Abstract
The interaction of nuclear receptors (NRs) with chemical compounds can cause dysregulation of endocrine signaling pathways, leading to adverse health outcomes due to the disruption of natural hormones. Thus, identifying possible ligands of NRs is a crucial task for understanding the adverse outcome pathway (AOP) for human toxicity as well as the development of novel drugs. However, the experimental assessment of novel ligands remains expensive and time-consuming. Therefore, an in silico approach with a wide range of applications instead of experimental examination is highly desirable. The recently developed novel molecular image-based deep learning (DL) method, DeepSnap-DL, can produce multiple snapshots from three-dimensional (3D) chemical structures and has achieved high performance in the prediction of chemicals for toxicological evaluation. In this study, we used DeepSnap-DL to construct prediction models of 35 agonist and antagonist allosteric modulators of NRs for chemicals derived from the Tox21 10K library. We demonstrate the high performance of DeepSnap-DL in constructing prediction models. These findings may aid in interpreting the key molecular events of toxicity and support the development of new fields of machine learning to identify environmental chemicals with the potential to interact with NR signaling pathways.
1. Introduction
Many chemical substances have potential harmful effects, causing the perturbation of endocrine homeostasis by interfering with various nuclear receptors (NRs) of hormones [1,2,3,4,5]. In the disruption of hormone pathways, structurally diverse groups of chemicals are known to interact primarily with ligand–NR bindings, which have the ability to substitute for natural ligands, ultimately resulting in proliferative, reproductive, and metabolic disorders [6,7,8,9,10,11,12]. NRs are a superfamily of ligand-dependent transcriptional factors containing n N-terminal transactivation domain, a flexible hinge region, and a C-terminal ligand-binding domain (LBD) [6,8,13]. NRs are classified mainly into two types according to their subcellular distribution in the absence of a ligand and their mechanisms: Type I steroid receptors, including the estrogen receptor (ER), androgen receptor (AR), progesterone receptor (PR), and glucocorticoid receptor (GR); and Type II nonsteroid receptors, including the thyroid receptor (TR alpha and beta), retinoic acid receptor (RAR alpha, beta, and gamma), retinoid X receptor (RXR), vitamin D receptor (VDR), peroxisome proliferator-activated receptor (PPAR alpha, beta, and gamma), liver X receptor (LXR), farnesoid X receptor (FXR), and pregane X receptor (PXR), [6,14,15]. In the absence of a ligand, the type I NR forms inactive complexes with chaperone proteins in the cytoplasm, whereas type II NR, regardless of the ligand-binding status, is located in the nucleus and binds to the DNA response elements of its target genes along with corepressors [6,14,16]. For these types of NRs, a number of allosteric modulators have been identified that can act as either agonist or antagonist by occupying the active pocket of the NR and blocking the recruitment of coactivators or corepressors to the transcriptional complex [11,17,18,19,20].
The perturbation of the NR signaling pathway due to the action of agonists or antagonists of chemical compounds is associated with various adverse health outcomes [19,21]. Although chemical hazard assessments have traditionally relied upon toxicity data from animal bioassays and epidemiological studies, there are some drawbacks to this testing method, such as high cost, lengthy test durations, and ethical concerns [5,22,23,24,25,26,27]. To resolve these issues, the in vitro high-throughput screening (HTS) assay has been developed as an alternative approach and improved by the Toxicity Forecaster (ToxCastTM) program run by the U.S. Environmental Protection Agency (EPA) [5,28,29,30] and The Toxicology in the 21st Century program (Tox21), an interagency federal collaboration launched by the consortium of the EPA, the U.S. Food and Drug Administration (FDA), the National Institutes of Health (NIH), and the National Toxicology Program (NTP) [5,31]. However, the HTS assay is not sufficient to screen all classes of chemicals, such as those still in molecular development and optimization phase, and thus cannot provide an accurate evaluation of the potential toxicity of chemicals in humans and the environment [5,32].
Recent technological advances have focused on in silico approaches, such as quantitative structure–activity relationship (QSAR), based on the assumption that similar structures are associated with similar biological activities, taking advantage of their ability to accurately predict the toxicologically discrete values of the chemical or biological properties of molecules [5,33,34,35,36,37]. However, the QSAR approach has the following disadvantages: (i) required skills and knowledge for feature extraction and selection, (ii) paucity of model interpretability, and (iii) low prediction performance due to the dependence on the choice of molecular descriptors and the prediction modeling algorithms [36,38,39,40]. To address these issues, a novel deep learning (DL)-based QSAR method, called DeepSnap-DL [41], was developed using molecular image files generated from the steric conformation of three-dimensional (3D) chemical structures, leveraging the increasing evidence of successful classification by convolutional neural networks (CNNs) through DL in toxicological fields [40,42,43]. This method has the following advantages. First, the feature(s) in the molecular images can be automatically extracted by CNNs. Second, high prediction performance can be expected as more detailed information of the chemical structure can be captured from different viewing directions along the x-, y-, and z-axes [41,44,45,46,47]. Third, determination and visualization of the conformer that is docked in the LBD of the receptor protein may reveal the critical conformation of the chemicals and domain of the receptor protein related to the adverse outcome.
In this study, using the DeepSnap-DL method, prediction models of 35 agonists and antagonists of NRs were constructed by 3D molecular structure representations using information of chemical compounds from the Tox21 10K library. The results obtained by the DeepSnap-DL method outperformed those of the methods that won the Tox21 data challenge. Therefore, our approach can be practically applied to build prediction models using a CNN for a large number of chemicals to determine their potential toxicity.
2. Results and Discussion
To build the prediction models of the agonists and antagonists of NRs, we downloaded the information of 35 NRs for the chemical structures and their activity scores from the Tox21 10K library. The mean number of chemicals was 7262 ± 267, and the highest and lowest numbers of the chemicals were respectively 7671 (progesterone receptor agonist: PR_ago, AID: 1347036), and 6735 (estrogen-related receptor agonist: ERR_ago, AID: 1259404) (Figure 1). Furthermore, we classified the datasets of these chemical compounds into two groups based on their activity scores—active chemicals were those with an activity score ≥ 40 and inactive chemicals had an activity score < 40. The mean number of active chemicals in the total chemicals was 0.0372 ± 0.0376, and the highest and lowest numbers of active chemicals were, respectively, 0.2052 (pregane X receptor agonist: PXR_ago, AID: 1347033) and 0.0022 (vitamin D receptor agonist: VDR_ago, AID: 743241) (Figure 1). These results indicate that the datasets are highly class imbalanced.
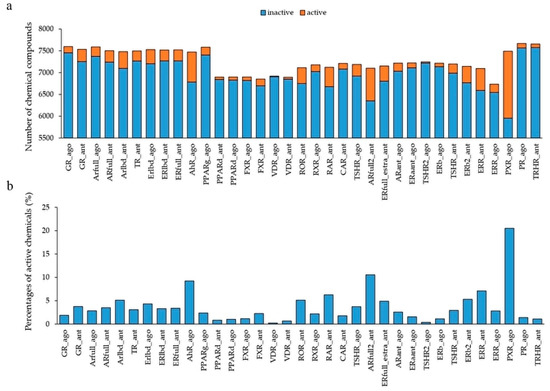
Figure 1.
Activity distribution of the Tox21 10K library against 35 NR agonists and antagonists used in the DeepSnap-deep learning (DL) approach. (a) Number of chemical compounds used in the modeling by the DeepSnap-DL, where orange and blue indicate active and inactive chemicals, respectively. (b) Percentage of active chemicals against total chemicals.
