Enhanced Stability of Lipid Structures by Dip-Pen Nanolithography on Block-Type MPC Copolymer
Abstract
1. Introduction
2. Results
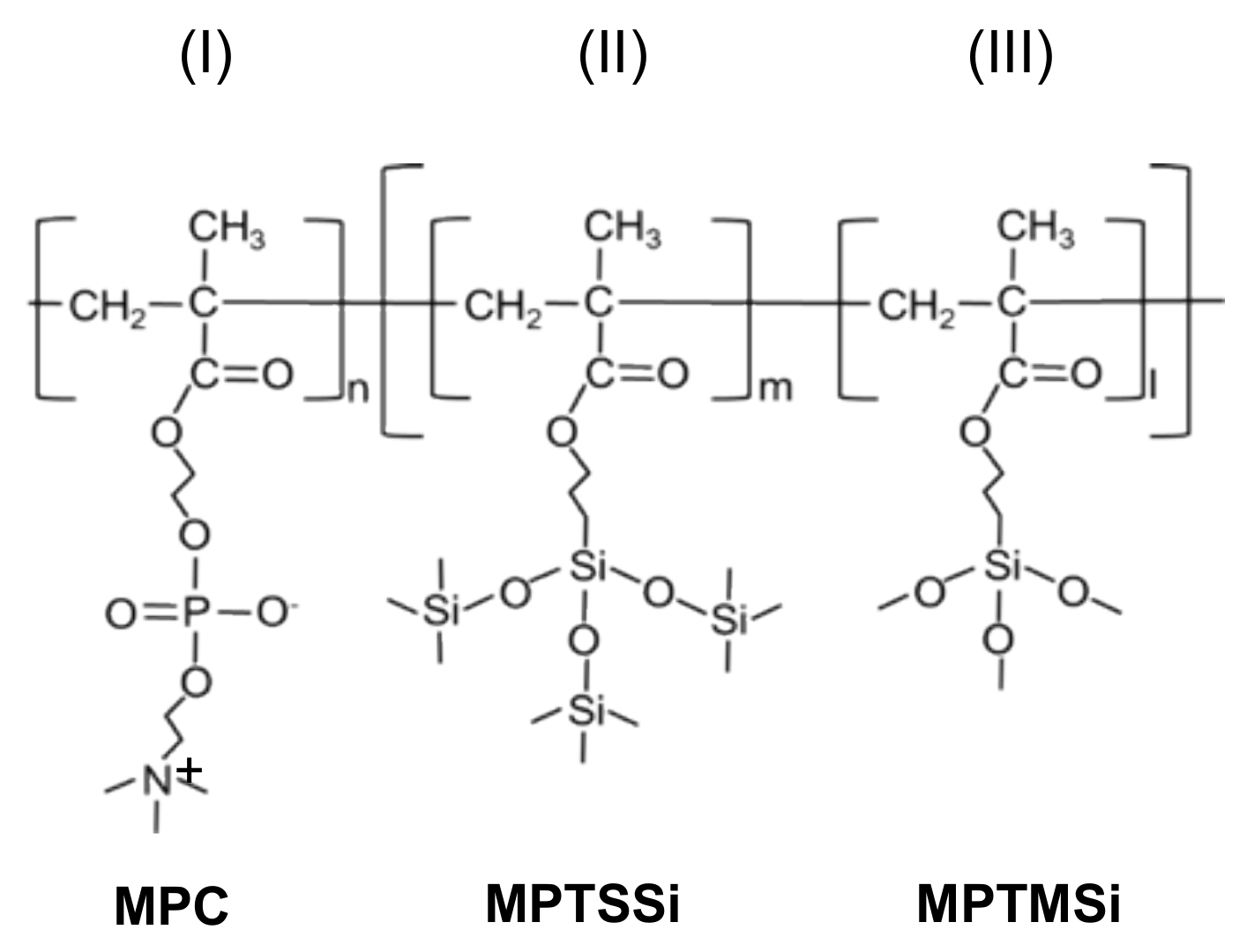
2.1. Characterization of Polymer Substrates
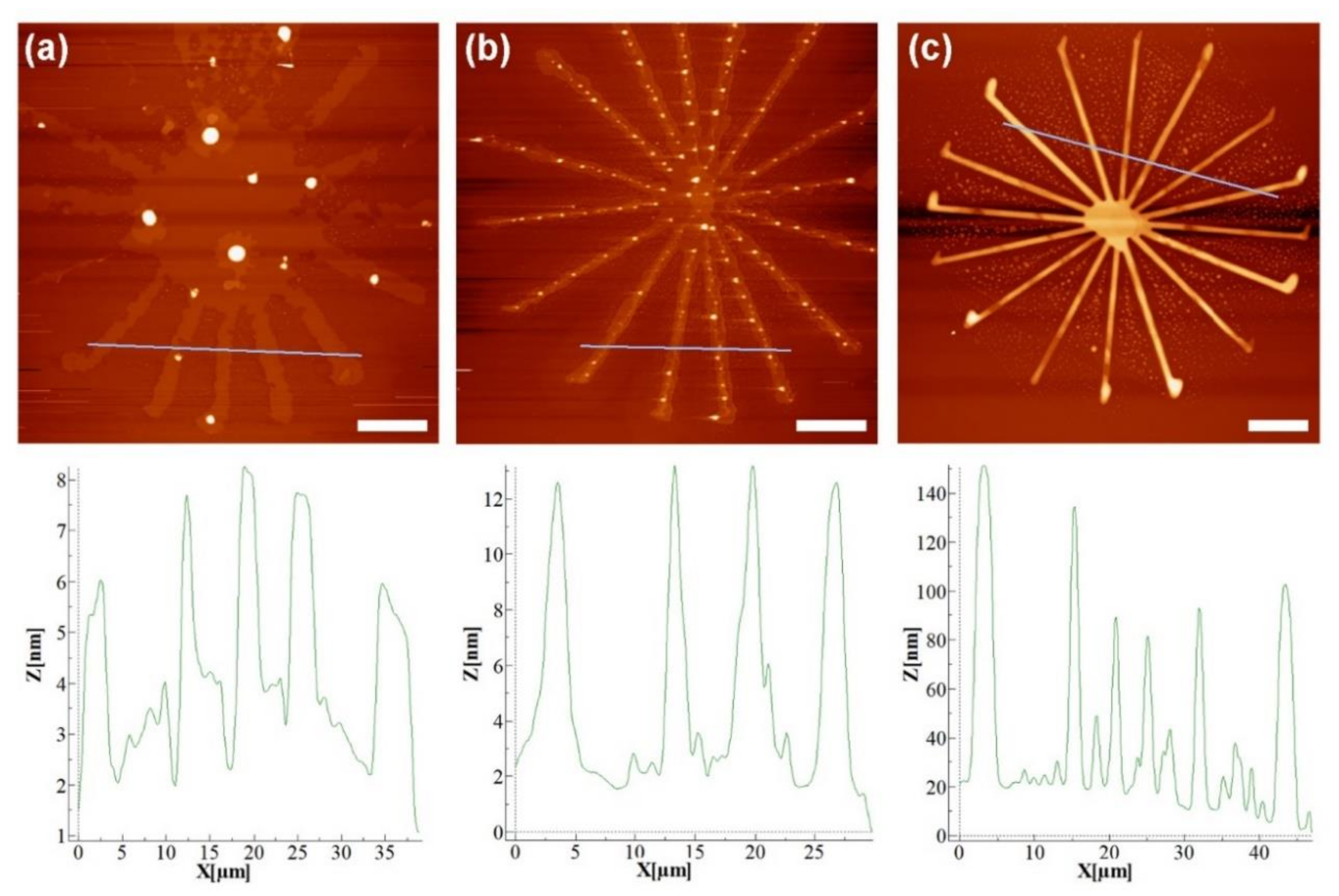
2.2. Comparison of Lipid Writing on the Different Substrates
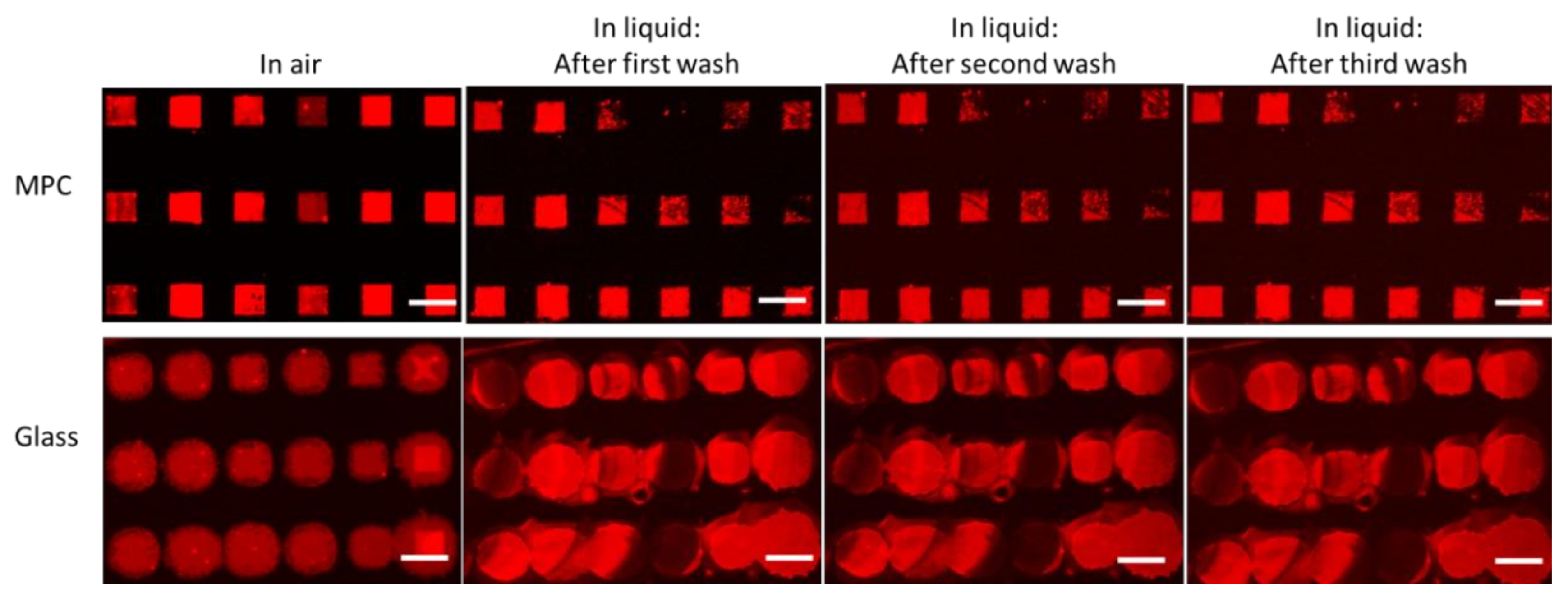
2.3. Stability of Lipid Arrays in Liquid
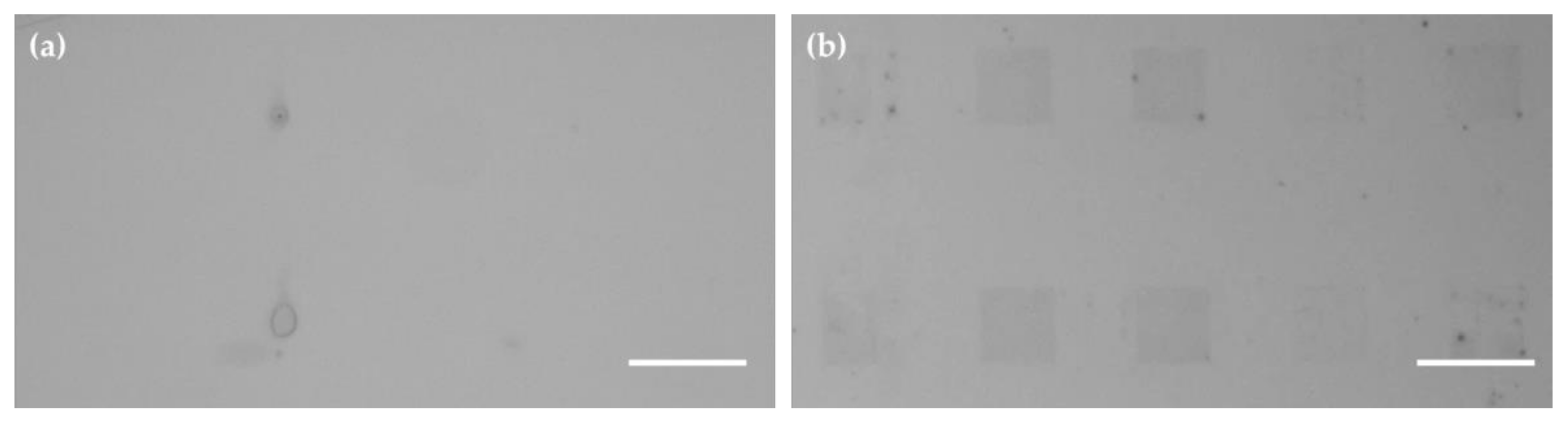
2.4. Multiplexed Immobilization of Antibodies
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of MPC Copolymer Substrates
4.3. Contact Angle Measurement
4.4. Atomic Force Microscopy
4.5. Writing of Lipid Structures and Functionalization
4.5.1. Writing of Lipid Structures by L-DPN
4.5.2. Multiplexed Immobilization of Antibodies
4.6. Optical Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gould, S.B. Membranes and evolution. Curr. Biol. 2018, 28, R381–R385. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Nikoleli, G.-P.; Nikolelis, D.; Siontorou, C.; Karapetis, S. Lipid Membrane Nanosensors for Environmental Monitoring: The Art, the Opportunities, and the Challenges. Sensors 2018, 18, 284. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Nikoleli, G.P.; Nikolelis, D.P.; Karapetis, S.K. Artificial lipid membranes: Past, present, and future. Membranes 2017, 7, 38. [Google Scholar] [CrossRef]
- Sugawara, M. Transmembrane Signaling with Lipid-Bilayer Assemblies as a Platform for Channel-Based Biosensing. Chem. Rec. 2018, 18, 433–444. [Google Scholar] [CrossRef]
- Suzuki, H.; Takeuchi, S. Microtechnologies for membrane protein studies. Anal. Bioanal. Chem. 2008, 391, 2695–2702. [Google Scholar] [CrossRef]
- Glazier, R.; Salaita, K. Supported lipid bilayer platforms to probe cell mechanobiology. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1465–1482. [Google Scholar] [CrossRef]
