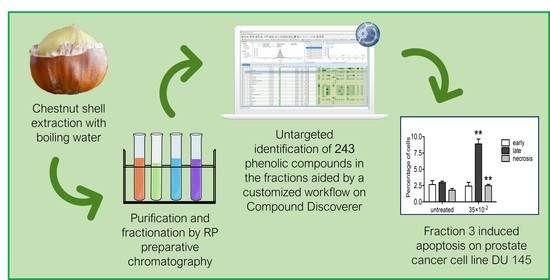
Untargeted Characterization of Chestnut (Castanea sativa Mill.) Shell Polyphenol Extract: A Valued Bioresource for Prostate Cancer Cell Growth Inhibition
Abstract
1. Introduction
2. Result and Discussion
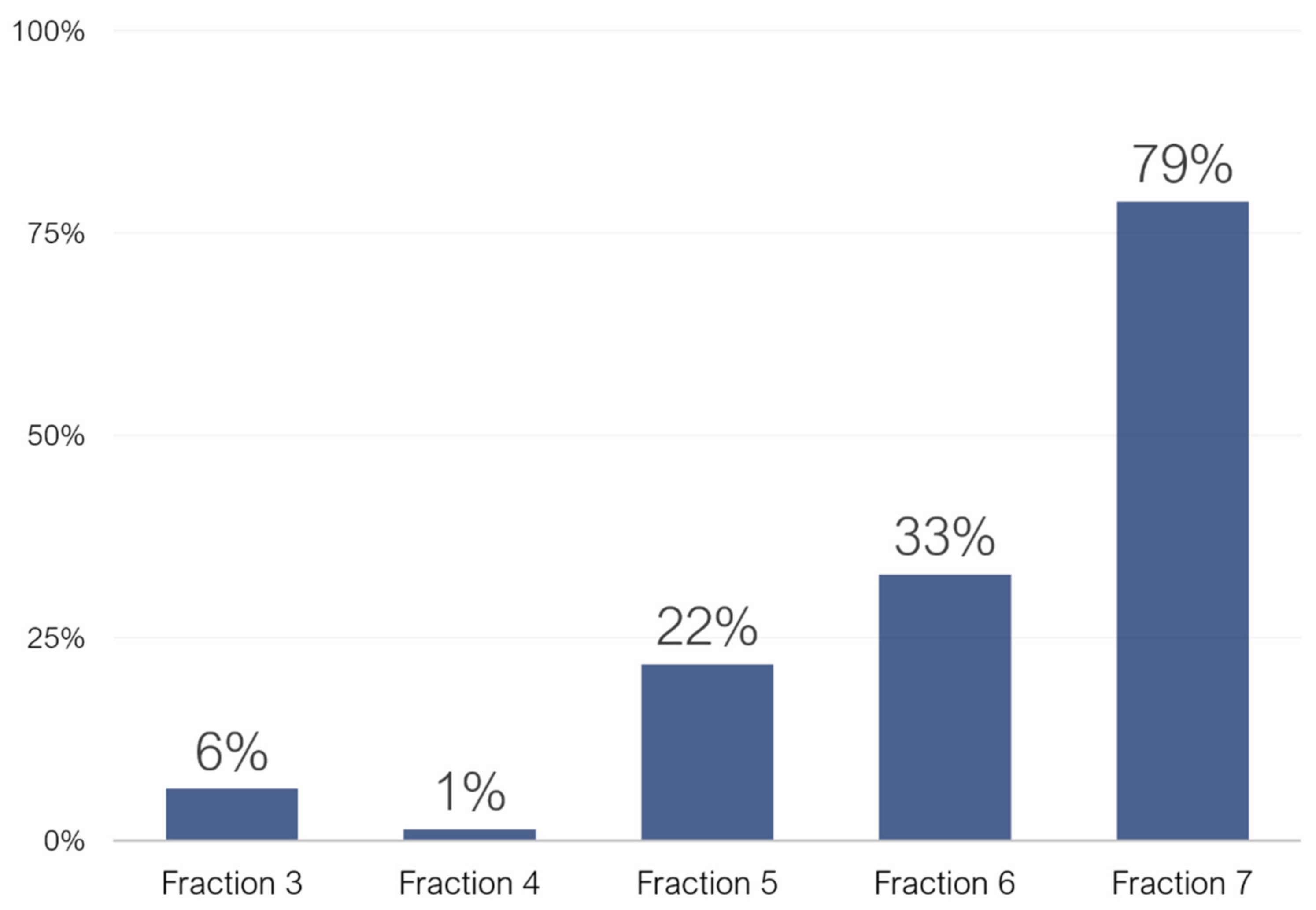
2.1. Untargeted Characterization of Chestnut Shell Extract Fractions
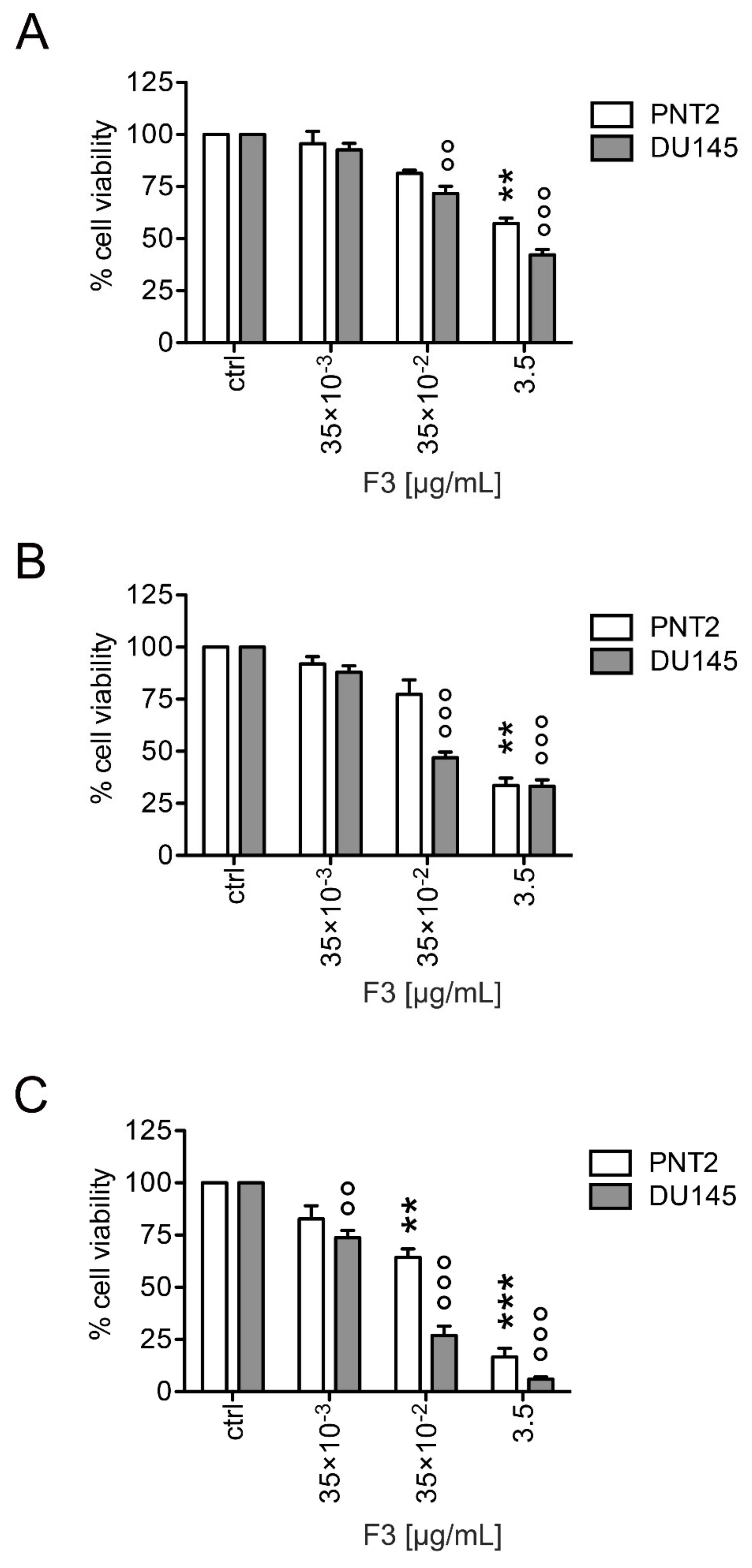
2.2. Fraction 3 Treatment Decreased Cell Viability in a Time- and Dose-Dependent Manner
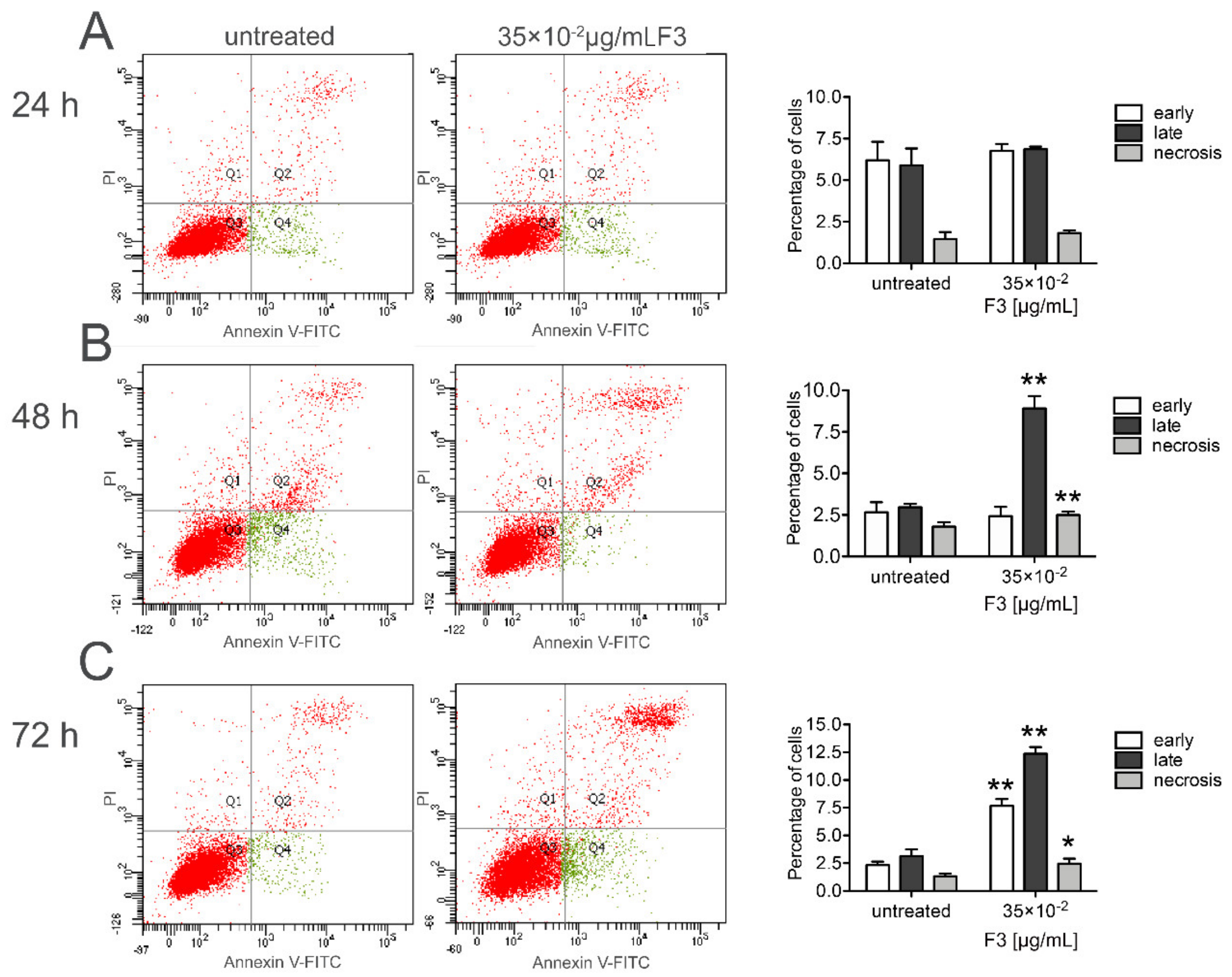
2.3. Fraction 3 Treatment-Induced Cell Death in DU 145 Cells Occurs Through Apoptosis
2.4. Fraction 3 Treatment Induced a Perturbation of DU145 Cell Cycle
3. Material and Methods
3.1. Chemicals and Reagents
3.2. Extract Preparation from Chestnut Shells
3.3. Total Phenolic Content
3.4. Purification of Phenolic Compounds by HPLC
3.5. UHPLC-MS/MS Analysis
3.6. Data Analysis and Phenolic Compound Validation
3.7. Cell Cultures
3.8. Assessment of Cell Viability by Trypan Blue Assay
3.9. Evaluation of Apoptosis by Flow Cytometry (FCM)
3.10. Cell Cycle Analysis by FCM
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Squillaci, G.; Apone, F.; Sena, L.M.; Carola, A.; Tito, A.; Bimonte, M.; De Lucia, A.; Colucci, G.; La Cara, F.; Morana, A. Chestnut (Castanea sativa Mill.) industrial wastes as a valued bioresource for the production of active ingredients. Process. Biochem. 2018, 64, 228–236. [Google Scholar] [CrossRef]
- Pinto, D.; Braga, N.; Silva, A.M.; Costa, P.; Delerue-Matos, C.; Rodrigues, F. Chestnut. In Valorization of Fruit Processing By-Products, 1st ed.; Galanakis, C.M., Ed.; Academic Press: Chania, Greece, 2019; Volume 1, pp. 127–144. [Google Scholar]