Next, the datasets were divided into Tra:Val:Test groups with a 4:4:1 ratio. The mean numbers of active and inactive chemicals were, respectively, 120.9 ± 124.1 and 3107.1 ± 153.1 in Tra, 120.8 ± 124.3 and 3106.8 ± 153.3 in Val, and 30.1 ± 30.9 and 271.8 ± 279.2 in Test (Table S1, Supplementary Materials). In addition, the highest and lowest numbers of the active chemicals were, respectively, 683 and 2 in Tra, 684 and 6 in Val, and 170 and 2 in Test (Table S1). The molecular images derived from the 3D chemical structures were generated using the DeepSnap approach at different angles along the x-, y-, and z-axes, i.e., (176°, 176°, 176°). A total of 27 images for one chemical compound was captured (Figure 2, Figure S1).
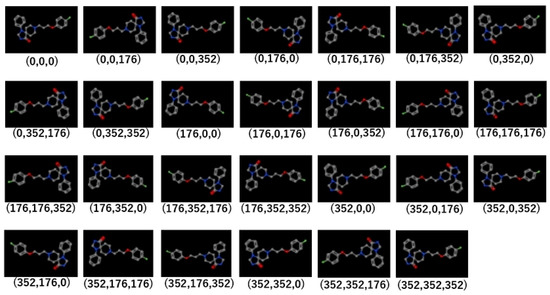
Figure 2.
Representative molecular images used in the DeepSnap-DL method. The parentheses below each image indicates the angles of depiction in DeepSnap.
Using these molecular images as input data into the DL, the prediction models of 35 NR agonists and antagonists were constructed using Tra, and validated with Val. The values of mean Loss (Val) and Acc (Val) were 0.0748 ± 0.0035 and 97.56 ± 0.09, respectively (Figure 3, Figure S4a, Supplementary Materials). In addition, the highest prediction performance on the Val dataset was observed in the thyroid-stimulating hormone receptor agonist (TSHR2_ago, AID: 1259393), for which the mean Loss (Val) and Acc (Val) were 0.0017 ± 0.0008 and 99.93 ± 0.02, respectively (Figure 3, Figure S4a). The prediction performance of these models was evaluated using Test based on five metrics, namely AUC, BAC, F, Acc (Test), and MCC. The results showed that the mean AUC, BAC, F, Acc (Test), and MCC were 0.8842 ± 0.0165, 0.8471 ± 0.0168, 0.3085 ± 0.0411, 82.73 ± 3.92, and 0.3536 ± 0.0377, respectively (Figure 4 and Figure 5, Figure S4a,b). In addition, the highest prediction performance on Test was observed in the thyroid-stimulating hormone receptor agonist (TSHR2_ago, AID: 1259393), with the mean AUC, BAC, F, Acc (Test), and MCC being 0.9994 ± 0.0006, 0.9997 ± 0.0003, 0.9286 ± 0.0714, 99.94 ± 0.06, and 0.9327 ± 0.0673, respectively (Figure 4 and Figure 5, Figure S4a,b).
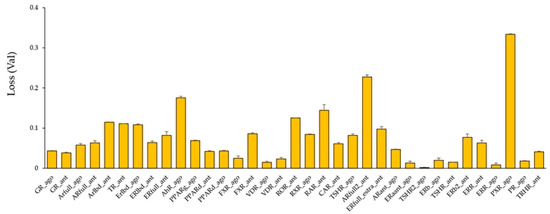
Figure 3.
Average Loss values in the validation (Val) dataset in models of 35 NR agonists and antagonists constructed by DeepSnap-DL. n = 2. Each bar indicates average of Loss (Val) ± standard error.
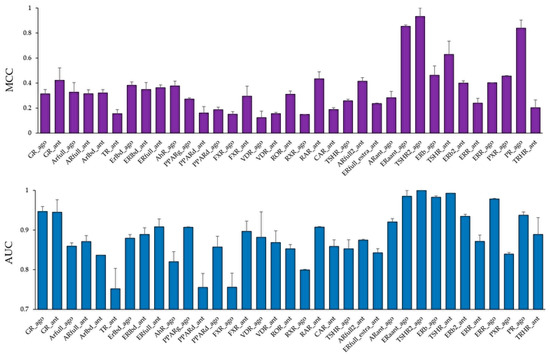
Figure 4.
Average Matthews correlation coefficient (MCC) (top) and area under the curve (AUC) (bottom) values in the Test dataset in the models of 35 NR agonists and antagonists constructed by DeepSnap-DL. n = 2. Each bar indicates average MCC and AUC ± standard error.

Figure 5.
Representative area under the curve of receiver operating characteristic curve (ROC_AUC) in the models of 35 NR agonists and antagonists constructed by DeepSnap-DL.
The Tox21 Data Challenge 2014 was designed to understand the interference of the chemical compounds derived from the Tox21 10K compound library in the biological pathway via crowdsourced data analysis by independent researchers. It used data generated from seven NR signaling pathway assays to construct prediction models for QSARs [48]. The BAC values of the three models constructed by the proposed DeepSnap-DL were 0.8361, 0.8204, and 0.8494, respectively, outperforming the Data Challenge models where the BACs of three models, namely AID:743053 (Arfull_ago), AID:743077 (Erlbd_ago), and AID:743140 (PPARg_ago), were 0.6500, 0.7147, and 0.7852, respectively. However, the best prediction model of AID:743122 (AhR_ago) had a BAC value of 0.8528 in the Data Challenge, whose BAC outperformed that in the DeepSnap-DL method (0.7785). Up to now, conflicting observations have been reported regarding whether DL performs better than conventional shallow machine learning (ML) methods, such as random forest, support vector machine, and gradient boosting decision tree [40,43,49,50,51,52,53]. Although some reports suggest that DL outperforms conventional ML methods owing to various improvements, the performance of DL in terms of QSAR may be affected by many factors, such as molecular descriptors, assay targets, chemical space, hyper-parameter optimization, DL architectures, input data size, and quality [40].
Furthermore, the DeepSnap-DL approach has the black box problem, that is, it lacks explainability and interpretability of the prediction models because the convolutional area on the image picture by CNN is not defined. This issue has been extensively studied, especially in the field of image recognition. These studies try to resolve the issue by calculating the gradient of the input image with respect to the output label and highlighting the target pixel as a recognition target when a slight change in a specific input pixel causes a large change in the output label. However, a simple calculation of the gradient generates a noisy highlight, so some improved methods have been proposed for sharpening [54,55,56,57,58,59]. In addition, in the DeepSnap-DL approach, the performance improves as data size increases, and performance deterioration is observed with insufficient data size or the presence of noise. However, simply increasing the sample size causes problems such as overfitting and increased calculation costs. To resolve the issues of the DeepSnap-DL approach, critical factors include specifying the image area and type required for effective feature extraction to reduce the input data volume, and clarification of the functional relationship of chemical substances with biological activity in vivo. Future applications may include screening of target molecules in specific pathological reactions.
To investigate whether the in vitro bioassays for agonist and antagonist mode in the Tox21 program affect the prediction performance of NRs, we compared prediction performances among four in vitro assays, namely, luciferase, beta-lactamase, cAMP, and intracellular calcium assays, using the results of 35 NR agonist and antagonist prediction models. In the Val dataset, the loss and accuracy values in the luciferase assay were significantly higher and lower, respectively, compared with that of the beta-lactamase assay (Figure 6a,b, p < 0.05 for both Loss (Val) and Acc (Val)).