- Arslan, A.; Fatih, Y. Biomimetic Lipid Membranes: Fundamentals, Applications, and Commercialization; Kök, F.N., Arslan Yildiz, A., Inci, F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; ISBN 978-3-030-11595-1. [Google Scholar]
- Kilic, A.; Kok, F.N. Biomimetic lipid bilayers on solid surfaces: Models for biological interactions. Surf. Innov. 2016, 4, 141–157. [Google Scholar] [CrossRef]
- Van Weerd, J.; Karperien, M.; Jonkheijm, P. Supported Lipid Bilayers for the Generation of Dynamic Cell-Material Interfaces. Adv. Healthc. Mater. 2015, 4, 2743–2779. [Google Scholar] [CrossRef]
- Piner, R.D.; Zhu, J.; Xu, F.; Hong, S.; Mirkin, C.A. “Dip-Pen” Nanolithography. Science 1999, 283, 661–663. [Google Scholar] [CrossRef]
- Liu, G.; Hirtz, M.; Fuchs, H.; Zheng, Z. Development of Dip-Pen Nanolithography (DPN) and Its Derivatives. Small 2019, 15, 1900564. [Google Scholar] [CrossRef] [PubMed]
- Lenhert, S.; Sun, P.; Wang, Y.; Fuchs, H.; Mirkin, C.A. Massively parallel dip-pen nanolithography of heterogeneous supported phospholipid multilayer patterns. Small 2007, 3, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Hirtz, M.; Sekula-Neuner, S.; Urtizberea, A.; Fuchs, H. Functional Lipid Assemblies by Dip-Pen Nanolithography and Polymer Pen Lithography. In Soft Matter Nanotechnology: From Structure to Function; Chen, X., Fuchs, H., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2015; pp. 161–186. ISBN 9783527682157. [Google Scholar]
- Nafday, O.A.; Lenhert, S. High-throughput optical quality control of lipid multilayers fabricated by dip-pen nanolithography. Nanotechnology 2011, 22, 225301. [Google Scholar] [CrossRef] [PubMed]
- Urtizberea, A.; Hirtz, M. A diffusive ink transport model for lipid dip-pen nanolithography. Nanoscale 2015, 7, 15618–15634. [Google Scholar] [CrossRef]
