Enzyme Mimetic Activity of ZnO-Pd Nanosheets Synthesized via a Green Route
Abstract
1. Introduction
2. Results and Discussion
2.1. UV–Vis Spectroscopy Analysis
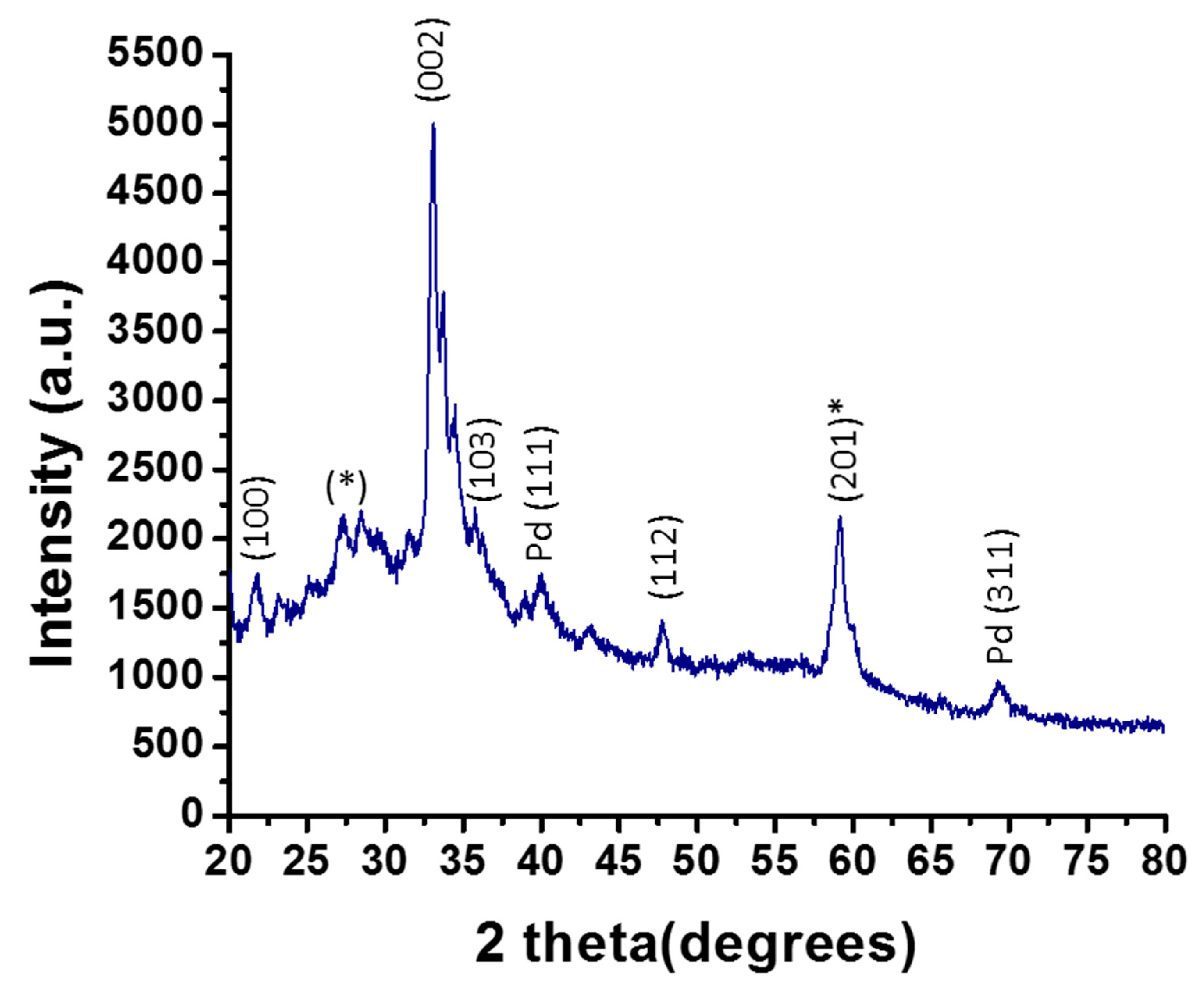
2.2. XRD Analysis
2.3. Field Emission Scanning Electron Microscope Analysis
2.4. TEM Analysis
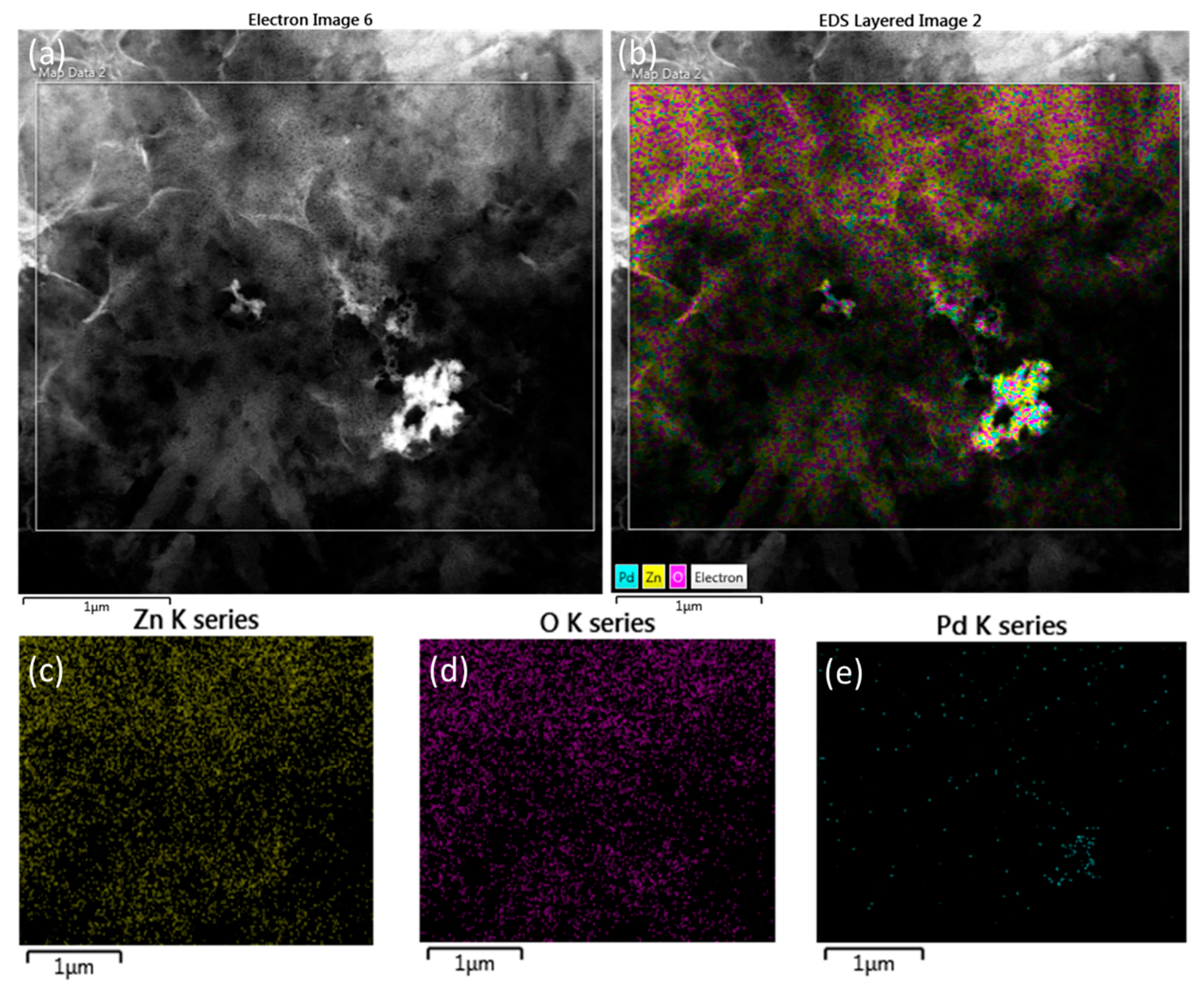
2.5. EDX Analysis