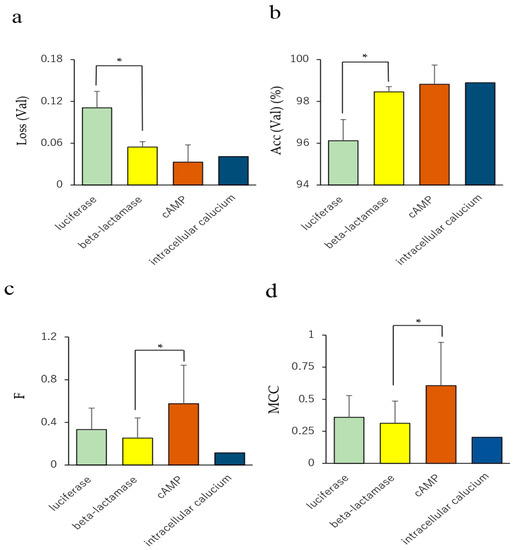
Figure 6.
Comparison of prediction performance among four in vitro assays. (a) Loss in the Val dataset, (b) accuracy in the Val dataset, (c) accuracy in the Test dataset. n = 14, 17, 3, and 1 for luciferase, beta-lactamase, cAMP, and intracellular calcium assays, respectively. Each bar indicates the average of the performance metric of the four in vitro assays with standard error. * p < 0.05 by Tukey–Kramer’s honestly significant difference test.
In addition, F and MCC in Test of the cAMP assay significantly increased compared with those of the beta-lactamase assay (Figure 6c,d, p < 0.05 for both F-measure and MCC). The BAC value in the Test dataset of the cAMP assay showed a moderate increase compared with that of the beta-lactamase assay (Figure S5c, Supplementary Materials, p < 0.09). These results indicate that the prediction performance of the NR agonists and antagonists in the Tox21 10K library may be affected by the choice of the in vitro assay method. There are several conflicting reports regarding the in vitro receptor-mediated activity. Chemicals such as bisphenol A (BSA) and its halogenated analogs (tetrabromo-BSA and tetrachloro-BSA) show weak TR antagonist activity but have a potential agonist-like effect at lower concentrations [60,61]. Thus, competitive agonists and antagonists of the steroids have long been known [62,63,64]. Among them, ligands exhibiting agonist and antagonist activity, called selective steroid receptor modulators (SSRMs), are known to show specificity on tissue or cell type [62,65,66,67,68,69]. In addition, a competitive antagonist, known as the passive antagonist, hinders the binding but induces the inactive state of NRs by modifying interaction with their corepressor and interfering with their nuclear translocation or DNA binding at saturated concentrations [62,70]. These reports suggest that the ligand of the steroid NRs can serve not only as competitive agonists and antagonists that affect binding to the NRs, but also as a unique allosteric modulator for subsequent molecular interactions. Therefore, classification of the chemicals in the Tox21 10K library may require more detailed insights of the molecular mechanisms of the NRs with chemical compounds and the conditions of in vitro bioassays.
3. Conclusions
In this study, we built prediction models of 35 NR agonists and antagonists using the DeepSnap-DL approach with information of the chemical structure and activity from the Tox21 10K library. Three prediction models outperformed the best performing models in the Tox21 Data Challenge 2014. These results suggest that the 3D chemical structure representation in the DeepSnap-DL approach may be useful for molecular image-based QSAR analysis, and the improvements to the DeepSnap-DL method may aid in achieving high-performing prediction models.
4. Materials and Methods
4.1. Data
In this study, the original datasets related to chemical structures and the corresponding agonist and antagonist scores were downloaded as reported previously [44,45,46,47], in the simplified molecular input line entry system (SMILES) format from the PubChem database. We used a keyword of the database search, namely “Tox21 bioassays”, and selected bioassays of the 35 from the NR signaling pathway for the identification of agonists/antagonists (Table 1). These bioassay data consisted of quantitative HTS (qHTS) data derived from two cell-based reporter gene assays, including beta-lactamase or luciferase reporter genes. The activity of these reporter genes is controlled by the binding of transcriptional factors induced or suppressed by an agonist/antagonist with response elements (REs) for ARs, ER-alpha, ER-beta, estrogen-related receptors (ERR), FXR, PPAR−gamma, PRs, retinoid-related orphan receptor gamma (ROR−gamma), RXR−alpha, RARs, GRs, TRs, thyroid-stimulating hormone receptors (TSHRs), aryl hydrocarbon receptors (AhRs), VDRs, constitutive androstane receptors (CARs), and PXRs. These receptors are stably integrated into cell lines, including human embryonic kidney 293 cells(HEK293 (AR, ER−alpha, ER−beta, ERR, and TSHR), HEK293H (PPAR−gamma, PPAR−delta, and HEK293T (ER−beta, FXR, PR, RXR−alpha, and VDR)), human breast cancer cells (MDA−MB (AR)), ovarian carcinoma cells (BG1 (ER−alpha)), Chinese hamster ovary cells CHO (ROR−gamma)), human cervical cancer cells (HeLa (GR)), rat pituitary tumor cells (GH3 (TR)), human hepatocellular carcinoma cells (HepG2 (AhR, CAR, PXR)), and C3H mouse embryo cells (C3RL4 (RXR−alpha)). Then, we can measure the ability to induce or inhibit RE-dependent transcription.

Table 1.
Nuclear receptors (NRs) and their bioassays used in this study.
The chemicals were derived from the Tox21 10K library, which contains approximately 8900 unique compounds gathered from commercial sources, such as pesticides, industrial and environmental chemicals, natural dietary supplement products, food additives, and drugs, by the NTP, the National Center for Advancing Translational Sciences (NCATS), and the EPA (Table 1) [71,72,73,74,75,76,77,78,79,80,81,82]. These compounds were dissolved in dimethyl sulfoxide (DMSO) as stock solutions, and compound plates with the different concentrations were prepared in the 1536-well plate format [71,72,73,80,83]. These cell lines of beta-lactamase reporter gene assay constitutively co-express a fusion protein comprised of the LBDs of the human NRs coupled to the DNA-binding domain (DBD) of the yeast transcription factor GAL4 [72,73,75,80]. When activated, these fusion proteins stimulate beta-lactamase reporter gene expression.
The cells were dispensed at 1500 to 5000 cells/5 (for antagonist mode) or 6 (for agonist mode) microL/well in 1536-well black wall/clear bottom plates [72,73,75,78,79,80]. After the cells were incubated at 37 °C for 5 to 6 h depending on the particular NR cell line to allow for cell attachment, 23 nL of the compounds at different concentrations were transferred to the assay plates. For the antagonist mode assay, the known agonist for each NR was added into the assay plates. For the agonist and antagonist mode assays, positive control compounds were dispensed into each other’s wells on the plates (Table 1) [72,73,75,78,79,80]. The plates were incubated for 16 to 18 h at 37 °C depending on the particular NR cell line. Then, a LiveBLAzerTM B/G FRET substrate (Invitrogen, Carlsbad, CA, USA) detection mix was added, and the plates were incubated at room temperature for 1.5 to 2 h. The fluorescence intensity (405 nm excitation, 460 and 530 nm emission) was measured using an Envision plate reader (PerkinElmer, Shelton, CT, USA). Data were expressed as the ratio of 460/530 nm emission values. To measure the luciferase reporter gene activity, 4 microL of ONE-GloTM Luciferase Assay reagent (Promega, Madison, WI, USA) were added to each plate, and the luminescence intensity was quantified by a ViewLux plate reader (PerkinElmer) after 30 min of incubation at room temperature. Data were expressed as relative luminescence units.
