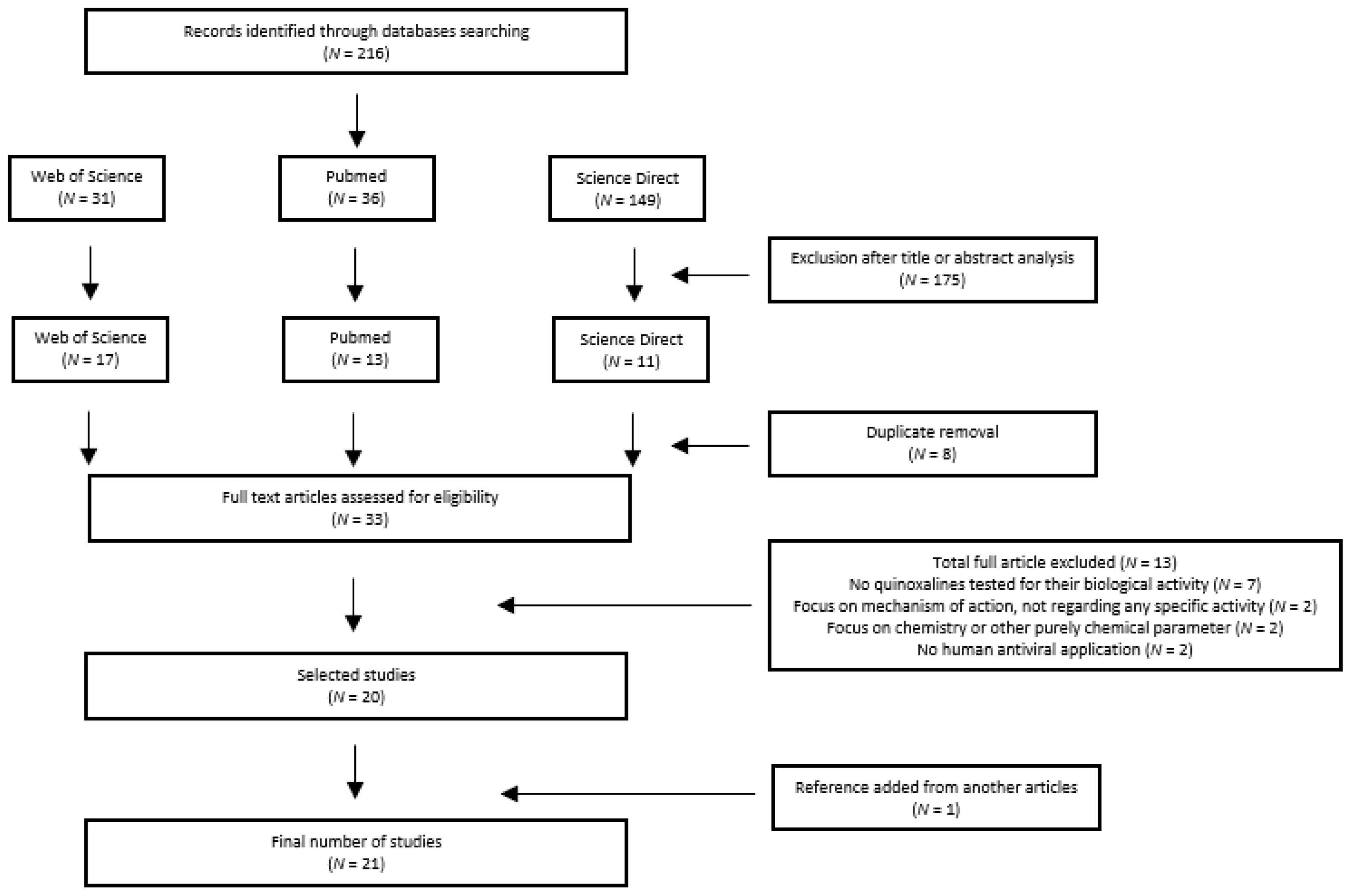
2.2. Data Sources and Searches
2.3. Study Selection
2.4. Data Extraction Process
3. Results
4. Discussion
4.1. Quinoxaline Derivatives Active against DNA Viruses
4.1.1. Quinoxaline Derivatives Active against Herpesviridae
4.1.2. Quinoxaline Derivatives Active against Poxviridae
4.1.3. Quinoxaline Derivatives Active against Hepadnaviridae
4.2. Quinoxaline Derivatives Active against ARN Viruses
4.2.1. Quinoxaline Derivatives Active against Picornaviridae
4.2.2. Quinoxaline Derivatives Active against Orthomyxoviridae
4.2.3. Quinoxaline Derivatives Active against Filoviridae
4.2.4. Quinoxaline Derivatives Active against Flaviviridae
4.2.5. Quinoxaline Derivatives Active against Rhabdoviridae
4.2.6. Quinoxaline Derivatives Active against Retroviridae
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Blair, W.; Cox, C. Current Landscape of Antiviral Drug Discovery. F1000Research 2016, 5, F1000. [Google Scholar] [CrossRef] [PubMed]
- Loughran, H.M.; Han, Z.; Wrobel, J.E.; Decker, S.E.; Ruthel, G.; Freedman, B.D.; Harty, R.N.; Reitz, A.B. Quinoxaline-based inhibitors of Ebola and Marburg VP40 egress. Bioorg. Med. Chem. Lett. 2016, 26, 3429–3435. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef] [PubMed]
- Pol, S.; Corouge, M.; Vallet-Pichard, A. Daclatasvir-sofosbuvir combination therapy with or without ribavirin for hepatitis C virus infection: From the clinical trials to real life. Hepatic Med. Evid. Res. 2016, 8, 21–26. [Google Scholar] [CrossRef]
- Shagufta; Ahmad, I. An insight into the therapeutic potential of quinazoline derivatives as anticancer agents. Med. Chem. Com. 2017, 8, 871–885. [Google Scholar] [CrossRef]
- Shagufta; Ahmad, I. Recent insight into the biological activities of the synthetic xanthone derivatives. Eur. J. Med. Chem. 2016, 116, 267–280. [Google Scholar] [CrossRef]
- Ahmad, I.; Shagufta. Recent developments in steroidal and nonsteroidal aromatase inhibitors for the chemoprevention of oestrogen-dependent breast cancer. Eur. J. Med. Chem. 2015, 102, 375–386. [Google Scholar] [CrossRef]
- Ahmad, I.; Shagufta, S. An important class of organic compounds with diverse biological activities. Int. J. Pharm. Sci. 2015, 7, 19–27. [Google Scholar]
- Deepika, Y.; Nath, P.S.; Sachin, K.; Shewta, S. Biological activity of quinoxaline derivatives. Int. J. Curr. Pharm. Res. 2011, 2, 33–46. [Google Scholar]
- Tariq, S.; Somakala, K.; Amir, M. Quinoxaline: An insight into the recent pharmacological advances. Eur. J. Med. Chem. 2018, 143, 542–557. [Google Scholar] [CrossRef]
- Cheeseman, G.W.H.; Cookson, R.F. Condensed Pyrazines. In The Chemistry of Heterocyclic Compounds; Weissberger, A., Taylor, E.C., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 1979; Volume 35, pp. 78–111. [Google Scholar]
- Pereira, J.A.; Pessoa, A.M.; Cordeiro, M.N.D.S.; Fernandes, R.; Prudencio, C.; Noronha, J.P.; Vieira, M. Quinoxaline, its derivative and applications: A state of the art review. Eur. J. Med. Chem. 2015, 97, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.J.; Ellman, J.A. The Chemistry of Heterocyclic Compounds; Wiley: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Aparicio, D.; Attanasi, O.A.; Filippone, P.; Ignacio, R.; Lillini, S.; Mantellini, F.; Palacios, F.; De Los Santos, J.M. Straightforward access to pyrazines, piperazinones, and quinoxalines by reactions of 1,2-diaza-1,3-butadienes with 1,2-diamines under solution, solvent-free, or solid-phase conditions. J. Org. Chem. 2006, 71, 5897–5905. [Google Scholar] [CrossRef] [PubMed]
- Kunkuma, V.; Bethala, L.A.P.D.; Bhongiri, Y.; Rachapudi, B.N.P.; Potharaju, S.S.P. An efficient synthesis of quinoxalines catalyzed by monoammonium salt of 12-tungstophosphoric acid. Eur. J. Med. Chem. 2011, 2, 495–498. [Google Scholar] [CrossRef][Green Version]
- Antoniotti, S.; Dunach, E. Direct and catalytic synthesis of quinoxaline derivatives from epoxides and ene-1,2-diamines. Tetrahedron Lett. 2002, 43, 3971–3973. [Google Scholar] [CrossRef]
- Cai, J.J.; Zou, J.P.; Pan, X.Q.; Zhang, W. Gallium triflate catalyzed synthesis of quinoxaline derivatives. Tetrahedron Lett. 2008, 49, 7386–7390. [Google Scholar] [CrossRef]