- Willems, N.; Urtizberea, A.; Verre, A.F.; Iliut, M.; Lelimousin, M.; Hirtz, M.; Vijayaraghavan, A.; Sansom, M.S.P. Biomimetic Phospholipid Membrane Organization on Graphene and Graphene Oxide Surfaces: A Molecular Dynamics Simulation Study. ACS Nano 2017, 11, 1613–1625. [Google Scholar] [CrossRef] [PubMed]
- Rivel, T.; Yesylevskyy, S.O.; Ramseyer, C. Structures of single, double and triple layers of lipids adsorbed on graphene: Insights from all-atom molecular dynamics simulations. Carbon 2017, 118, 358–369. [Google Scholar] [CrossRef]
- Blachon, F.; Harb, F.; Munteanu, B.; Piednoir, A.; Fulcrand, R.; Charitat, T.; Fragneto, G.; Pierre-Louis, O.; Tinland, B.; Rieu, J.-P. Nanoroughness Strongly Impacts Lipid Mobility in Supported Membranes. Langmuir 2017, 33, 2444–2453. [Google Scholar] [CrossRef]
- Gavutis, M.; Navikas, V.; Rakickas, T.; Vaitekonis, Š.; Valiokas, R. Lipid dip-pen nanolithography on self-assembled monolayers. J. Micromech. Microeng. 2016, 26, 025016. [Google Scholar] [CrossRef]
- Rebaud, S.; Maniti, O.; Girard-Egrot, A.P. Tethered bilayer lipid membranes (tBLMs): Interest and applications for biological membrane investigations. Biochimie 2014, 107, 135–142. [Google Scholar] [CrossRef]
- Andersson, J.; Köper, I.; Knoll, W. Tethered Membrane Architectures—Design and Applications. Front. Mater. 2018, 5, 55. [Google Scholar] [CrossRef]
- Nagahashi, K.; Teramura, Y.; Takai, M. Stable surface coating of silicone elastomer with phosphorylcholine and organosilane copolymer with cross-linking for repelling proteins. Colloids Surf. B Biointerfaces 2015, 134, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Ausserré, D.; Valignat, M.-P. Surface enhanced ellipsometric contrast (SEEC) basic theory and lambda/4 multilayered solutions. Opt. Express 2007, 15, 8329–8339. [Google Scholar] [CrossRef] [PubMed]