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Montone, C.M.; Chiozzi, R.Z.; Lagana, A. Recent trends and analytical challenges in plant bioactive peptide separation, identification and validation. Anal. Bioanal. Chem. 2018, 410, 3425–3444. [Google Scholar] [CrossRef] [PubMed]
- Liberti, A.; Goretti, G.; Russo, M.V. PCDD and PCDF formation in the combustion of vegetable wastes. Chemosphere 1983, 12, 661–663. [Google Scholar] [CrossRef]
- Vazquez, G.; Freire, M.S.; Santos, J.; Antorrena, G.; González-Álvarez, J. Optimisation of Polyphenols Extraction from Chestnut Shell by Response Surface Methodology. Waste Biomass Valoriz. 2010, 1, 219–225. [Google Scholar] [CrossRef]
- Braga, N.; Rodrigues, F.; Oliveira, M.B.P.P. Castanea sativa by-products: A review on added value and sustainable application. Nat. Prod. Res. 2014, 29, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, G.; González-Álvarez, J.; Santos, J.; Freire, M.S.; Antorrena, G. Evaluation of potential applications for chestnut (Castanea sativa) shell and eucalyptus (Eucalyptus globulus) bark extracts. Ind. Crop. Prod. 2009, 29, 364–370. [Google Scholar] [CrossRef]
- Comandini, P.; Lerma-García, M.J.; Simó-Alfonso, E.F.; Toschi, T.G. Tannin analysis of chestnut bark samples (Castanea sativa Mill.) by HPLC-DAD–MS. Food Chem. 2014, 157, 290–295. [Google Scholar] [CrossRef]
- Vella, F.M.; Laratta, B.; La Cara, F.; Morana, A. Recovery of bioactive molecules from chestnut (Castanea sativa Mill.) by-products through extraction by different solvents. Nat. Prod. Res. 2017, 32, 1022–1032. [Google Scholar] [CrossRef]
- Youn, U.-Y.; Shon, M.-S.; Kim, G.-N.; Katagiri, R.; Harata, K.; Ishida, Y.; Lee, S.-C. Antioxidant and anti-adipogenic activities of chestnut (Castanea crenata) byproducts. Food Sci. Biotechnol. 2016, 25, 1169–1174. [Google Scholar] [CrossRef]