- Thakuria, H.; Das, G. One-pot efficient green synthesis of 1,4-dihydroquinoxaline-2,3-dione derivatives. J. Chem. Sci. 2006, 118, 425–428. [Google Scholar] [CrossRef]
- Gris, J.; Glisoni, R.; Fabian, L.; Fernandez, B.; Moglioni, A.G. Synthesis of potential chemotherapic quinoxalinone derivatives by biocatalysis or microwave-assisted Hinserg reaction. Tetrahedron Lett. 2008, 49, 1053–1056. [Google Scholar] [CrossRef]
- Rostamizadeh, S.; Jafari, S. The synthesis of quinoxalines under microwave irradiation Indian. J. Heterocycl. Chem. 2001, 10, 303–304. [Google Scholar]
- Nageswar, Y.V.D.; Reddy, K.H.V.; Ramesh, K.; Murthy, S.N. Recent developments in the synthesis of quinoxaline derivatives by green synthetic approaches. Org. Prep. Proc. Int. 2013, 45, 1–27. [Google Scholar] [CrossRef]
- Harmenberg, J.; Akesson-Johansson, A.; Graslund, A.; Malmfors, T.; Bergman, J.; Wahren, B.; Akerfeldt, S.; Lundblad, L.; Cox, S. The mechanism of action of the anti-herpes virus compound 2,3-dimethyl-6(2-dimethylaminoethyl)-6H-indolo-(2,3-b)quinoxaline. Antiviral. Res. 1991, 15, 193–204. [Google Scholar] [CrossRef]
- Montana, M.; Mathias, F.; Terme, T.; Vanelle, P. Antitumoral activity of quinoxaline derivatives: A systematic review. Eur. J. Med. Chem. 2019, 163, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Cogo, J.; Cantizani, J.; Cotillo, I.; Pereira sangi, D.; Gonçalves Corrêa, A.; Ueda-Nakamura, T.; Prado Dias Filho, B.; Martin, J.J.; Vataru Nakamura, C. Quinoxaline derivatives as potential antitrypanosomal and antileishmanialagents. Bioorg. Med. Chem. 2018, 7, 4065–4072. [Google Scholar] [CrossRef]
- González, M.; Cerecetto, H. Quinoxaline derivatives: A patent review (2006--present). Expert Opin. Ther. Pat. 2012, 22, 1289–1302. [Google Scholar] [CrossRef]
- Kleim, J.P.; Bender, R.; Billhardt, U.M.; Meichsner, C.; Riess, G.; Rösner, M.; Winkler, I.; Paessens, A. Activity of a novel quinoxaline derivative against human immunodeficiency virus type 1 reverse transcriptase and viral replication. Antimicrob. Agents Chemother. 1993, 37, 1659–1664. [Google Scholar] [CrossRef][Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Open Med. 2009, 3, 123–130. [Google Scholar]
- Tanis, S.P.; Strohbach, J.W.; Parker, T.T.; Moon, M.W.; Thaisrivongs, S.; Perrault, W.R.; Hopkins, T.A.; Knechtel, M.L.; Oien, N.L.; Wieber, J.L.; et al. The design and development of 2-aryl-2-hydroxy ethylamine substituted 1H,7H-pyrido[1,2,3-de]quinoxaline-6-carboxamides as inhibitors of human cytomegalovirus polymerase. Bioorg. Med. Chem. Lett. 2010, 20, 1994–2000. [Google Scholar] [CrossRef]
- Henen, M.A.; El Bialy, S.A.A.; Goda, F.E.; Nasr, M.N.A.; Eisa, H.M. [1,2,4] Triazolo [4,3-a] quinoxaline: Synthesis, antiviral, and antimicrobial activities. Med. Chem. Res. 2012, 9, 2368–2378. [Google Scholar] [CrossRef]
- El-Tombary, A.A.; El-Hawash, S.A. Synthesis, antioxidant, anticancer and antiviral activities of novel quinoxaline hydrazone derivatives and their acyclic C-nucleosides. Med. Chem. 2014, 10, 521–532. [Google Scholar] [CrossRef] [PubMed]
- El-Zahabi, H.S.A. Synthesis, Characterization, and Biological Evaluation of Some Novel Quinoxaline Derivatives as Antiviral Agents. Arch. Pharm. (Weinheim) 2017, 350, e1700028. [Google Scholar] [CrossRef] [PubMed]
- Galal, S.A.; Abdelsamie, A.S.; Tokuda, H.; Suzuki, N.; Lida, A.; Elhefnawi, M.M.; Ramadan, R.A.; Atta, M.H.; El Diwani, H.I. Part I: Synthesis, cancer chemopreventive activity and molecular docking study of novel quinoxaline derivatives. Eur. J. Med. Chem. 2011, 46, 327–340. [Google Scholar] [CrossRef]
- Manta, S.; Gkaragkouni, D.N.; Kaffesaki, E.; Gkizis, P.; Hadjipavlou-Litina, D.; Pontiki, E.; Balzarini, J.; Dehaen, W.; Komiotis, D. A novel and easy two-step, microwave-assisted method for the synthesis of halophenyl pyrrolo[2,3-b]quinoxalines via their pyrrolo precursors. Eval. Their Bioactivity Tetrahedron Lett. 2014, 55, 1873–1876. [Google Scholar] [CrossRef]
- Fakhreldin, Y.; El-Kousy, S.M.; El Ashry, M. Synthesis and novel chemical reaction for a new class of 3-(1′,2′-dihydroxyeth-1′-yl)-1-phenylpyrazolo [3,4-b] quinoxaline series “C-nucleosides” as antiviral agents. Life Sci. 2012, 9, 234–239. [Google Scholar]
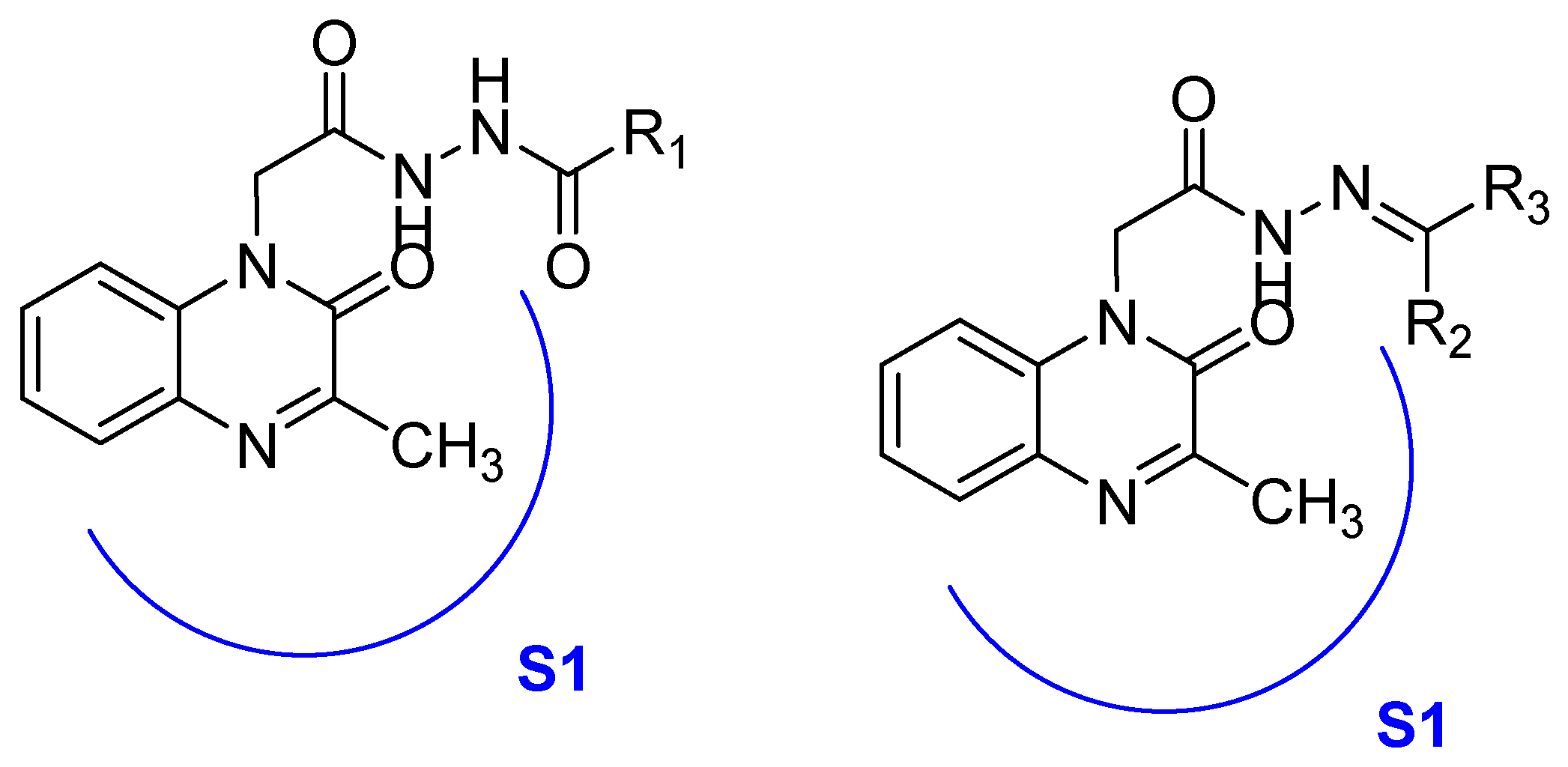
