Potential of Flavonoid-Inspired Phytomedicines against COVID-19
Abstract
1. Introduction
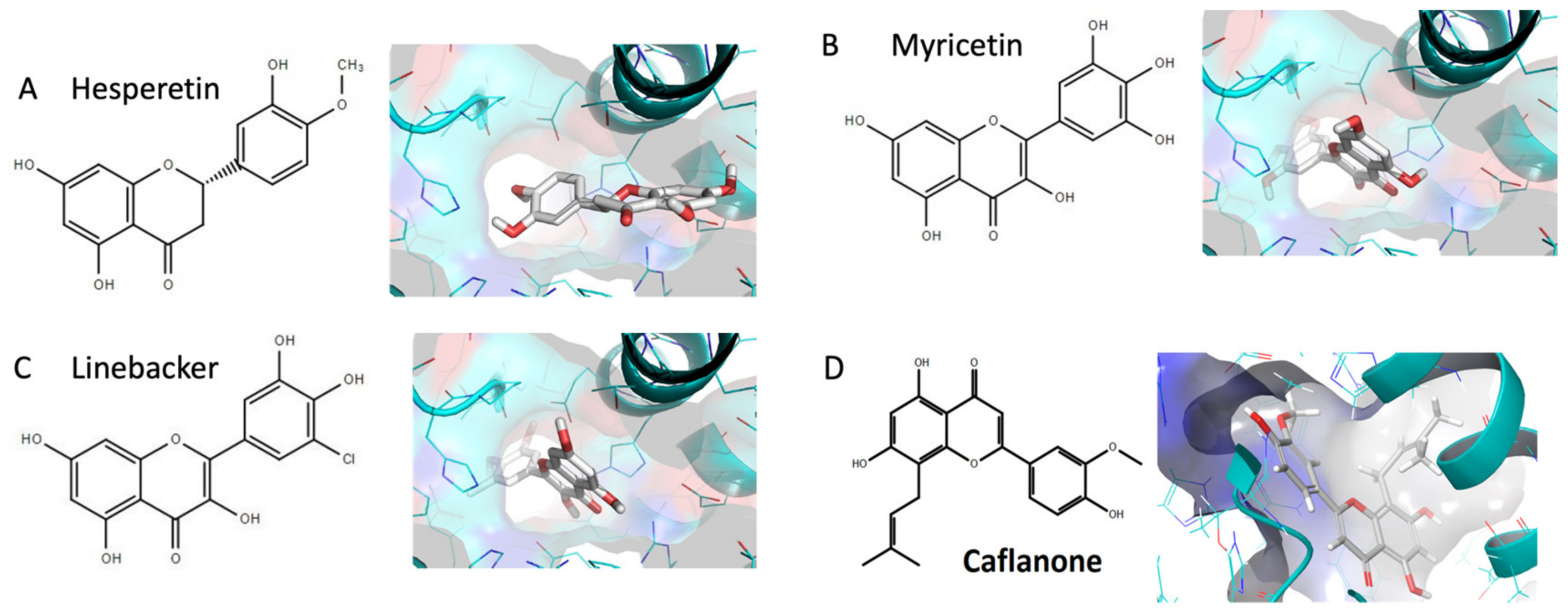
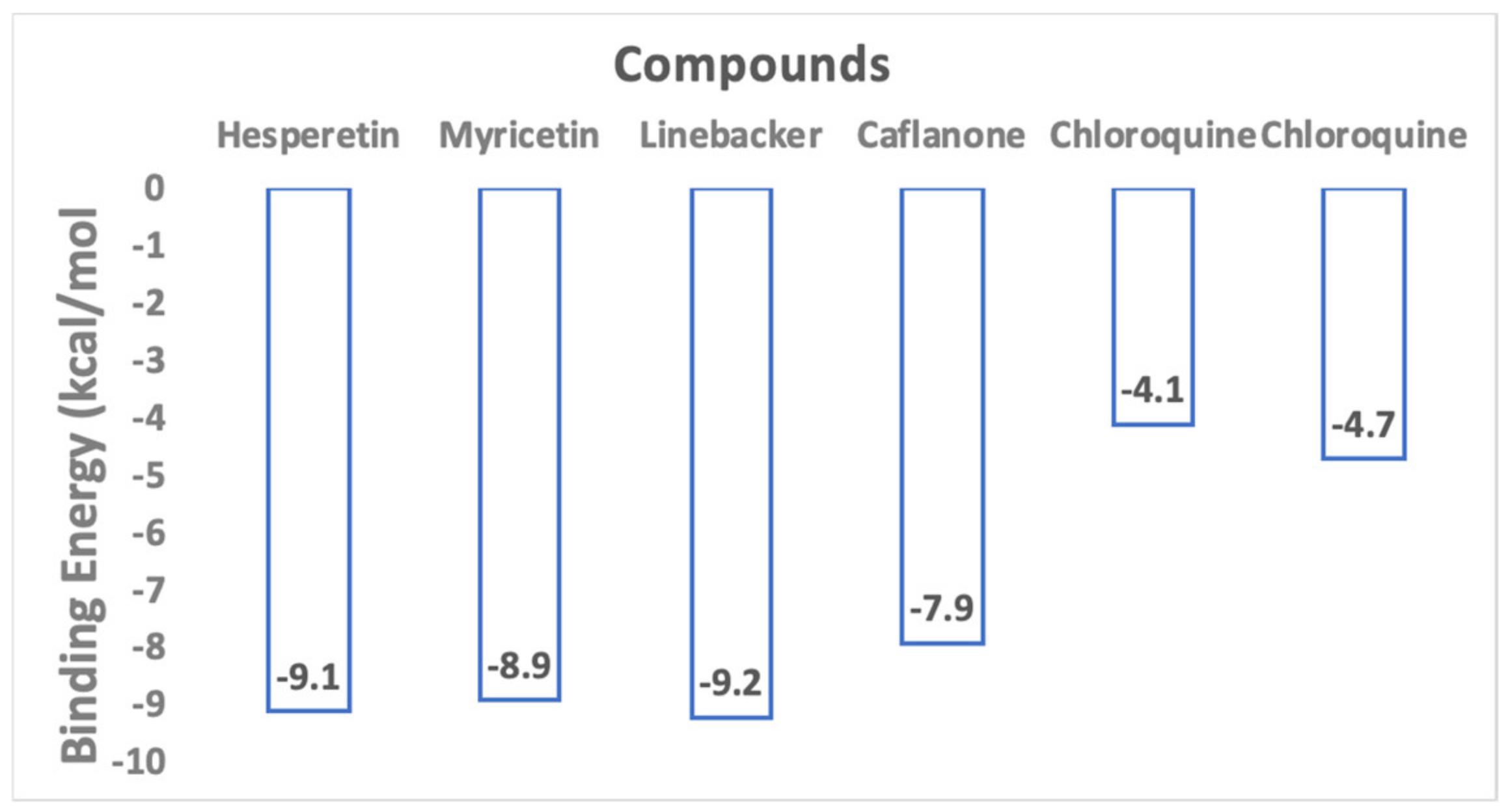
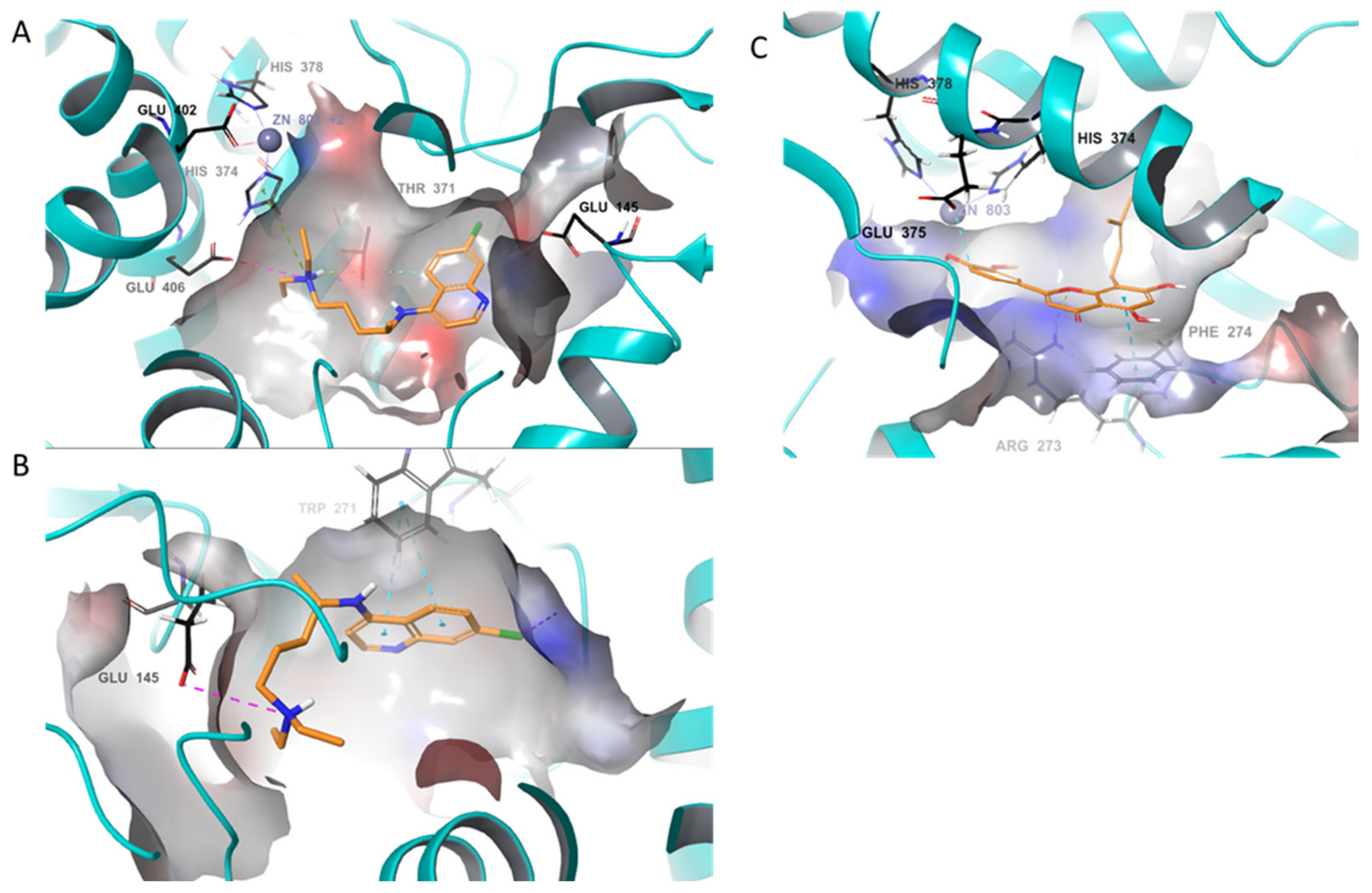
2. Results and Discussions
3. Conclusions
4. Materials and Methods
4.1. In-Silico Studies
4.2. In Vitro Studies
4.2.1. Inhibition of Hcov-OC43 Human Coronavirus
4.2.2. Kinase Inhibition Assay
4.2.3. Proinflammatory Cytokine Inhibition
4.3. Smart Nanoparticles Studies
Author Contributions
Funding
Conflicts of Interest
References
- Ledford, H. How does COVID-19 kill? Uncertainty is hampering doctors’ ability to choose treatments. Nature 2020, 580, 311–312. [Google Scholar] [CrossRef] [PubMed]
- Maxmen, A. More than 80 clinical trials launch to test coronavirus treatments. Nature 2020, 578, 347–348. [Google Scholar] [CrossRef]
- Chan, D.M. WHO Traditional Medicine Strategy: 2014–2023; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Cui, H.-T.; Li, Y.-T.; Guo, L.-Y.; Liu, X.-G.; Wang, L.-S.; Jia, J.-W.; Liao, J.-B.; Miao, J.; Zhang, Z.-Y.; Wang, L.; et al. Traditional Chinese medicine for treatment of coronavirus disease 2019: A review. Tradit. Med. Res. 2020, 16, 1708–1717. [Google Scholar]
- Tih, F. WHO to Study Madagascar’s Drug to Treat COVID-19. Available online: https://www.aa.com.tr/en/africa/who-to-study-madagascars-drug-to-treat-covid-19-/1840971 (accessed on 15 May 2019).