4.2. qHTS Data Analysis
The Tox21 10k library can be grouped into clusters with similar activity that share similar annotated models of action according to PubChem activity scores. In the qHTS of the Tox21 program, to identify the chemical compounds in both potential agonist and antagonist modes, the PubChem activity scores were determined from 0% to 100% by normalizing each titration point relative to the positive control compound (agonist mode: 100%, antagonist mode: 0%) and DMSO-only wells (agonist mode: 0%, antagonist mode: -100%) according to the following equation: % Activity = [(Vcompound − Vdmso)/(Vpos − Vdmso)] × 100, where Vcompound, Vdmso, and Vpos denote the compound well values, the median values of the DMSO-only wells, and the median value of the positive control well, respectively.
The datasets were then corrected using compound-free control plates, i.e., DMSO-only plates, at the beginning and end of the compound plate measurement [72,73,75,78,79,80]. The half maximum inhibition values (IC50) and the maximum response values for each compound were calculated by fitting the concentration–response curves of each compound to a four-parameter Hill equation [84,85].
The PubCem activity scores of the agonists and antagonists were grouped into three classes, namely (1) 0, (2) 1–39, and (3) 40–100, which represent inactive, inconclusive, and active compounds, respectively. In this study, compounds with activity scores of 40–100 or 0–39 were defined as active or inactive, respectively. The dataset includes some similar chemical compounds, but with different activity scores for different ID (identification) numbers due to the presence of possible stereoisomers or salts. Therefore, chemical compounds with indefinite activity criteria, nonorganic compounds, and/or inaccurate SMILES were eliminated.
4.3. DeepSnap
We then applied a 3D conformational import from the SMILES format using molecular operating environment (MOE) 2018 software (MOLSIS Inc., Tokyo, Japan) to generate the chemical database. Here, the neutralization of the protonation state and the coordinating washed species were used by the external program, CORINA classic software (Figure S1, Supplementary Materials) [86]. The resulting 3D structures were then saved in an SDF file format. Using the SDF files prepared by the MOE application, the 3D chemical structures were depicted as 3D ball-and-stick models with different colors corresponding to different atoms by Jmol, an open-source Java viewer software (version number, manufacturer, city, state abbreviation, country) for 3D molecular modeling of chemical structures [44,45,46,47]. These 3D chemical structures produce different images depending on the direction. The 3D chemical models were captured automatically as snapshots with user-defined angle increments with respect to the x-, y-, and z-axes. In this study, one angle increment was used, i.e., (176°, 176°, 176°). Other parameters for the DeepSnap depiction process were set based on previous studies as follows: image pixel size: 256 × 256; molecule number per SDF file to split into: 100; zoom factor (%): 100; atom size for van der Waals radius (%): 23; bond radius (mÅ): 14.5; minimum bond distance: 0.4; and bond tolerance: 0.8 [44,45,46,47]. The snapshots saved as 256 × 256 pixel resolution PNG files (RGB) were divided into three types of datasets: training (Tra), validation (Val), and test (Test) (Figure S1, Figure 2).
4.4. Preparation of Dataset
Three groups of datasets were prepared by dividing the data into Tra, Val, and Test groups. The data were first split into 11 groups, and the two dataset groups (4:4:1_01 and 4:4:1_02) were then built in accordance with the ratio of Tra:Val:Test = 4:4:1. A prediction model was created using the Tra and Val datasets. Then, the prediction performance was evaluated using the Test dataset (4:4:1_01) (Figure S2, Supplementary Materials). For a subsequent analysis, the remaining Test dataset was selected from the group not used in the first analysis. The model was then built, and its probability calculation was examined in the same manner (4:4:1_02). Finally, two tests were performed and the average was calculated (Figure S2).
4.5. Deep Learning
All the two-dimensional (2D) PNG images produced by DeepSnap were resized by utilizing the NVIDIA DL GPU Training System (DIGITS) version 4.0.0 software (NVIDIA, Santa Clara, CA, USA), on four-GPU systems, Tesla-V100-PCIE (31.7 GB), with a resolution of 256 × 256 pixels as input data, as previously reported [44,45,46,47]. The prediction model was pre-trained as transfer learning [44,45,46,47] by the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) 2012 dataset [87], which includes 1000 classes, such as animal (40%), device (12%), container (9%), consumer goods (6%), and equipment (4%). The ILSVRC 2012 dataset was divided as 1.2 million Tra, 50,000 Val, and 1 million Test datasets extracted from ImageNet [88]. To rapidly train and fine-tune the highly accurate CNNs using the input Tra and Val datasets based on the image classification and building the pre-trained prediction model, we used a pre-trained open-source DL model, Caffe, and the open-source software on the CentOS Linux distribution 7.3.1611. In this study, the deep CNN architecture was GoogLeNet, which is a complex network inspired by LeNet and implemented with a novel module called “Inception”, which facilitates batch normalization, image distortions, and RMSprop; concatenates different filter sizes and dimensions into a single new filter; and introduces sparsity and multiscale information in one block (Figure S3, Supplementary Materials). The network is a 22-layer deep CNN, comprising two convolutional layers, two types of pooling layers (four max pools and one avg pool), and nine Inception modules, each module having six convolution layers and one pooling layer, with 4 million parameters (Figure S3) [89,90,91].
In the DeepSnap-DL method, the prediction models were constructed by training datasets using 30 epochs with 1 snapshot interval in each epoch, 1 validation interval in each epoch, 1 random seed, a stochastic gradient descent-type solver, a learning rate of 0.006, and a batch size of 108 in DL. Among the epochs, the lowest Loss value in the Val dataset (Loss (Val)), which is the error rate between the results obtained from the validation data and the corresponding labeled dataset, was selected for subsequent examination of prediction using the Test dataset.
4.6. Evaluation of the Predictive Model
Through two tests conducted on the Test datasets for the experiments, with Tra:Val:Test = 4:4:1 in the DL prediction model, we analyzed the probability of the prediction results with the lowest minimum Loss (Val) value among 30 examined epochs. We calculated the probabilities for each image of one molecule captured at different angles with respect to the x-, y-, and z-axes using DeepSnap-DL. The medians of each of these predicted values were used as the representative values for target molecules as previously reported [44,45,46,47]. The performance of each model in predicting the NR agonists and antagonists was evaluated in terms of the following metrics: area under the curve of receiver operating characteristic curve (ROC_AUC); balanced accuracy (BAC); accuracy (Acc), which is the percentage of correct answers based on the results obtained from the validation dataset and the corresponding labeled dataset; F-measure; and Matthews correlation coefficient (MCC) calculated using JMP Pro 14, which is a statistical discovery software (SAS Institute Inc., Cary, NC, USA), as previously reported [44,45,46,47]. These performance metrics are defined as follows:
where TP, FN, TN, and FP denote true positive, false negative, true negative, and false positive, respectively. To determine the optimal cutoff point for the definition of TP, FN, TN, and FP, the method of maximizing sensitivity (1–specificity), which is called the Youden index [92,93], was adopted using JMP Pro software. The index has a value ranging from 0 to 1, where 1 represents maximum effectiveness and 0 represents minimum effectiveness.