- Carta, A.; Sanna, G.; Briguglio, I.; Madeddu, S.; Vitale, G.; Piras, S.; Corona, P.; Peana, A.T.; Laurini, E.; Fermeglia, M.; et al. Quinoxaline derivatives as new inhibitors of coxsackievirus B5. Eur. J. Med. Chem. 2018, 145, 559–569. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Cho, E.J.; Leavitt, J.; Ma, L.C.; Montelione, G.T.; Anslyn, E.V.; Krug, R.M.; Ellington, A.; Robertus, J.D. Synthesis and evaluation of quinoxaline derivatives as potential influenza NS1A protein inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 3007–3011. [Google Scholar] [CrossRef]
- Carta, A.; Briguglio, I.; Piras, S.; Corona, P.; Boatto, G.; Nieddu, M.; Giunchedi, P.; Marongiu, M.E.; Giliberti, G.; Iuliano, F.; et al. Quinoline tricyclic derivatives. Design, synthesis and evaluation of the antiviral activity of three new classes of RNA-dependent RNA polymerase inhibitors. Bioorg. Med. Chem. 2011, 19, 7070–7084. [Google Scholar] [CrossRef] [PubMed]
- Briguglio, I.; Loddo, R.; Laurini, E.; Fermeglia, M.; Piras, S.; Corona, P.; Giunchedi, P.; Gavini, E.; Sanna, G.; Giliberti, G.; et al. Synthesis, cytotoxicity and antiviral evaluation of new series of imidazo[4,5-g]quinoline and pyrido[2,3-g]quinoxalinone derivatives. Eur. J. Med. Chem. 2015, 105, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Summa, V.; Ludmerer, S.W.; McCauley, J.A.; Fandozzi, C.; Burlein, C.; Claudio, G.; Coleman, P.J.; Dimuzio, J.M.; Ferrara, M.; Di Filippo, M.; et al. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob. Agents Chemother. 2012, 56, 4161–4167. [Google Scholar] [CrossRef]
- Guo, Z.; Black, S.; Hu, Y.; McMonagle, P.; Ingravallo, P.; Chase, R.; Curry, S.; Asante-Appiah, E. Unraveling the structural basis of grazoprevir potency against clinically relevant substitutions in hepatitis C virus NS3/4A protease from genotype 1a. J. Biol. Chem. 2017, 292, 6202–6212. [Google Scholar] [CrossRef]
- Rusere, L.N.; Matthew, A.N.; Lockbaum, G.J.; Jahangir, M.; Newton, A.; Petropoulos, C.J.; Huang, W.; Kurt Yilmaz, N.; Schiffer, C.A.; Ali, A. Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants. ACS Med. Chem. Lett. 2018, 9, 691–696. [Google Scholar] [CrossRef]
- Matthew, A.N.; Zephyr, J.; Hill, C.J.; Jahangir, M.; Newton, A.; Petropoulos, C.J.; Huang, W.; Kurt Yilmaz, N.; Schiffer, C.A.; Ali, A. Hepatitis C Virus NS3/4A Protease Inhibitors Incorporating Flexible P2 Quinoxalines Target Drug Resistant Viral Variants. J. Med. Chem. 2017, 60, 5699–5716. [Google Scholar] [CrossRef]
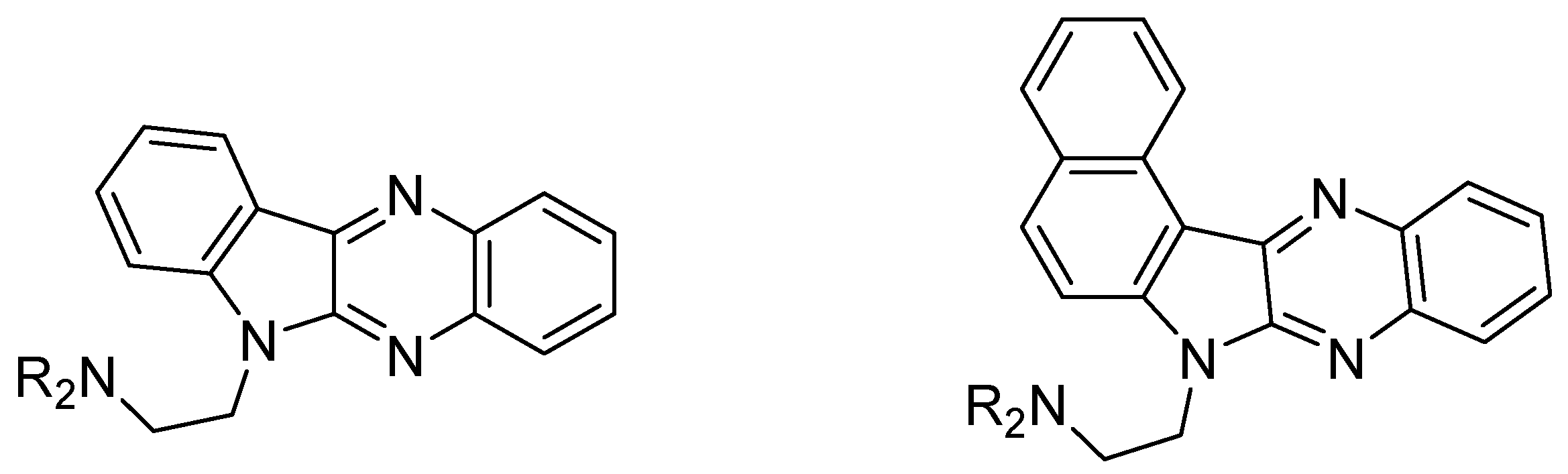
- Shibinskaya, M.O.; Karpenko, A.S.; Lyakhov, S.A.; Andronati, S.A.; Zholobak, N.M.; Spivak, N.Y.; Samochina, N.A.; Shafran, L.M.; Zubritsky, M.J.; Galat, V.F. Synthesis and biological activity of 7H-benzo[4,5]indolo[2,3-b]-quinoxaline derivatives. Eur. J. Med. Chem. 2011, 46, 794–798. [Google Scholar] [CrossRef] [PubMed]
- Shibinskaya, M.O.; Lyakhov, S.A.; Mazepa, A.V.; Andronati, S.A.; Turov, A.V.; Zholobak, N.M.; Spivak, N.Y. Synthesis, cytotoxicity, antiviral activity and interferon inducing ability of 6-(2-aminoethyl)-6H-indolo[2,3-b]quinoxalines. Eur. J. Med. Chem. 2010, 45, 1237–1243. [Google Scholar] [CrossRef]
- Fabian, L.; Taverna Porro, M.; Gómez, N.; Salvatori, M.; Turk, G.; Estrin, D.; Moglioni, A. Design, synthesis and biological evaluation of quinoxaline compounds as anti-HIV agents targeting reverse transcriptase enzyme. Eur. J. Med. Chem. 2020, 188, 111987. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.B.; Patel, B.D.; Pannecouque, C.; Bhatt, H.G. Design, synthesis and anti-HIV activity of novel quinoxaline derivatives. Eur. J. Med. Chem. 2016, 117, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, P.; Wu, J.F.; Yang, L.M.; Wang, R.R.; Pang, W.; Li, Y.G.; Shen, Y.M.; Zheng, Y.T.; Li, X. Design, synthesis and anti-HIV-1 evaluation of hydrazide-based peptidomimetics as selective gelatinase inhibitors. Bioorg. Med. Chem. 2016, 24, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Nathanson, N.; González-Scarano, F. Patterns of infection. Viral Pathogenesis 2016, 81–94. [Google Scholar]
- Ryu, W.S. Discovery and Classification. Mol. Virol. Human Pathog. Viruses 2017, 3–20. [Google Scholar]
- Villareal, E.C. Current and potential therapies for the treatment of herpes-virus infections. Prog. Drug Res. 2003, 60, 263–307. [Google Scholar]