- Zakaryan, H.; Arabyan, E.; Oo, A.; Zandi, K. Flavonoids: Promising natural compounds against viral infections. Arch. Virol. 2017, 162, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020, 35, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Moore, J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020, 55, 105960. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Guerrero, L.; Castillo, J.; Quiñones, M.; Garcia-Vallvé, S.; Arola, L.; Pujadas, G.; Muguerza, B. Inhibition of Angiotensin-Converting Enzyme Activity by Flavonoids: Structure-Activity Relationship Studies. PLoS ONE 2012, 7, e49493. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.; Ibeh, U.; Decosmo, K.; Bih, N.; Yasmin-Karim, S.; Toyang, N.; Lowe, H.; Ngwa, W. Flavonoid Derivative of Cannabis Demonstrates Therapeutic Potential in Preclinical Models of Metastatic Pancreatic Cancer. Front. Oncol. 2019, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Sisk, J.M.; Frieman, M.B.; Machamer, C.E. Coronavirus S. protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J. Gen. Virol. 2018, 99, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Poon, L.L.M.; Ng, I.H.Y.; Luk, W.; Sia, S.-F.; Wu, M.H.S.; Chan, K.-H.; Yuen, K.-Y.; Gordon, S.; Guan, Y.; et al. Cytokine Responses in Severe Acute Respiratory Syndrome Coronavirus-Infected Macrophages In Vitro: Possible Relevance to Pathogenesis. J. Virol. 2005, 79, 7819–7826. [Google Scholar] [CrossRef]
- Yang, N.; Ping, M.; Lang, J.; Zhang, Y.; Deng, J.; Ju, X.; Zhang, G.; Jiang, C. Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry. J. Biol. Chem. 2012, 28, 8457–8467. [Google Scholar] [CrossRef]
- Li, M.; Chen, L.; Xiong, C.; Li, X. The ACE2 expression of maternal-fetal interface and fetal organs indicates potential risk of vertical transmission of SARS-COV-2. BioRxiv 2020, 15, e0230295. [Google Scholar]
- Thilakarathna, S.H.; Vasantha Rupasinghe, H.P. Flavonoid bioavailability and attempts for bioavailability enhancement. Nutrients 2013, 5, 3367–3387. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Kostarelos, K. Nanoscale nights of COVID-19. Nat. Nanotechnol. 2020, 5, 343–344. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, W.; Qu, X.; Wu, H.; Qu, L.; Zhang, X.; Mäkilä, E.; Salonen, J.; Zhu, Y.; Yang, Z.; et al. Photothermal-responsive nanosized hybrid polymersome as versatile therapeutics codelivery nanovehicle for effective tumor suppression. Proc. Natl. Acad. Sci. USA 2019, 116, 7744–7749. [Google Scholar] [CrossRef] [PubMed]
- Mayr, N.A.; Hu, K.S.; Liao, Z.; Viswanathan, A.N.; Wall, T.J.; Amendola, B.E.; Calaguas, M.J.; Palta, J.R.; Yue, N.J.; Rengan, R.; et al. International outreach: What is the responsibility of ASTRO and the major international radiation oncology societies? Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Mueller, R.; Yasmin-Karim, S.; Hesser, J.; Ngwa, W. Nanoparticle Drones to Label, Kill and Track Circulating Tumor Cells During Radiotherapy. Available online: https://w3.aapm.org/meetings/2019am/programinfo/programabs.php?sid=7980&aid=44735. (accessed on 18 May 2020).