- Hirtz, M.; Corso, R.R.; Sekula-Neuner, S.; Fuchs, H. Comparative Height Measurements of Dip-Pen Nanolithography-Produced Lipid Membrane Stacks with Atomic Force, Fluorescence, and Surface Enhanced Ellipsometric Contrast Microscopy. Langmuir 2011, 27, 11605–11608. [Google Scholar] [CrossRef] [PubMed]
- Dadfar, S.M.M.; Sekula-Neuner, S.; Bog, U.; Trouillet, V.; Hirtz, M. Site-Specific Surface Functionalization via Microchannel Cantilever Spotting (µCS): Comparison between Azide-Alkyne and Thiol-Alkyne Click Chemistry Reactions. Small 2018, 14, 1800131. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Urtizberea, A.; Ghosh, S.; Bog, U.; Rainer, Q.; Lenhert, S.; Fuchs, H.; Hirtz, M. Polymer Pen Lithography with Lipids for Large-Area Gradient Patterns. Langmuir 2017, 33, 8739–8748. [Google Scholar] [CrossRef] [PubMed]
- Hirtz, M.; Greiner, A.M.; Landmann, T.; Bastmeyer, M.; Fuchs, H. Click-Chemistry Based Multi-Component Microarrays by Quill-Like Pens. Adv. Mater. Interfaces 2014, 1, 1300129. [Google Scholar] [CrossRef]
- Atwater, J.; Mattes, D.S.; Streit, B.; von Bojničić-Kninski, C.; Loeffler, F.F.; Breitling, F.; Fuchs, H.; Hirtz, M. Combinatorial Synthesis of Macromolecular Arrays by Microchannel Cantilever Spotting (µCS). Adv. Mater. 2018, 30, 1801632. [Google Scholar] [CrossRef]
- Green, N.M. Avidin. Adv. Protein Chem. 1975, 29, 85–133. [Google Scholar]
- Wilchek, M.; Bayer, E.A. Introduction to avidin-biotin technology. In Methods in Enzymology—Vol. 184: Avidin-Biotin Technology; Elsevier: Amsterdam, The Netherlands, 1990; pp. 5–13. [Google Scholar]
- Lenhert, S.; Brinkmann, F.; Laue, T.; Walheim, S.; Vannahme, C.; Klinkhammer, S.; Xu, M.; Sekula, S.; Mappes, T.; Schimmel, T.; et al. Lipid multilayer gratings. Nat. Nanotechnol. 2010, 5, 275–279. [Google Scholar] [CrossRef]