- Brideau, R.J.; Knechtel, M.L.; Huang, A.; Vaillancourt, V.A.; Vera, E.E.; Oien, N.L.; Hopkins, T.A.; Wieber, J.L.; Wilkinson, K.F.; Rush, B.D.; et al. Broad-spectrum antiviral activity of PNU-183792, a 4-oxo-dihydroquinoline, against human and animal herpesviruses. Antiviral Res. 2002, 54, 19–28. [Google Scholar] [CrossRef]
- Faulds, D.; Heel, R.C. Ganciclovir. A review of its antiviral activity, pharmacokinetic properties and therapeutic efficacy in cytomegalovirus infections. Drugs 1990, 39, 597–638. [Google Scholar] [CrossRef]
- Lalezari, J. Drug-resistant herpes: Cidofovir gel in limbo. Interview with Jay Lalezari, M.D. Interview by John S. James. AIDS Treat. News 1997, 283, 5–6. [Google Scholar]
- Kolb, T.M.; Davis, M.A. The tumor promoter 12-O-tetradecanoylphorbol 13-acetate (TPA) provokes a prolonged morphologic response and ERK activation in Tsc2-null renal tumor cells. Toxicol. Sci. 2004, 81, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Louten, J. Essential Human Virology, 1st ed.; Elsevier, Academic Press: Cambridge, MA, USA, 2016; p. 344. [Google Scholar]
- Badri, T.; Gandhi, G.R. Molluscum Contagiosum. Treasure Island (FL): StatPearls Publishing. January 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK441898/ (accessed on 5 May 2020).
- Poltronieri, P.; Sun, B.; Mallardo, M. RNA Viruses: RNA Roles in Pathogenesis, Coreplication and Viral Load. Curr. Genomics 2015, 16, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Gaaloul, I.; Riabi, S.; Harrath, R.; Hunter, T.; Hamda, K.B.; Ghzala, A.B.; Huber, S.; Aouni, M. Coxsackievirus B detection in cases of myocarditis, myopericarditis, pericarditis and dilated cardiomyopathy in hospitalized patients. Mol. Med. Rep. 2014, 10, 2811–2818. [Google Scholar] [CrossRef]
- Lee, C.J.; Huang, Y.C.; Yang, S.; Tsao, K.C.; Chen, C.J.; Hsieh, Y.C.; Chiu, C.H.; Lin, T.Y. Clinical features of coxsackievirus A4, B3 and B4 infections in children. PLoS ONE 2014, 9, e87391. [Google Scholar] [CrossRef]
- Hale, B.G.; Randall, R.E.; Ortín, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef]
- Yin, C.; Khan, J.A.; Swapna, G.V.; Ertekin, A.; Krug, R.M.; Tong, L.; Montelione, G.T. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J. Biol. Chem. 2007, 282, 20584–20592. [Google Scholar] [CrossRef]
- Min, J.Y.; Krug, R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 7100–7105. [Google Scholar] [CrossRef]
- Mulangu, S.; Dodd, L.E.; Davey, R.T., Jr.; Tshiani Mbaya, O.; Proschan, M.; Mukadi, D.; Lusakibanza Manzo, M.; Nzolo, D.; Tshomba Oloma, A.; Ibanda, A.; et al. Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381, 2293–2303. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of High-Consequence Pathogens and Pathology (DHCPP), Viral Special Pathogens Branch (VSPB). Available online: https://www.cdc.gov/vhf/ebola/treatment/index.html (accessed on 5 May 2020).
- World Health Organisation. Hepatitis C. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 5 May 2020).
- Liverton, N.J.; Holloway, M.K.; McCauley, J.A.; Rudd, M.T.; Butcher, J.W.; Carroll, S.S.; DiMuzio, J.; Fandozzi, C.; Gilbert, K.F.; Mao, S.S.; et al. Molecular modeling-based approach to potent P2-P4 macrocyclic inhibitors of hepatitis C NS3/4A protease. J. Am. Chem. Soc. 2008, 130, 4607–4609. [Google Scholar] [CrossRef]
- Romano, K.P.; Ali, A.; Aydin, C.; Soumana, D.; Ozen, A.; Deveau, L.M.; Silver, C.; Cao, H.; Newton, A.; Petropoulos, C.J.; et al. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 2012, 8, e1002832. [Google Scholar] [CrossRef] [PubMed]
- Shah, U.; Jayne, C.; Chackalamannil, S.; Velázquez, F.; Guo, Z.; Buevich, A.; Howe, J.A.; Chase, R.; Soriano, A.; Agrawal, S.; et al. Novel Quinoline-Based P2-P4 Macrocyclic Derivatives As Pan-Genotypic HCV NS3/4a Protease Inhibitors. ACS Med. Chem. Lett. 2014, 5, 264–269. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quiroz, E.; Moreno, N.; Peralta, P.H.; Tesh, R.B. A human case of encephalitis associated with vesicular stomatitis virus (Indiana serotype) infection. Am. J. Trop. Med. Hyg. 1988, 39, 312–314. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Allergy and Infectious Diseases. Antiretroviral Drug Discovery and Development. Available online: https://www.niaid.nih.gov/diseases-conditions/antiretroviral-drug-development (accessed on 5 May 2020).