BAC = (sensitivity + specificity)/2, where
Sensitivity = ΣTPs / (ΣTPs + ΣFNs),
Specificity = ΣTNs / (ΣTNs + ΣFPs),
Accuracy = (TP + TN) / (TP + FP + TN + FN),
F-measure = 2 × Recall × Precision / (Recall + Precision), where
Precision = TP / (TP + FP),
Recall = TP / (TP + FN),
4.7. Statistical Analysis
Differences in prediction performance of in vitro assays in terms of loss (Val), Acc (Val), and Acc (Test), were analyzed by Tukey–Kramer’s honestly significant difference test with JMP Pro 14 [94]. Results with p < 0.05 were considered statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/1420-3049/25/12/2764/s1, Figure S1. DeepSnap-DL procedure, Figure S2. Schematic illustrating how preparation of Tra/Val/Test datasets. Figure S3. Schematic view of GoogeLeNet archirecture. (a) Total layers used in this study. (b) Inception model within the GoogLeNet. Figure S4a. Average Accuracy values in Val and Test datasets in models of 35 NRs agonist/antagonist by the DeepSnap-DL. N = 2. Figure S4b. Average F and BAC values in Test dataset in models of 35 NRs agonist/antagonist by the DeepSnap-DL. N = 2. Figure S5. Comparison of prediction performances among four in vitro assays. (a) Loss in the Val dataset, (b) accuracy in the Val dataset, (c) accuracy in the Test dataset. Table S1: NRs and chemical compounds used in this study.
Author Contributions
Y.U. initiated and supervised the work, designed the experiments, collected the information about chemical compounds, and edited the manuscript. Y.M. performed the computer analysis and the statistical analysis, and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded in part by grants from the Ministry of Economy, Trade and Industry, AI-SHIPS (AI-based Substances Hazardous Integrated Prediction System), Japan, project (20180314ZaiSei8).
Acknowledgments
The environmental setting was supported by Shunichi Sasaki and Kota Kurosaki.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
| Acc (Test) | accuracy in the test dataset |
| AhR | aryl hydrocarbon receptor |
| AOP | adverse outcome pathway |
| AR | androgen receptor |
| AUC | area under the curve |
| Acc (Val) | accuracy in the validation dataset |
| BAC | balanced accuracy |
| BSA | bisphenol A |
| CAR | constitutive androstane receptor |
| CNN | convolutional neural network |
| DBD | DNA-binding domain |
| DIGITS | deep learning GPU training system |
| DL | deep learning |
| DMSO | dimethyl sulfoxide |
| ER | estrogen receptor |
| ERR | estrogen-related receptor |
| F | F-measure |
| FN | false negative |
| FP | false positive |
| FXR | farnesoid X receptor |
| GR | glucocorticoid receptor |
| LBD | ligand-binding domain |
| Loss (Val) | loss in the validation dataset |
| LXR | liver X receptor |
| MCC | Matthews correlation coefficient |
| ML | machine learning |
| MOE | molecular operating environment |
| NR | nuclear receptor |
| PPAR | peroxisome proliferator-activated receptor |
| PR | progesterone receptor |
| PXR | pregane X receptor |
| qHTS | quantitative high-throughput screening |
| QSAR | quantitative structure–activity relationship |
| RAR | retinoic acid receptor |
| RE | response element |
| RXR | retinoid X receptor |
| ROC | receiver operating characteristic |
| SE | standard error |
| SMILES | simplified molecular input line entry system |
| SSRM | selective steroid receptor modulator |
| TN | true negative |
| Tox21 | Toxicology in the 21st Century |
| TP | true positive |
| TR | thyroid receptor |
| TSHR | thyroid-stimulating hormone receptor |
| VDR | vitamin D receptor |
References
- Hall, J.M.; Greco, C.W. Perturbation of Nuclear Hormone Receptors by Endocrine Disrupting Chemicals: Mechanisms and Pathological Consequences of Exposure. Cells 2019, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Leso, V.; Ercolano, M.L.; Cioffi, D.L.; Iavicoli, I. Occupational Chemical Exposure and Breast Cancer Risk According to Hormone Receptor Status: A Systematic Review. Cancers (Basel) 2019, 11, 1882. [Google Scholar] [CrossRef] [PubMed]
- Tarnow, P.; Tralau, T.; Luch, A. Chemical activation of estrogen and aryl hydrocarbon receptor signaling pathways and their interaction in toxicology and metabolism. Expert Opin. Drug Metab. Toxicol. 2019, 15, 219–229. [Google Scholar] [CrossRef] [PubMed]
- McArdle, M.E.; Freeman, E.L.; Staveley, J.P.; Ortego, L.S.; Coady, K.K.; Weltje, L.; Weyers, A.; Wheeler, J.R.; Bone, A.J. Critical Review of Read-Across Potential in Testing for Endocrine-Related Effects in Vertebrate Ecological Receptors. Environ. Toxicol. Chem. 2020, 39, 739–753. [Google Scholar] [CrossRef]
- Mansouri, K.; Kleinstreuer, N.; Abdelaziz, A.M.; Alberga, D.; Alves, V.M.; Andersson, P.L.; Andrade, C.H.; Bai, F.; Balabin, I.; Ballabio, D.; et al. CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity. Environ. Health Perspect. 2020, 128, 27002. [Google Scholar] [CrossRef]
- Grimaldi, M.; Boulahtouf, A.; Delfosse, V.; Thouennon, E.; Bourguet, W.; Balaguer, P. Reporter Cell Lines for the Characterization of the Interactions between Human Nuclear Receptors and Endocrine Disruptors. Front. Endocrinol. (Lausanne) 2015, 6, 62. [Google Scholar] [CrossRef]
- Dallel, S.; Tauveron, I.; Brugnon, F.; Baron, S.; Lobaccaro, J.M.A.; Maqdasy, S. Liver X Receptors: A Possible Link between Lipid Disorders and Female Infertility. Int. J. Mol. Sci. 2018, 19, 2177. [Google Scholar] [CrossRef]
- Mazaira, G.I.; Zgajnar, N.R.; Lotufo, C.M.; Daneri-Becerra, C.; Sivils, J.C.; Soto, O.B.; Cox, M.B.; Galigniana, M.D. The Nuclear Receptor Field: A Historical Overview and Future Challenges. Nucl. Receptor Res. 2018, 5, 101320. [Google Scholar] [CrossRef]
- Watanabe, M.; Kakuta, H. Retinoid X Receptor Antagonists. Int. J. Mol. Sci. 2018, 19, 2354. [Google Scholar] [CrossRef]
- Jackson, E.N.; Thatcher, S.E.; Larian, N.; English, V.; Soman, S.; Morris, A.J.; Weng, J.; Stromberg, A.; Swanson, H.I.; Pearson, K.; et al. Effects of Aryl Hydrocarbon Receptor Deficiency on PCB-77-Induced Impairment of Glucose Homeostasis during Weight Loss in Male and Female Obese Mice. Environ. Health Perspect. 2019, 127, 77004. [Google Scholar] [CrossRef]
- Meijer, F.A.; Leijten-van de Gevel, I.A.; de Vries, R.M.J.M.; Brunsveld, L. Allosteric small molecule modulators of nuclear receptors. Mol. Cell Endocrinol. 2019, 485, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Makar, S.; Swetha, R.; Gutti, G.; Singh, S.K. Estrogen signaling: An emanating therapeutic target for breast cancer treatment. Eur. J. Med. Chem. 2019, 177, 116–143. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Smieško, M. Ligand Pathways in Nuclear Receptors. J. Chem. Inf. Model. 2019, 59, 3100–3109. [Google Scholar] [CrossRef]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef] [PubMed]
- Veras Ribeiro Filho, H.; Tambones, I.L.; Mariano Gonçalves Dias, M.; Bernardi Videira, N.; Bruder, M.; Amorim Amato, A.; Migliorini Figueira, A.C. Modulation of nuclear receptor function: Targeting the protein-DNA interface. Mol. Cell Endocrinol. 2019, 484, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Tecalco-Cruz, A.C. Molecular pathways involved in the transport of nuclear receptors from the nucleus to cytoplasm. J. Steroid Biochem. Mol. Biol. 2018, 178, 36–44. [Google Scholar] [CrossRef]
- Baker, J.D.; Ozsan, I.; Rodriguez Ospina, S.; Gulick, D.; Blair, L.J. Hsp90 Heterocomplexes Regulate Steroid Hormone Receptors: From Stress Response to Psychiatric Disease. Int. J. Mol. Sci. 2018, 20, 79. [Google Scholar] [CrossRef]
- Gabler, M.; Kramer, J.; Schmidt, J.; Pollinger, J.; Weber, J.; Kaiser, A.; Löhr, F.; Proschak, E.; Schubert-Zsilavecz, M.; Merk, D. Allosteric modulation of the farnesoid X receptor by a small molecule. Sci. Rep. 2018, 8, 6846. [Google Scholar] [CrossRef]