- Echegaray, N.; Gómez, B.; Barba, F.J.; Franco, D.; Estévez, M.; Carballo, J.; Marszałek, K.; Lorenzo, J.M. Chestnuts and by-products as source of natural antioxidants in meat and meat products: A review. Trends Food Sci. Technol. 2018, 82, 110–121. [Google Scholar] [CrossRef]
- Barreira, J.C.; Ferreira, I.C.F.R.; Oliveira, M.B.P.P.; Pereira, J.A. Antioxidant Potential of Chestnut (Castanea sativa L.) and Almond (Prunus dulcis L.) By-products. Food Sci. Technol. Int. 2010, 16, 209–216. [Google Scholar] [CrossRef]
- Barreira, J.C.; Ferreira, I.C.F.R.; Oliveira, M.; Pereira, J.A.; Oliveira, M.B.P.P. Antioxidant activities of the extracts from chestnut flower, leaf, skins and fruit. Food Chem. 2008, 107, 1106–1113. [Google Scholar] [CrossRef]
- Lee, J.H.; Khor, T.O.; Shu, L.; Su, Z.-Y.; Fuentes, F.; Kong, A.-N. Dietary phytochemicals and cancer prevention: Nrf2 signaling, epigenetics, and cell death mechanisms in blocking cancer initiation and progression. Pharmacol. Ther. 2012, 137, 153–171. [Google Scholar] [CrossRef]
- Dashwood, R.H. Frontiers in Polyphenols and Cancer Prevention. J. Nutr. 2007, 137, 267S–269S. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, H.; Farzaei, M.H.; Khodarahmi, R.; Farzei, M.H. Polyphenols and their benefits: A review. Int. J. Food Prop. 2017, 1–42. [Google Scholar] [CrossRef]
- Vauzour, D.; Rodriguez-Mateos, A.; Corona, G.; Concha, M.J.O.; Spencer, J.P. Polyphenols and Human Health: Prevention of Disease and Mechanisms of Action. Nutrients 2010, 2, 1106–1131. [Google Scholar] [CrossRef] [PubMed]