- Hirtz, M.; Brglez, J.; Fuchs, H.; Niemeyer, C.M.C.M. Selective Binding of DNA Origami on Biomimetic Lipid Patches. Small 2015, 11, 5752–5758. [Google Scholar] [CrossRef]
- Sekula, S.; Fuchs, J.; Weg-Remers, S.; Nagel, P.; Schuppler, S.; Fragala, J.; Theilacker, N.; Franzreb, M.; Wingren, C.; Ellmark, P.; et al. Multiplexed lipid dip-pen nanolithography on subcellular scales for the templating of functional proteins and cell culture. Small 2008, 4, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Sekula-Neuner, S.; Maier, J.; Oppong, E.; Cato, A.C.B.; Hirtz, M.; Fuchs, H. Allergen Arrays for Antibody Screening and Immune Cell Activation Profiling Generated by Parallel Lipid Dip-Pen Nanolithography. Small 2012, 8, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Oppong, E.; Hedde, P.N.; Sekula-Neuner, S.; Yang, L.; Brinkmann, F.; Dörlich, R.M.; Hirtz, M.; Fuchs, H.; Nienhaus, G.U.; Cato, A.C.B. Localization and Dynamics of Glucocorticoid Receptor at the Plasma Membrane of Activated Mast Cells. Small 2014, 10, 1991–1998. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Hirtz, M.; Sekula-Neuner, S.; Laue, T.; Fuchs, H.; Willaert, R.G. Dip-pen nanolithography-assisted protein crystallization. J. Am. Chem. Soc. 2015, 137, 154–157. [Google Scholar] [CrossRef]
- Oberhansl, S.; Castaño, A.G.; Lagunas, A.; Prats-Alfonso, E.; Hirtz, M.; Albericio, F.; Fuchs, H.; Samitier, J.; Martinez, E. Mesopattern of immobilised bone morphogenetic protein-2 created by microcontact printing and dip-pen nanolithography influence C2C12 cell fate. RSC Adv. 2014, 4, 56809–56815. [Google Scholar] [CrossRef]
- Navikas, V.; Gavutis, M.; Rakickas, T.; Valiokas, R. Scanning Probe-Directed Assembly and Rapid Chemical Writing Using Nanoscopic Flow of Phospholipids. ACS Appl. Mater. Interfaces 2019, 11, 28449–28460. [Google Scholar] [CrossRef] [PubMed]