- Azhagiya Singam, E.R.; Tachachartvanich, P.; La Merrill, M.A.; Smith, M.T.; Durkin, K.A. Structural Dynamics of Agonist and Antagonist Binding to the Androgen Receptor. J. Phys. Chem. B 2019, 123, 7657–7666. [Google Scholar] [CrossRef]
- D’Aniello, E.; Iannotti, F.A.; Falkenberg, L.G.; Martella, A.; Gentile, A.; De Maio, F.; Ciavatta, M.L.; Gavagnin, M.; Waxman, J.S.; Di Marzo, V.; et al. In Silico Identification and Experimental Validation of (-)-Muqubilin A, a Marine Norterpene Peroxide, as PPARα/γ-RXRα Agonist and RARα Positive Allosteric Modulator. Mar. Drugs 2019, 17, 110. [Google Scholar] [CrossRef]
- Fay, K.A.; Villeneuve, D.L.; Swintek, J.; Edwards, S.W.; Nelms, M.D.; Blackwell, B.R.; Ankley, G.T. Differentiating Pathway-Specific From Nonspecific Effects in High-Throughput Toxicity Data: A Foundation for Prioritizing Adverse Outcome Pathway Development. Toxicol. Sci. 2018, 163, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Clippinger, A.J.; Allen, D.; Behrsing, H.; BéruBé, K.A.; Bolger, M.B.; Casey, W.; DeLorme, M.; Gaça, M.; Gehen, S.C.; Glover, K.; et al. Pathway-based predictive approaches for non-animal assessment of acute inhalation toxicity. Toxicol. In Vitr. 2018, 52, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Dal Negro, G.; Eskes, C.; Belz, S.; Bertein, C.; Chlebus, M.; Corvaro, M.; Corvi, R.; Dhalluin, S.; Halder, M.; Harvey, J.; et al. One science-driven approach for the regulatory implementation of alternative methods: A multi-sector perspective. Regul. Toxicol. Pharmacol. 2018, 99, 33–49. [Google Scholar] [CrossRef]
- Sewell, F.; Gellatly, N.; Beaumont, M.; Burden, N.; Currie, R.; de Haan, L.; Hutchinson, T.H.; Jacobs, M.; Mahony, C.; Malcomber, I.; et al. The future trajectory of adverse outcome pathways: A commentary. Arch. Toxicol. 2018, 92, 1657–1661. [Google Scholar] [CrossRef]
- Terron, A.; Bennekou, S.H. Towards a regulatory use of alternative developmental neurotoxicity testing (DNT). Toxicol. Appl. Pharmacol. 2018, 354, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Prior, H.; Casey, W.; Kimber, I.; Whelan, M.; Sewell, F. Reflections on the progress towards non-animal methods for acute toxicity testing of chemicals. Regul. Toxicol. Pharmacol. 2019, 102, 30–33. [Google Scholar] [CrossRef]
- Thomas, R.S.; Bahadori, T.; Buckley, T.J.; Cowden, J.; Deisenroth, C.; Dionisio, K.L.; Frithsen, J.B.; Grulke, C.M.; Gwinn, M.R.; Harrill, J.A.; et al. The Next Generation Blueprint of Computational Toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 2019, 169, 317–332. [Google Scholar] [CrossRef]
- Kavlock, R.; Chandler, K.; Houck, K.; Hunter, S.; Judson, R.; Kleinstreuer, N.; Knudsen, T.; Martin, M.; Padilla, S.; Reif, D.; et al. Update on EPA’s ToxCast program: Providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 2012, 25, 1287–1302. [Google Scholar] [CrossRef]
- Judson, R.; Houck, K.; Martin, M.; Knudsen, T.; Thomas, R.S.; Sipes, N.; Shah, I.; Wambaugh, J.; Crofton, K. In vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast programme. Basic Clin. Pharmacol. Toxicol. 2014, 115, 69–76. [Google Scholar] [CrossRef]
- Kleinstreuer, N.C.; Yang, J.; Berg, E.L.; Knudsen, T.B.; Richard, A.M.; Martin, M.T.; Reif, D.M.; Judson, R.S.; Polokoff, M.; Dix, D.J.; et al. Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol. 2014, 32, 583–591. [Google Scholar] [CrossRef]
- Tice, R.R.; Austin, C.P.; Kavlock, R.J.; Bucher, J.R. Improving the human hazard characterization of chemicals: A Tox21 update. Environ. Health Perspect. 2013, 121, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.S.; Magpantay, F.M.; Chickarmane, V.; Haskell, C.; Tania, N.; Taylor, J.; Xia, M.; Huang, R.; Rotroff, D.M.; Filer, D.L.; et al. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci. 2015, 148, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Fourches, D.; Ash, J. 4D-quantitative structure-activity relationship modeling: Making a comeback. Expert Opin. Drug Discov. 2019, 14, 1227–1235. [Google Scholar] [CrossRef]
- Hisaki, T.; Kaneko, M.A.N.; Hirota, M.; Matsuoka, M.; Kouzuki, H. Integration of read-across and artificial neural network-based QSAR models for predicting systemic toxicity: A case study for valproic acid. J. Toxicol. Sci. 2020, 45, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kleinstreuer, N.C.; Fourches, D. Hierarchical Quantitative Structure-Activity Relationship Modeling Approach for Integrating Binary, Multiclass, and Regression Models of Acute Oral Systemic Toxicity. Chem. Res. Toxicol. 2020, 33, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, I.L.; Gómez-Nieto, M.Á. Building Highly Reliable Quantitative Structure-Activity Relationship Classification Models Using the Rivality Index Neighborhood Algorithm with Feature Selection. J. Chem. Inf. Model. 2020, 60, 133–151. [Google Scholar] [CrossRef]
- Santos, K.L.B.D.; Cruz, J.N.; Silva, L.B.; Ramos, R.S.; Neto, M.F.A.; Lobato, C.C.; Ota, S.S.B.; Leite, F.H.A.; Borges, R.S.; Silva, C.H.T.P.D.; et al. Identification of Novel Chemical Entities for Adenosine Receptor Type 2A Using Molecular Modeling Approaches. Molecules 2020, 25, 1245. [Google Scholar] [CrossRef]
- Danishuddin Khan, A.U. Descriptors and their selection methods in QSAR analysis: Paradigm for drug design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar] [CrossRef]
- Dutt, R.; Madan, A.K. Development and application of novel molecular descriptors for predicting biological activity. Med. Chem. Res. 2017, 26, 1988–2006. [Google Scholar] [CrossRef]
- Idakwo, G.; Thangapandian, S.; Luttrell, J., 4th; Zhou, Z.; Zhang, C.; Gong, P. Deep Learning-Based Structure-Activity Relationship Modeling for Multi-Category Toxicity Classification: A Case Study of 10K Tox21 Chemicals With High-Throughput Cell-Based Androgen Receptor Bioassay Data. Front. Physiol. 2019, 10, 1044. [Google Scholar] [CrossRef]
- Uesawa, Y. Quantitative structure-activity relationship analysis using deep learning based on a novel molecular image input technique. Bioorg. Med. Chem. Lett. 2018, 28, 3400–3403. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Hochreiter, S. DeepTox: Toxicity Prediction using Deep Learning. Front. Environ. Sci. 2016, 3, 80. [Google Scholar] [CrossRef]
- Wu, K.; Wei, G.W. Quantitative Toxicity Prediction Using Topology Based Multitask Deep Neural Networks. J. Chem. Inf. Model. 2018, 58, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, Y.; Uesawa, Y. Optimization of a Deep-Learning Method Based on the Classification of Images Generated by Parameterized Deep Snap a Novel Molecular-Image-Input Technique for Quantitative Structure-Activity Relationship (QSAR) Analysis. Front. Bioeng. Biotechnol. 2019, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, Y.; Uesawa, Y. Prediction Model with High-Performance Constitutive Androstane Receptor (CAR) Using DeepSnap-Deep Learning Approach from the Tox21 10K Compound Library. Int. J. Mol. Sci. 2019, 20, 4855. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, Y.; Uesawa, Y. DeepSnap-Deep Learning Approach Predicts Progesterone Receptor Antagonist Activity with High Performance. Front. Bioeng. Biotechnol. 2020, 7, 485. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Hosaka, T.; Ogaito, A.; Yoshinari, K.; Uesawa, Y. Prediction Model of Aryl Hydrocarbon Receptor Activation by a Novel QSAR Approach, DeepSnap-Deep Learning. Deep Learning. Molecules 2020, 25, 1317. [Google Scholar] [CrossRef]
- Available online: https://tripod.nih.gov/tox21/challenge/index.jsp (accessed on 12 June 2020).