Sample Availability: No samples available. |



| Parameter | Inclusion | Exclusion | |
|---|---|---|---|
| 1 | Language | English, French | Any other language |
| 2 | Type of study | Biological activity, In vitro and/or in vivo studies | Exclusively in silico, articles that focus only on synthesis or other purely chemical parameters |
| 3 | Type of publication | Original manuscripts | Reviews, book chapters, posters, table of contents, personal opinions, indexes, conference abstracts, letters |
| 4 | Search terms | Merely citing keywords in text | |
| 5 | Mechanism of action | Articles that evaluate the biological activity of quinoxaline derivatives | Articles that concern non-human or animal viruses |
| Activity | Reference |
|---|---|
| DNA viruses | |
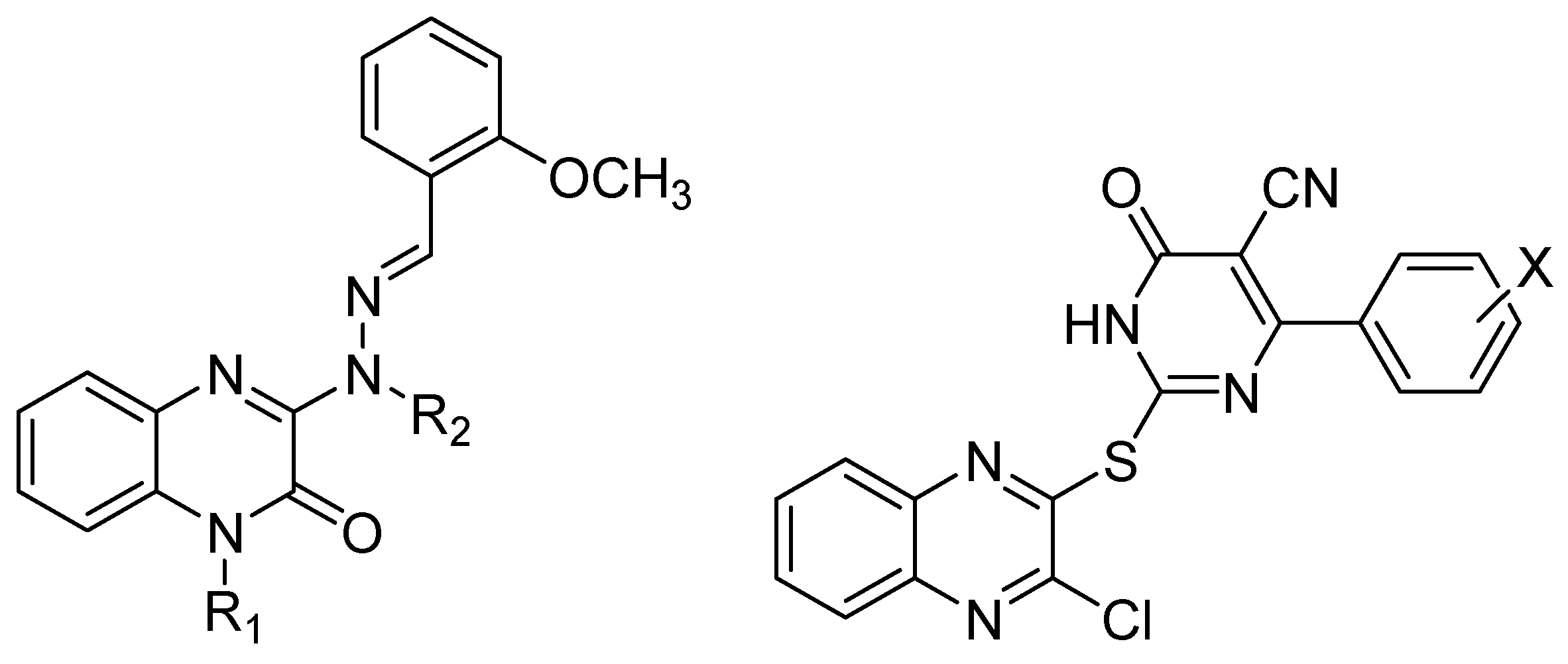
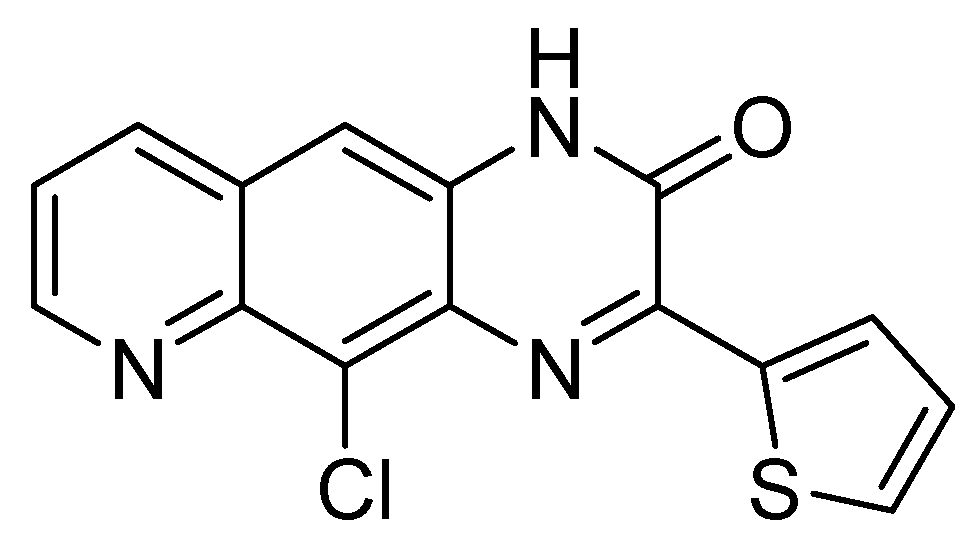
| Herpesviridae | [28,29,30,31,32] |
| Poxviridae | [33] |
| Hepadnaviridae | [34] |
| RNA viruses | |
| Picornaviridae | [35] |
| Orthomyxoviridae | [36] |
| Filoviridae | [2] |
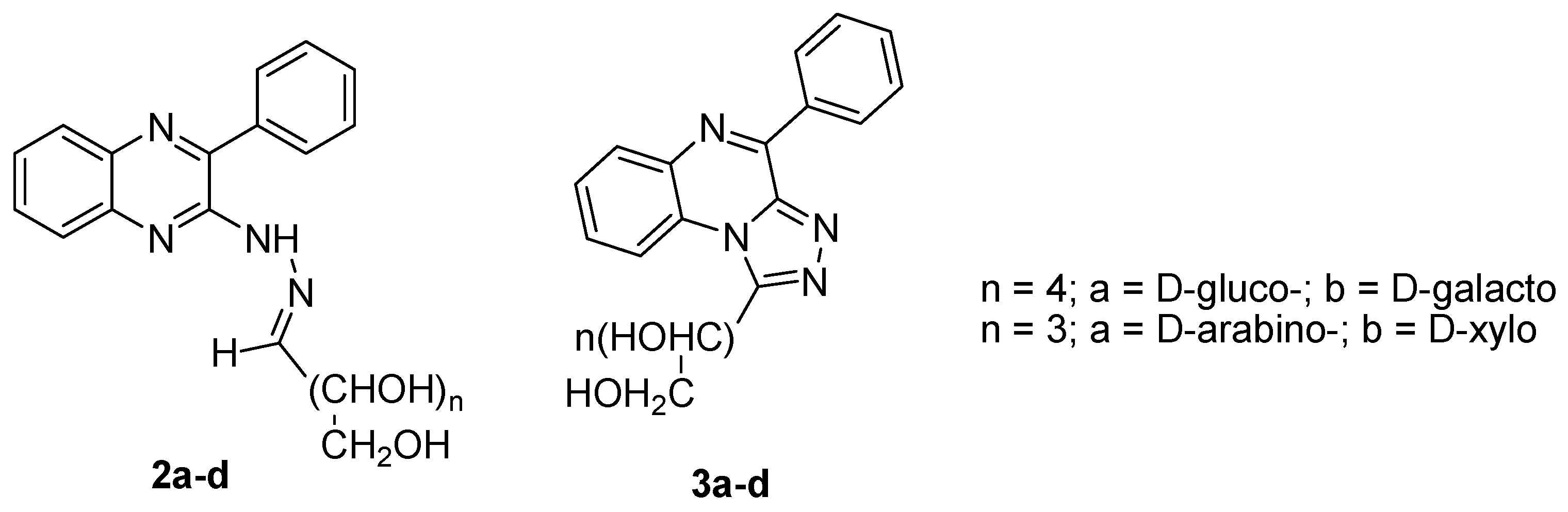
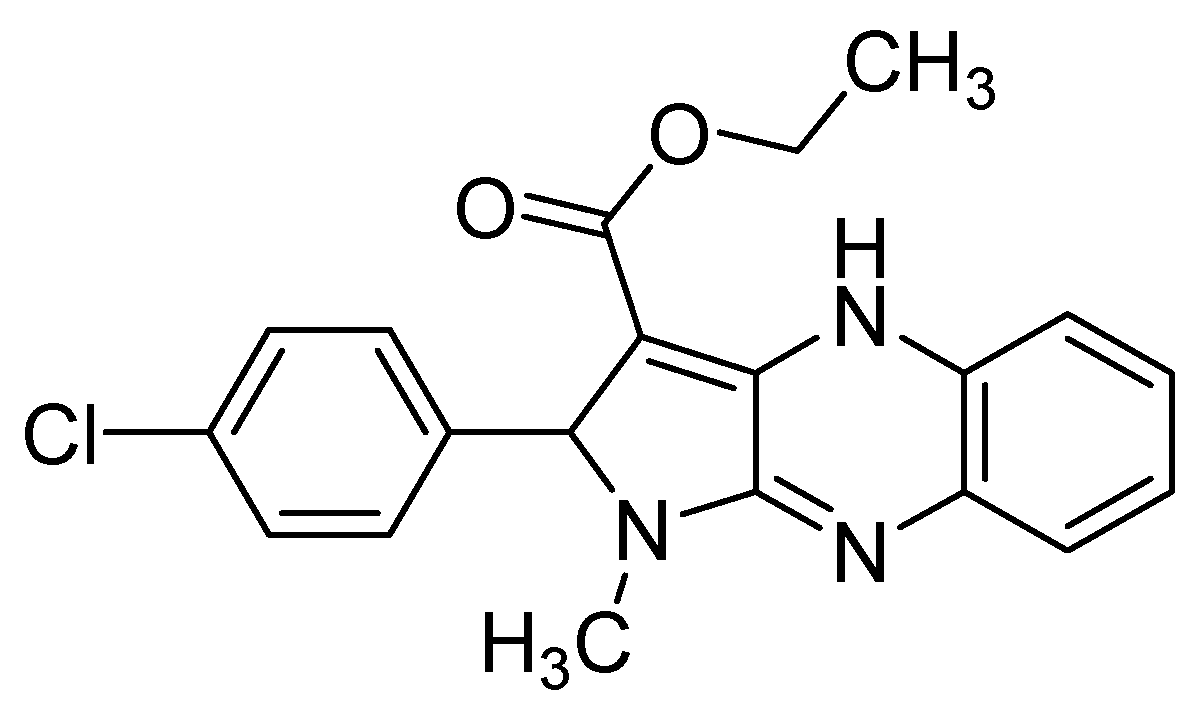