- Lowry, T.W.; Hariri, H.; Prommapan, P.; Kusi-Appiah, A.; Vafai, N.; Bienkiewicz, E.A.; Van Winkle, D.H.; Stagg, S.M.; Lenhert, S. Quantification of Protein-Induced Membrane Remodeling Kinetics In Vitro with Lipid Multilayer Gratings. Small 2016, 12, 506–515. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, J.C.; Tseng, P.Y.; Tsai, W.S.; Liao, M.Y.; Lu, S.H.; Frank, C.W.; Chen, J.S.; Wu, H.C.; Chang, Y.C. Antibody conjugated supported lipid bilayer for capturing and purification of viable tumor cells in blood for subsequent cell culture. Biomaterials 2013, 34, 5191–5199. [Google Scholar] [CrossRef] [PubMed]
- Rädler, J.O.; Strey, H.; Sackmann, E. Phenomenology and Kinetics of Lipid Bilayer Spreading on Hydrophilic Surfaces. Langmuir 1995, 11, 4539–4548. [Google Scholar] [CrossRef]
- Howland, M.C.; Sapuri-Butti, A.R.; Dixit, S.S.; Dattelbaum, A.M.; Shreve, A.P.; Parikh, A.N. Phospholipid morphologies on photochemically patterned silane monolayers. J. Am. Chem. Soc. 2005, 127, 6752–6765. [Google Scholar] [CrossRef]
- Nissen, J.; Gritsch, S.; Wiegand, G.; Rädler, J.O. Wetting of phospholipid membranes on hydrophilic surfaces—Concepts towards self-healing membranes. Eur. Phys. J. B 1999, 10, 335–344. [Google Scholar] [CrossRef]
- Sanii, B.; Parikh, A.N. Surface-energy dependent spreading of lipid monolayers and bilayers. Soft Matter. 2007, 3, 974. [Google Scholar] [CrossRef]
- Parikh, A.N. Membrane-substrate interface: Phospholipid bilayers at chemically and topographically structured surfaces. Biointerphases 2008, 3, FA22. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Ishihara, K.; Watanabe, A.; Nakabayashi, N. Interaction between phospholipids and biocompatible polymers containing a phosphorylcholine moiety. In The Biomaterials: Silver Jubilee Compendium; Elsevier: Amsterdam, The Netherlands, 1991; Volume 12, pp. 69–72. ISBN 9780080451541. [Google Scholar]