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Steijaert, M.; Wegner, J.K.; Ceulemans, H.; Clevert, D.A.; Hochreiter, S. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Sci. 2018, 9, 5441–5451. [Google Scholar] [CrossRef]
- Xu, Y.; Pei, J.; Lai, L. Deep Learning Based Regression and Multiclass Models for Acute Oral Toxicity Prediction with Automatic Chemical Feature Extraction. J. Chem. Inf. Model. 2017, 57, 2672–2685. [Google Scholar] [CrossRef]
- Ambe, K.; Ishihara, K.; Ochibe, T.; Ohya, K.; Tamura, S.; Inoue, K.; Yoshida, M.; Tohkin, M. In Silico Prediction of Chemical-Induced Hepatocellular Hypertrophy Using Molecular Descriptors. Toxicol. Sci. 2018, 162, 667–675. [Google Scholar] [CrossRef]
- Fernandez, M.; Ban, F.; Woo, G.; Hsing, M.; Yamazaki, T.; LeBlanc, E.; Rennie, P.S.; Welch, W.J.; Cherkasov, A. Toxic Colors: The Use of Deep Learning for Predicting Toxicity of Compounds Merely from Their Graphic Images. J. Chem. Inf. Model. 2018, 58, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Madore, M.; Glover, K.P.; Feasel, M.G.; Wallqvist, A. Assessing Deep and Shallow Learning Methods for Quantitative Prediction of Acute Chemical Toxicity. Toxicol. Sci. 2018, 164, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Terry, J.G.; Wang, J.; Nath, V.; Bermudez, C.; Bao, S.; Parvathaneni, P.; Carr, J.J.; Landman, B.A. Coronary Calcium Detection using 3D Attention Identical Dual Deep Network Based on Weakly Supervised Learning. Proc. SPIE Int. Soc. Opt. Eng. 2019, 10949, 1094917. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, J.; Gilmer, J.; Muelly, M.; Goodfellow, I.; Hardt, M.; Kim, B. Sanity Checks for Saliency Maps. arXiv 2018, arXiv:1810.03292v2. Available online: https://arxiv.org/abs/1810.03292 (accessed on 12 June 2020).
- Fiosina, J.; Fiosins, M.; Bonn, S. Explainable Deep Learning for Augmentation of Small RNA Expression Profiles. J. Comput. Biol. 2019, 27, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Böhle, M.; Eitel, F.; Weygandt, M.; Ritter, K. Layer-Wise Relevance Propagation for Explaining Deep Neural Network Decisions in MRI-Based Alzheimer’s Disease Classification. Front. Aging Neurosci. 2019, 11, 194. [Google Scholar] [CrossRef] [PubMed]
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. SmoothGrad: Removing noise by adding noise. arXiv 2017, arXiv:1706.03825v1. Available online: https://arxiv.org/abs/1706.03825 (accessed on 12 June 2020).
- Goh, G.S.W.; Lapuschkin, S.; Weber, L.; Samek, W.; Binder, A. Understanding Integrated Gradients with SmoothTaylor for Deep Neural Network Attribution. arXiv 2020, arXiv:2004.10484v1. Available online: https://arxiv.org/abs/2004.10484 (accessed on 12 June 2020).
- Paul-Friedman, K.; Martin, M.; Crofton, K.M.; Hsu, C.W.; Sakamuru, S.; Zhao, J.; Xia, M.; Huang, R.; Stavreva, D.A.; Soni, V.; et al. Limited Chemical Structural Diversity Found to Modulate Thyroid Hormone Receptor in the Tox21 Chemical Library. Environ. Health Perspect. 2019, 127, 97009. [Google Scholar] [CrossRef]
- Zhang, J.; Li, T.; Wang, T.; Yuan, C.; Zhong, S.; Guan, T.; Li, Z.; Wang, Y.; Yu, H.; Luo, Q.; et al. Estrogenicity of halogenated bisphenol A: In vitro and in silico investigations. Arch. Toxicol. 2018, 92, 1215–1223. [Google Scholar] [CrossRef]
- Smirnova, O.V. Competitive Agonists and Antagonists of Steroid Nuclear Receptors: Evolution of the Concept or Its Reversal. Biochemistry (Moscow) 2015, 80, 1227–1234. [Google Scholar] [CrossRef]
- Liu, S.; Han, S.J.; Smith, C.L. Cooperative activation of gene expression by agonists and antagonists mediated by estrogen receptor heteroligand dimer complexes. Mol. Pharmacol. 2013, 83, 1066–1077. [Google Scholar] [CrossRef]
- Dotzlaw, H.; Papaioannou, M.; Moehren, U.; Claessens, F.; Baniahmad, A. Agonist-antagonist induced coactivator and corepressor interplay on the human androgen receptor. Mol. Cell Endocrinol. 2003, 213, 79–85. [Google Scholar] [CrossRef]
- Arnal, J.F.; Lenfant, F.; Metivier, R.; Flouriot, G.; Henrion, D.; Adlanmerini, M.; Fontaine, C.; Gourdy, P.; Chambon, P.; Katzenellenbogen, B.; et al. Membrane and Nuclear Estrogen Receptor Alpha Actions: From Tissue Specificity to Medical Implications. Physiol. Rev. 2017, 97, 1045–1087. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, K.L.; Farman, H.; Henning, P.; Lionikaite, V.; Movérare-Skrtic, S.; Wu, J.; Ryberg, H.; Koskela, A.; Gustafsson, J.Å.; Tuukkanen, J.; et al. The role of membrane ERα signaling in bone and other major estrogen responsive tissues. Sci. Rep. 2016, 6, 29473. [Google Scholar] [CrossRef] [PubMed]
- Furuya, K.; Yamamoto, N.; Ohyabu, Y.; Morikyu, T.; Ishige, H.; Albers, M.; Endo, Y. Mechanism of the tissue-specific action of the selective androgen receptor modulator S-101479. Biol. Pharm. Bull. 2013, 36, 442–451. [Google Scholar] [CrossRef]
- Arao, Y.; Hamilton, K.J.; Ray, M.K.; Scott, G.; Mishina, Y.; Korach, K.S. Estrogen receptor α AF-2 mutation results in antagonist reversal and reveals tissue selective function of estrogen receptor modulators. Proc. Natl. Acad. Sci. USA 2011, 108, 14986–14991. [Google Scholar] [CrossRef] [PubMed]
- Spillman, M.A.; Manning, N.G.; Dye, W.W.; Sartorius, C.A.; Post, M.D.; Harrell, J.C.; Jacobsen, B.M.; Horwitz, K.B. Tissue-specific pathways for estrogen regulation of ovarian cancer growth and metastasis. Cancer Res. 2010, 70, 8927–8936. [Google Scholar] [CrossRef] [PubMed]