| Flaviviridae | [31,37,38,39,40,41,42] |
| Rhabdoviridae | [43,44] |
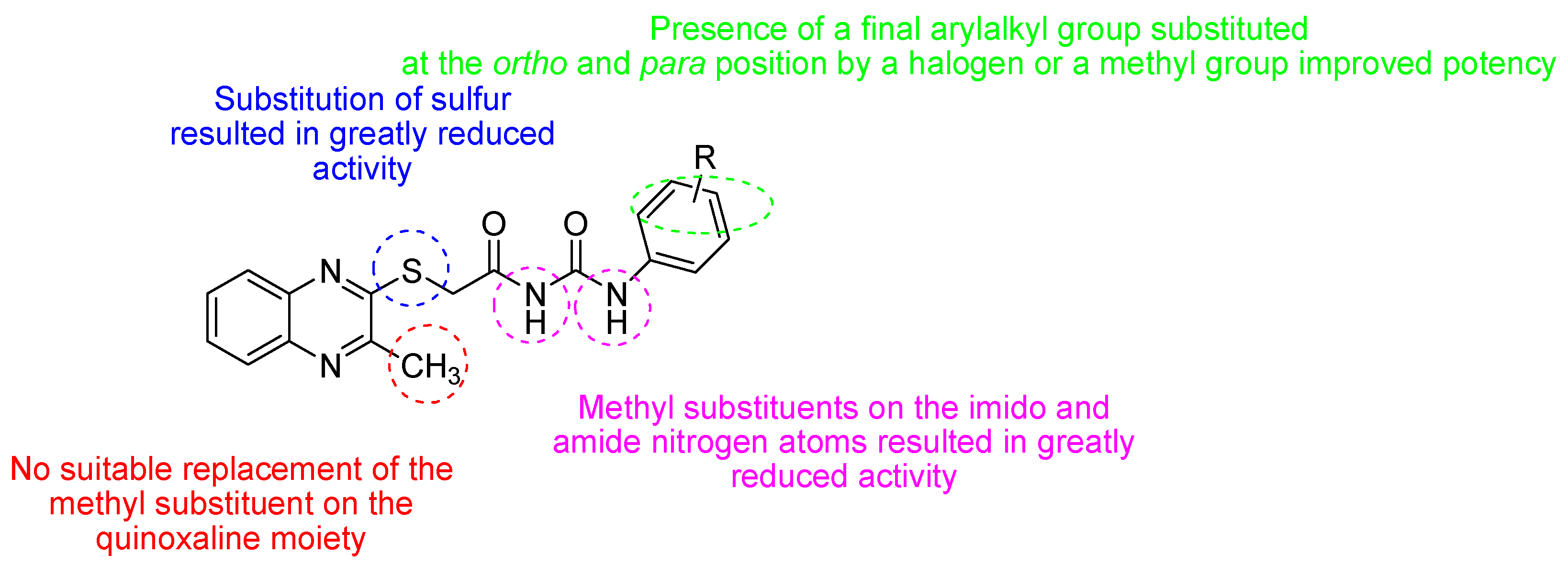
| Retroviridae | [45,46,47] |
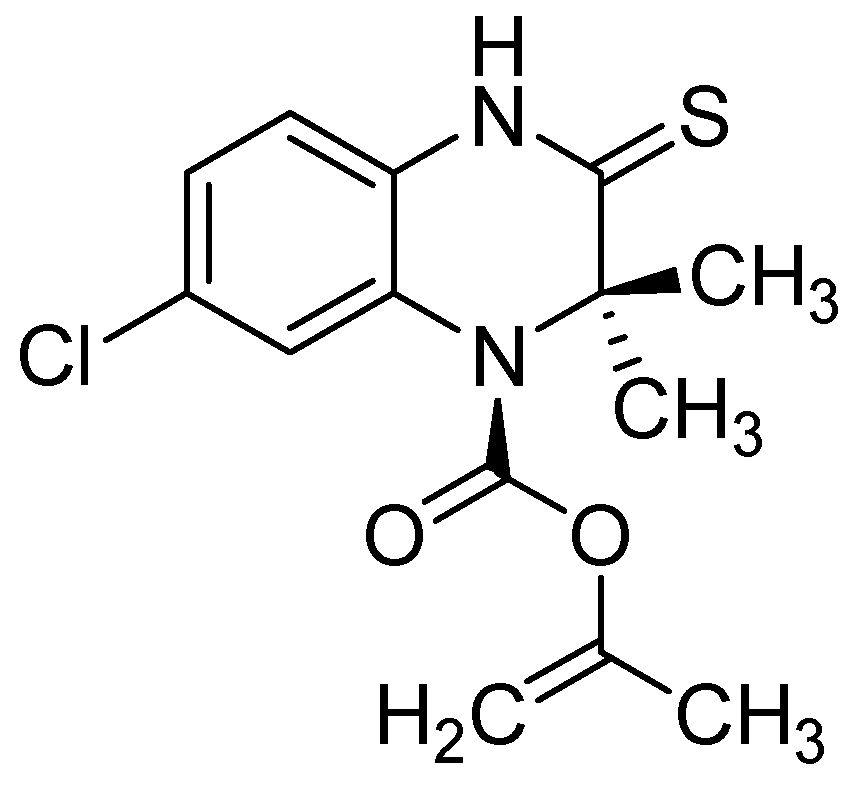
| Compound | Structure | EC50 (µM) |
|---|---|---|
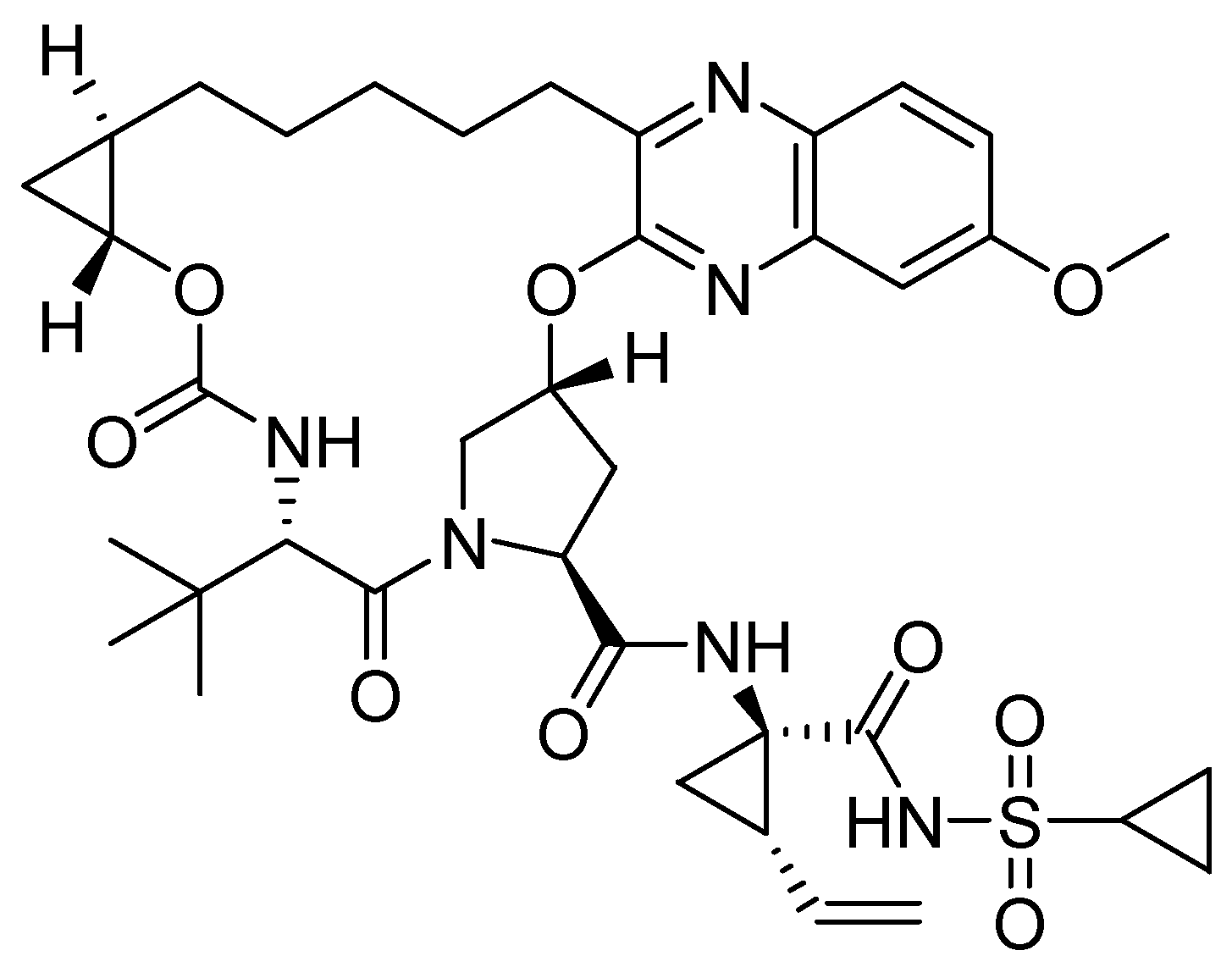
| 4 | 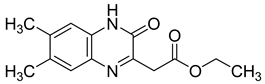 | <0.05 |
| 5 | 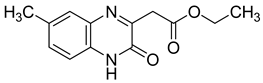 | >30 |
| 6 | 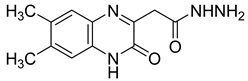 | >1.2 |
| 7 | 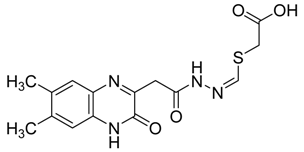 | >30 |
| 8 | 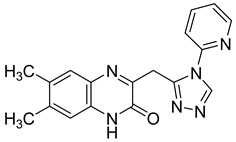 | <0.05 |
| Ganciclovir | - | 0.059 |
| Compound | Structure | HCMV pol Activity IC50 (nM) | HCMV pra Activity IC50 (nM) |
|---|---|---|---|
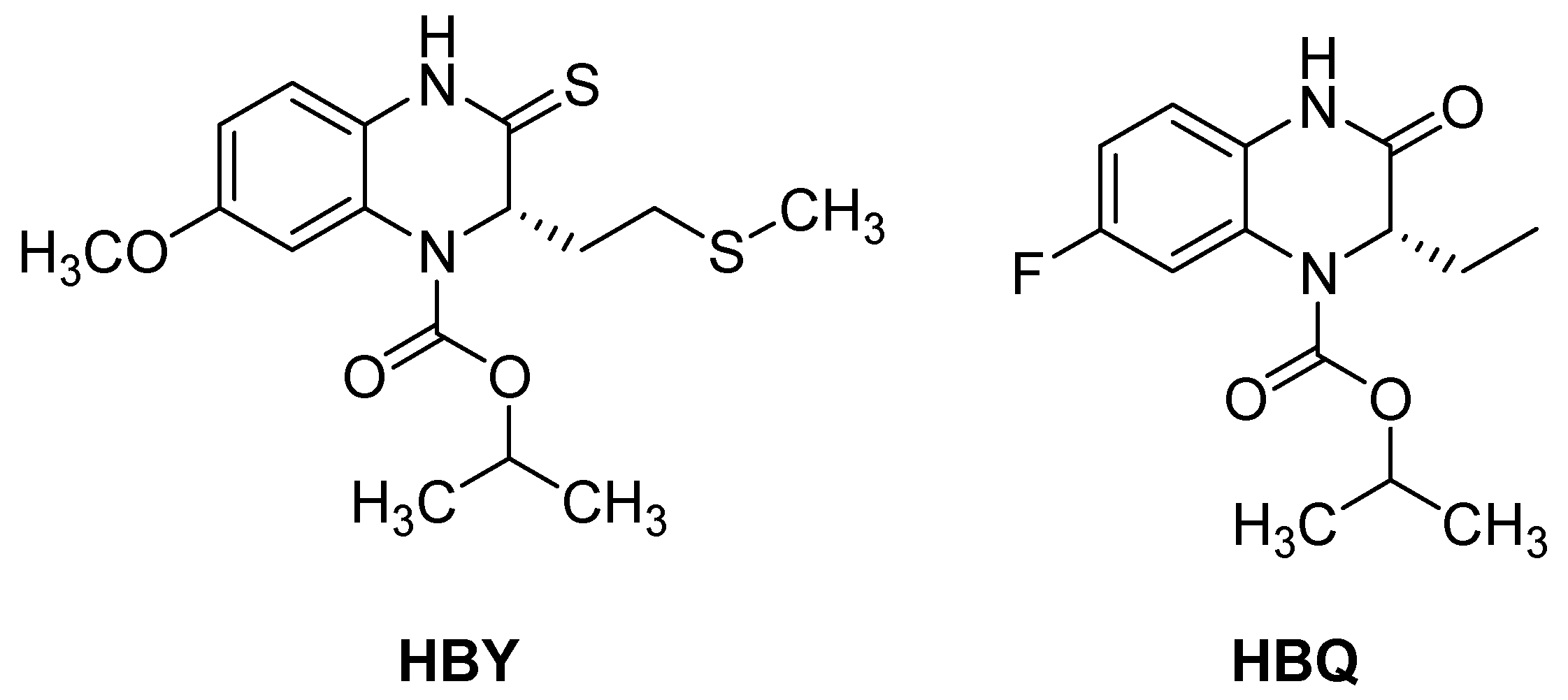
| 9 |  | 620 | 100 |
| 10 | 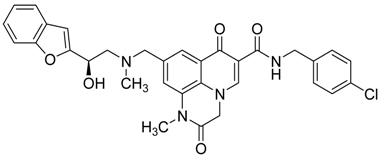 | 4 | 1 |
| Ganciclovir | - | 1300 | |
| Acyclovir | - | >20,000 | |
| Foscarnet | 2500 | - | |
| Aphidicolin | - | 487 | - |
| Compound | Structure | EC50 CBV5 (µM) | CC50 Vero-76 cells (µM) |
|---|---|---|---|
| 11 | 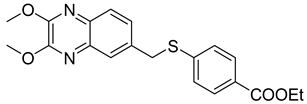 | 0.09 ± 0.01 | >100 |