- Ngwa, W.; Kumar, R.; Moreau, M.; Dabney, R.; Herman, A. Nanoparticle Drones to Target Lung Cancer with Radiosensitizers and Cannabinoids. Front. Oncol. 2017, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Korideck, H.; Ngwa, W.; Berbeco, R.I.; Makrigiorgos, G.M.; Sridhar, S. Third generation gold nanoplatform optimized for radiation therapy. Transl. Cancer. Res. 2013, 2, 1–6. [Google Scholar]
- Hu, T.Y.; Frieman, M.; Wolfram, J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 2020, 15, 247–249. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Mueller, J.L.; Tan, C.; Brooks, J.F.; Zikherman, J.; Cheng, W. Synthetic Liposomal Mimics of Biological Viruses for the Study of Immune Responses to Infection and Vaccination. Bioconjug. Chem. 2020, 31, 685–697. [Google Scholar] [CrossRef]
- Minuesa, G.; Albanese, S.K.; Xie, W.; Kazansky, Y.; Worroll, D.; Chow, A.; Schurer, A.; Park, S.M.; Rotsides, C.Z.; Taggart, J.; et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia. Nat. Commun. 2019, 10, 2691. [Google Scholar] [CrossRef]
- Springer, T.I.; Reid, T.E.; Gies, S.L.; Feix, J.B. Interactions of the effector ExoU from pseudomonas aeruginosa with short-chain phosphatidylinositides provide insights into ExoU targeting to host membranes. J. Biol. Chem. 2019, 294, 19012–19021. [Google Scholar] [CrossRef]
- Anastassiadis, T.; Deacon, S.W.; Devarajan, K.; Ma, H.; Peterson, J.R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1039. [Google Scholar] [CrossRef]
- Herter-Sprie, G.S.; Korideck, H.; Christensen, C.L.; Herter, J.M.; Rhee, K.; Berbeco, R.I.; Bennett, D.G.; Akbay, E.A.; Kozono, D.; Mak, R.H.; et al. Image-guided radiotherapy platform using single nodule conditional lung cancer mouse models. Nat. Commun. 2014, 5, 5870. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds caflanone is available from the authors. |

| Bioactivity | EC50/IC50 (µM) |
|---|---|
| hCov-OC43 beta virus | 0.42 |
| ABL-2 | 0.27 |
| AXL | <5.0 |
| Cathepsin L | 3.28 |
| IL-1β | 2.4 |
| IL-6 | 9.1 |
| IL-8 | 9.9 |
| Mip-1α | 8.9 |
| TNF-α | 8.7 |
| CK2a2 | 0.038 |
| JAK2 | 1.85 |
| MNK2 | 0.549 |
| PI4Kiiiβ | 0.136 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngwa, W.; Kumar, R.; Thompson, D.; Lyerly, W.; Moore, R.; Reid, T.-E.; Lowe, H.; Toyang, N. Potential of Flavonoid-Inspired Phytomedicines against COVID-19. Molecules 2020, 25, 2707. https://doi.org/10.3390/molecules25112707
Ngwa W, Kumar R, Thompson D, Lyerly W, Moore R, Reid T-E, Lowe H, Toyang N. Potential of Flavonoid-Inspired Phytomedicines against COVID-19. Molecules. 2020; 25(11):2707. https://doi.org/10.3390/molecules25112707
Chicago/Turabian StyleNgwa, Wilfred, Rajiv Kumar, Daryl Thompson, William Lyerly, Roscoe Moore, Terry-Elinor Reid, Henry Lowe, and Ngeh Toyang. 2020. "Potential of Flavonoid-Inspired Phytomedicines against COVID-19" Molecules 25, no. 11: 2707. https://doi.org/10.3390/molecules25112707
APA StyleNgwa, W., Kumar, R., Thompson, D., Lyerly, W., Moore, R., Reid, T.-E., Lowe, H., & Toyang, N. (2020). Potential of Flavonoid-Inspired Phytomedicines against COVID-19. Molecules, 25(11), 2707. https://doi.org/10.3390/molecules25112707