- Sorice, A.; Siano, F.; Capone, F.; Guerriero, E.; Picariello, G.; Budillon, A.; Ciliberto, G.; Paolucci, M.; Costantini, S.; Volpe, M.G. Potential Anticancer Effects of Polyphenols from Chestnut Shell Extracts: Modulation of Cell Growth, and Cytokinomic and Metabolomic Profiles. Molecules 2016, 21, 1411. [Google Scholar] [CrossRef]
- Prochazkova, K.; Boušová, I.; Wilhelmova, N. Antioxidant and prooxidant properties of flavonoids. Fitoterapia 2011, 82, 513–523. [Google Scholar] [CrossRef]
- Sroka, Z. Antioxidative and Antiradical Properties of Plant Phenolics. Zeitschrift für Naturforschung C 2005, 60, 833–843. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazúr, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Boil. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Cerrato, A.; Cannazza, G.; Capriotti, A.L.; Citti, C.; La Barbera, G.; Laganà, A.; Montone, C.M.; Piovesana, S.; Cavaliere, C. A new software-assisted analytical workflow based on high-resolution mass spectrometry for the systematic study of phenolic compounds in complex matrices. Talanta 2020, 209, 120573. [Google Scholar] [CrossRef] [PubMed]
- Cacciola, N.A.; Squillaci, G.; D’Apolito, M.; Petillo, O.; Veraldi, F.; LaCara, F.; Peluso, G.; Margarucci, S.; Morana, A. Castanea sativa Mill. Shells Aqueous Extract Exhibits Anticancer Properties Inducing Cytotoxic and Pro-Apoptotic Effects. Molecules 2019, 24, 3401. [Google Scholar] [CrossRef] [PubMed]
- Veeresham, C. Natural products derived from plants as a source of drugs. J. Adv. Pharm. Technol. Res. 2012, 3, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef] [PubMed]
- Gironi, F.; Piemonte, V. Temperature and solvent effects on polyphenol extraction process from chestnut tree wood. Chem. Eng. Res. Des. 2011, 89, 857–862. [Google Scholar] [CrossRef]