| 12 | 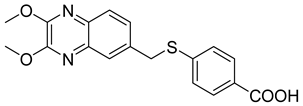 | 0.06 ± 0.01 | >65 |
| 13 |  | 0.3 ± 0.05 | >100 |
| Compound | Structure | % Binding at 50 µM | % Intercalation at 50 µM |
|---|---|---|---|
| 14 | 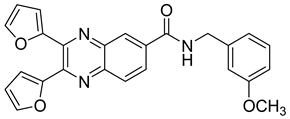 | 74.0 | 4.5 |
| 15 | 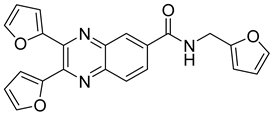 | 79.5 | 5.9 |
| Compound | Structure | %RT Inhibition | MT2 IC50 (µM) | Selectivity Index (SI) | |
|---|---|---|---|---|---|
| 100 µM | 10 µM | ||||
| 18 | 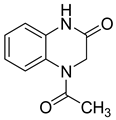 | 56 | 7 | NE | NE |
| 19 | 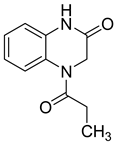 | 99 | 91 | 0.63 (0.53–0.76) | 31,798 |
| 20 | 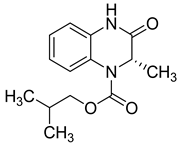 | 64 | 37 | 64 (38–103) | NE |
| 21 | 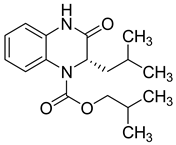 | 51 | 22 | 67 (54–87) | 74 |
| 22 | 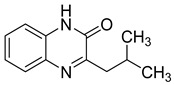 | 73 | 15 | 23 (21–25) | NE |
| Nevirapine | - | 98 | 91 | 0.13 (0.09–0.17) | 14,353 |
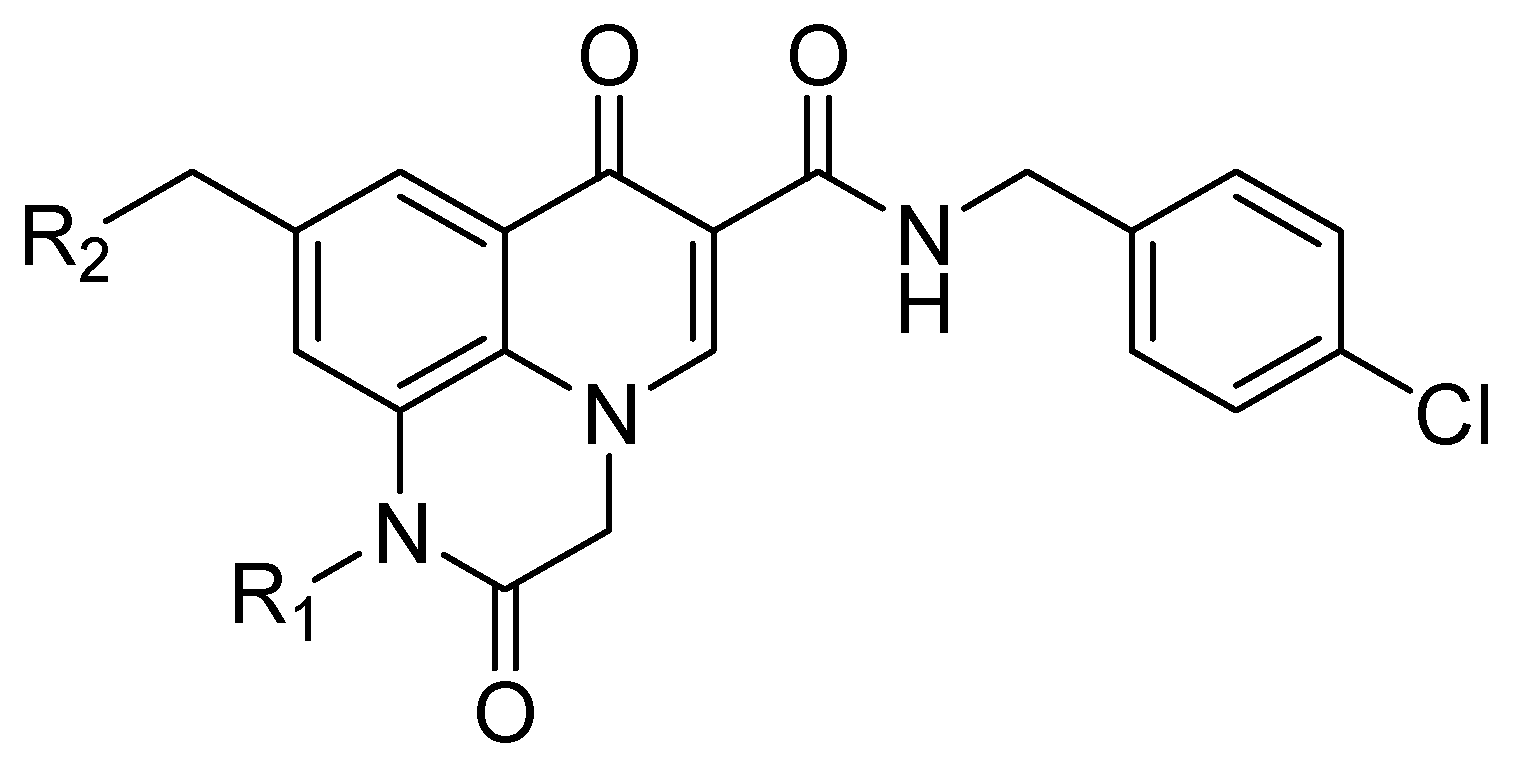
| Compound | Structure | Strain IIIB IC50 (µg/mL) | Vero IC50 (µg/mL) |
|---|---|---|---|
| 23 |  | >11.78 | >100 |
| 24 | 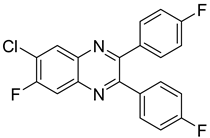 | >15.45 | >100 |