- Nagasawa, D.; Azuma, T.; Noguchi, H.; Uosaki, K.; Takai, M. Role of Interfacial Water in Protein Adsorption onto Polymer Brushes as Studied by SFG Spectroscopy and QCM. J. Phys. Chem. C 2015, 119, 17193–17201. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lynch, M.; Huff, J.L.; Mosher, C.; Vengasandra, S.; Ding, G.; Henderson, E. Microfabricated quill-type surface patterning tools for the creation of biological micro/nano arrays. Biomed. Microdevices 2004, 6, 117–123. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the MPC copolymer are available from the authors. |

| Substrate | Rq (nm) | Contact Angle (°) |
|---|---|---|
| Glass | 1.05 ± 0.08 | 27.5 ± 1.2 [26] |
| SiOx (SEEC) [25] | 0.22 ± 0.02 | 61.9 ± 3.3 [27] |
| Block-type MPC copolymer | 0.43 ± 0.07 | 25.5 ± 1.8 |
| Substrate | Average Line Width (µm) |
|---|---|
| Glass | 6.8 ± 0.7 |
| SiOx (SEEC) | 5.5 ± 0.2 |
| Block-type MPC copolymer | 2.5 ± 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.-Y.; Kumar, R.; Takai, M.; Hirtz, M. Enhanced Stability of Lipid Structures by Dip-Pen Nanolithography on Block-Type MPC Copolymer. Molecules 2020, 25, 2768. https://doi.org/10.3390/molecules25122768
Liu H-Y, Kumar R, Takai M, Hirtz M. Enhanced Stability of Lipid Structures by Dip-Pen Nanolithography on Block-Type MPC Copolymer. Molecules. 2020; 25(12):2768. https://doi.org/10.3390/molecules25122768
Chicago/Turabian StyleLiu, Hui-Yu, Ravi Kumar, Madoka Takai, and Michael Hirtz. 2020. "Enhanced Stability of Lipid Structures by Dip-Pen Nanolithography on Block-Type MPC Copolymer" Molecules 25, no. 12: 2768. https://doi.org/10.3390/molecules25122768
APA StyleLiu, H.-Y., Kumar, R., Takai, M., & Hirtz, M. (2020). Enhanced Stability of Lipid Structures by Dip-Pen Nanolithography on Block-Type MPC Copolymer. Molecules, 25(12), 2768. https://doi.org/10.3390/molecules25122768