- Fernández-Agulló, A.; Freire, M.S.; Antorrena, G.; Pereira, J.A.; González-Álvarez, J. Effect of the Extraction Technique and Operational Conditions on the Recovery of Bioactive Compounds from Chestnut (Castanea sativa) Bur and Shell. Sep. Sci. Technol. 2014, 49, 267–277. [Google Scholar] [CrossRef]
- Kuo, P.; Hsu, Y.; Lin, T.; Lin, C. The antiproliferative activity of prodelphinidin B-2 3′-O-gallate from green tea leaf is through cell cycle arrest and Fas-mediated apoptotic pathway in A549 cells. Food Chem. Toxicol. 2005, 43, 315–323. [Google Scholar] [CrossRef]
- Santulli, C.; Brizi, C.; Durante, M.; Micucci, M.; Budriesi, R.; Chiarini, A.; Frosini, M. Apoptotic-induced Effects of Castanea sativa Bark Extract in Human SH-SY5Y Neuroblastoma Cells. Nat. Prod. Commun. 2018, 13. [Google Scholar] [CrossRef]
- Elmore, S.A. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, C.; Singh, R.P.; Agarwal, R. Grape seed extract induces apoptotic death of human prostate carcinoma DU145 cells via caspases activation accompanied by dissipation of mitochondrial membrane potential and cytochrome c release. Carcinogenesis 2002, 23, 1869–1876. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- La Barbera, G.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Samperi, R.; Chiozzi, R.Z.; Lagana, A. Comprehensive polyphenol profiling of a strawberry extract (Fragaria × ananassa) by ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry. Anal. Bioanal. Chem. 2017, 409, 2127–2142. [Google Scholar] [CrossRef] [PubMed]