| Compound | Structure | Gelatinase IC50 (µM) | C8166 CC50 (µM) |
|---|---|---|---|
| 25 | 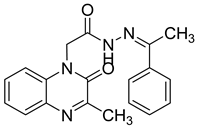 | 9.39 ± 0.7 | >100 |
| 26 | 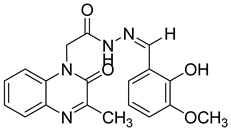 | 7.17 ± 0.95 | >100 |
| LY52 | 5.64 ± 0.6 | >100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montana, M.; Montero, V.; Khoumeri, O.; Vanelle, P. Quinoxaline Derivatives as Antiviral Agents: A Systematic Review. Molecules 2020, 25, 2784. https://doi.org/10.3390/molecules25122784
Montana M, Montero V, Khoumeri O, Vanelle P. Quinoxaline Derivatives as Antiviral Agents: A Systematic Review. Molecules. 2020; 25(12):2784. https://doi.org/10.3390/molecules25122784
Chicago/Turabian StyleMontana, Marc, Vincent Montero, Omar Khoumeri, and Patrice Vanelle. 2020. "Quinoxaline Derivatives as Antiviral Agents: A Systematic Review" Molecules 25, no. 12: 2784. https://doi.org/10.3390/molecules25122784
APA StyleMontana, M., Montero, V., Khoumeri, O., & Vanelle, P. (2020). Quinoxaline Derivatives as Antiviral Agents: A Systematic Review. Molecules, 25(12), 2784. https://doi.org/10.3390/molecules25122784