- Schoch, G.A.; D’Arcy, B.; Stihle, M.; Burger, D.; Bär, D.; Benz, J.; Thoma, R.; Ruf, A. Molecular switch in the glucocorticoid receptor: Active and passive antagonist conformations. J. Mol. Biol. 2010, 395, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Titus, S.; Neumann, S.; Zheng, W.; Southall, N.; Michael, S.; Klumpp, C.; Yasgar, A.; Shinn, P.; Thomas, C.J.; Inglese, J.; et al. Quantitative high-throughput screening using a live-cell cAMP assay identifies small-molecule agonists of the TSH receptor. J. Biomol. Screen 2008, 13, 120–127. [Google Scholar] [CrossRef]
- Huang, R.; Sakamuru, S.; Martin, M.T.; Reif, D.M.; Judson, R.S.; Houck, K.A.; Casey, W.; Hsieh, J.H.; Shockley, K.R.; Ceger, P.; et al. Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci. Rep. 2014, 4, 5664. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Cho, M.H.; Sakamuru, S.; Shinn, P.; Houck, K.A.; Dix, D.J.; Judson, R.S.; Witt, K.L.; Kavlock, R.J.; et al. Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ. Health Perspect. 2011, 119, 1142–1148. [Google Scholar] [CrossRef]
- Chen, Y.; Sakamuru, S.; Huang, R.; Reese, D.H.; Xia, M. Identification of compounds that modulate retinol signaling using a cell-based qHTS assay. Toxicol. In Vitro 2016, 32, 287–296. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Sakamuru, S.; Zhao, J.; Shahane, S.A.; Attene-Ramos, M.; Zhao, T.; Austin, C.P.; Simeonov, A. Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat. Commun. 2016, 7, 10425. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.; Zhao, J.; Wang, H.; Xia, M. Quantitative High-Throughput Luciferase Screening in Identifying CAR Modulators. Methods Mol. Biol. 2016, 1473, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.T.; Hsieh, J.H.; Zhao, J.; Huang, R.; Xia, M.; Martin, N.; Gao, X.; Dixon, D.; Auerbach, S.S.; Witt, K.L.; et al. Development of Novel Cell Lines for High-Throughput Screening to Detect Estrogen-Related Receptor Alpha Modulators. SLAS Discov. 2017, 22, 720–731. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Sakamuru, S.; Zhao, J.; Lynch, C.; Zhao, T.; Zhu, H.; Austin, C.P.; Simeonov, A. Expanding biological space coverage enhances the prediction of drug adverse effects in human using in vitro activity profiles. Sci. Rep. 2018, 8, 3783. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.; Zhao, J.; Huang, R.; Kanaya, N.; Bernal, L.; Hsieh, J.H.; Auerbach, S.S.; Witt, K.L.; Merrick, B.A.; Chen, S.; et al. Identification of Estrogen-Related Receptor α Agonists in the Tox21 Compound Library. Endocrinology 2018, 159, 744–753. [Google Scholar] [CrossRef]
- Lynch, C.; Zhao, J.; Sakamuru, S.; Zhang, L.; Huang, R.; Witt, K.L.; Merrick, B.A.; Teng, C.T.; Xia, M. Identification of Compounds That Inhibit Estrogen-Related Receptor Alpha Signaling Using High-Throughput Screening Assays. Molecules 2019, 24, 841. [Google Scholar] [CrossRef]
- Lynch, C.; Mackowiak, B.; Huang, R.; Li, L.; Heyward, S.; Sakamuru, S.; Wang, H.; Xia, M. Identification of Modulators That Activate the Constitutive Androstane Receptor From the Tox21 10K Compound Library. Toxicol. Sci. 2019, 167, 282–292. [Google Scholar] [CrossRef]
- Wei, Z.; Sakamuru, S.; Zhang, L.; Zhao, J.; Huang, R.; Kleinstreuer, N.C.; Chen, Y.; Shu, Y.; Knudsen, T.B.; Xia, M. Identification and Profiling of Environmental Chemicals That Inhibit the TGFβ/SMAD Signaling Pathway. Chem. Res. Toxicol. 2019, 32, 2433–2444. [Google Scholar] [CrossRef]
- Xia, M.; Huang, R.; Guo, V.; Southall, N.; Cho, M.H.; Inglese, J.; Austin, C.P.; Nirenberg, M. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc. Natl. Acad. Sci. USA 2009, 106, 2412–2417. [Google Scholar] [CrossRef]
- Inglese, J.; Auld, D.S.; Jadhav, A.; Johnson, R.L.; Simeonov, A.; Yasgar, A.; Zheng, W.; Austin, C.P. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. USA 2006, 103, 11473–11478. [Google Scholar] [CrossRef]
- Wang, Y.; Jadhav, A.; Southal, N.; Huang, R.; Nguyen, D.T. A grid algorithm for high throughput fitting of dose-response curve data. Curr. Chem. Genomics 2010, 4, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Molecular Networks GmbH, Nürnberg, Germany. Available online: https://www.mn-am.com/products/corina (accessed on 12 June 2020).
- Available online: http://image-net.org/challenges/LSVRC/2012/browse-synsets (accessed on 12 June 2020).
- Available online: http://www.image-net.org/index (accessed on 12 June 2020).
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, Y.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. arXiv 2014, arXiv:1409.4842v1. [Google Scholar]
- Yang, Y.; Yan, L.F.; Zhang, X.; Han, Y.; Nan, H.Y.; Hu, Y.C.; Hu, B.; Yan, S.L.; Zhang, J.; Cheng, D.L.; et al. Glioma Grading on Conventional MR Images: A Deep Learning Study With Transfer Learning. Front. Neurosci. 2018, 12, 804. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, H.E.; Choi, Y.H.; Lee, S.J.; Jeon, J.S. CNN-based diagnosis models for canine ulcerative keratitis. Sci. Rep. 2019, 9, 14209. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.H.; Chun, S.M.; Kim, J.C.; Shin, H.I. Obesity cutoff values in Korean men with motor complete spinal cord injury: Body mass index and waist circumference. Spinal Cord 2019, 57, 110–116. [Google Scholar] [CrossRef]
- Liang, K.; Wang, C.; Yan, F.; Wang, L.; He, T.; Zhang, X.; Li, C.; Yang, W.; Ma, Z.; Ma, A.; et al. HbA1c Cutoff Point of 5.9% Better Identifies High Risk of Progression to Diabetes among Chinese Adults: Results from a Retrospective Cohort Study. J. Diabetes Res. 2018, 2018, 7486493. [Google Scholar] [CrossRef]
- Available online: https://www.jmp.com/support/help/en/15.1/index.shtml#page/jmp/example-of-all-pairs-tukey-hsd-test.shtml (accessed on 12 June 2020).
Sample Availability: Samples of the compounds are available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).