- Cavaliere, C.; Antonelli, M.; Capriotti, A.L.; La Barbera, G.; Montone, C.M.; Piovesana, S.; Laganà, A. A Triple Quadrupole and a Hybrid Quadrupole Orbitrap Mass Spectrometer in Comparison for Polyphenol Quantitation. J. Agric. Food Chem. 2019, 67, 4885–4896. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cacciola, N.A.; Cerrato, A.; Capriotti, A.L.; Cavaliere, C.; D’Apolito, M.; Montone, C.M.; Piovesana, S.; Squillaci, G.; Peluso, G.; Laganà, A. Untargeted Characterization of Chestnut (Castanea sativa Mill.) Shell Polyphenol Extract: A Valued Bioresource for Prostate Cancer Cell Growth Inhibition. Molecules 2020, 25, 2730. https://doi.org/10.3390/molecules25122730
Cacciola NA, Cerrato A, Capriotti AL, Cavaliere C, D’Apolito M, Montone CM, Piovesana S, Squillaci G, Peluso G, Laganà A. Untargeted Characterization of Chestnut (Castanea sativa Mill.) Shell Polyphenol Extract: A Valued Bioresource for Prostate Cancer Cell Growth Inhibition. Molecules. 2020; 25(12):2730. https://doi.org/10.3390/molecules25122730
Chicago/Turabian StyleCacciola, Nunzio Antonio, Andrea Cerrato, Anna Laura Capriotti, Chiara Cavaliere, Maria D’Apolito, Carmela Maria Montone, Susy Piovesana, Giuseppe Squillaci, Gianfranco Peluso, and Aldo Laganà. 2020. "Untargeted Characterization of Chestnut (Castanea sativa Mill.) Shell Polyphenol Extract: A Valued Bioresource for Prostate Cancer Cell Growth Inhibition" Molecules 25, no. 12: 2730. https://doi.org/10.3390/molecules25122730
APA StyleCacciola, N. A., Cerrato, A., Capriotti, A. L., Cavaliere, C., D’Apolito, M., Montone, C. M., Piovesana, S., Squillaci, G., Peluso, G., & Laganà, A. (2020). Untargeted Characterization of Chestnut (Castanea sativa Mill.) Shell Polyphenol Extract: A Valued Bioresource for Prostate Cancer Cell Growth Inhibition. Molecules, 25(12), 2730. https://doi.org/10.3390/molecules25122730