2.6. FTIR Analysis
2.7. Peroxidase Mimetic Activity
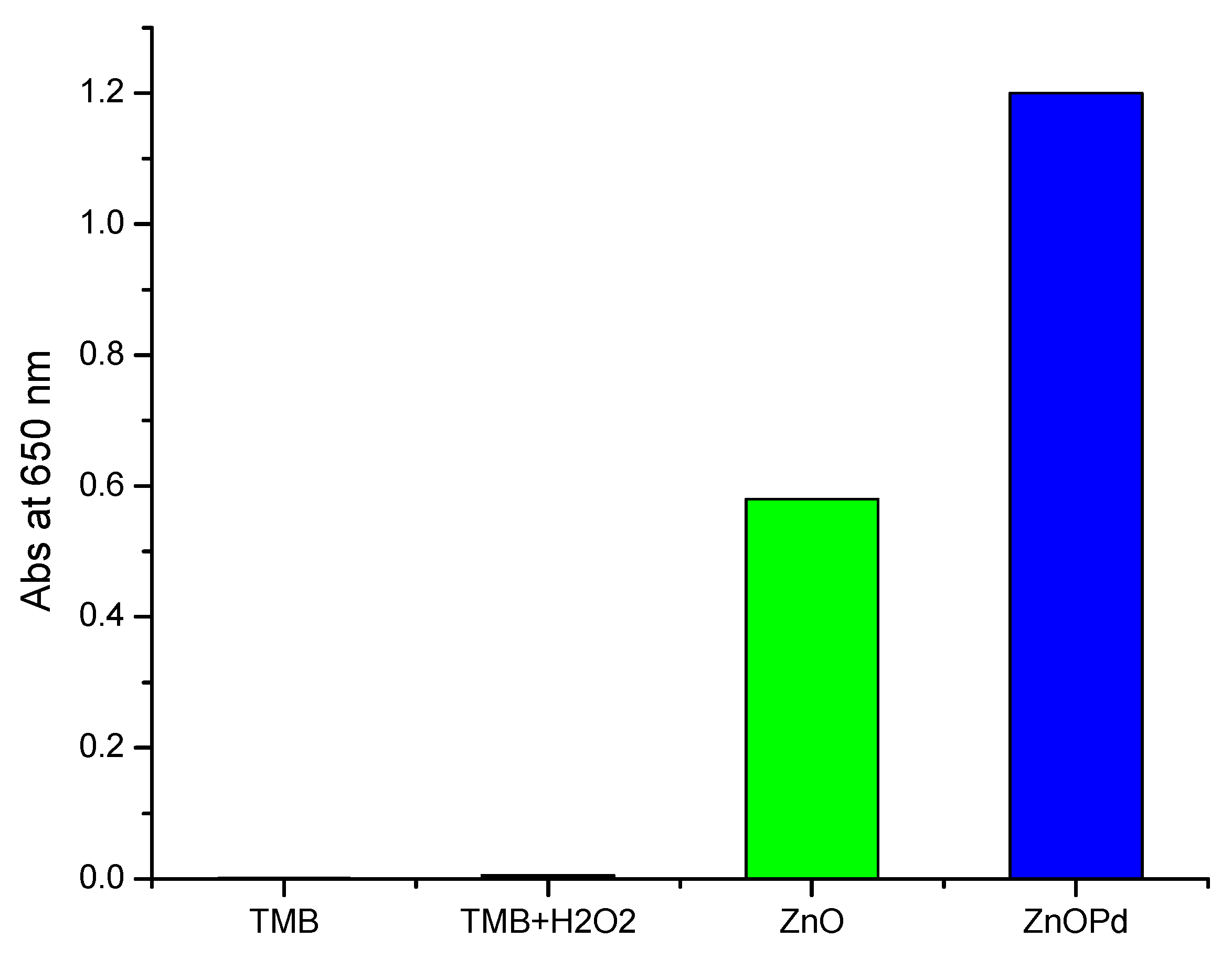
2.8. Comparison of Peroxidase Mimetic Activity of ZnO and ZnO-Pd Nanosheets
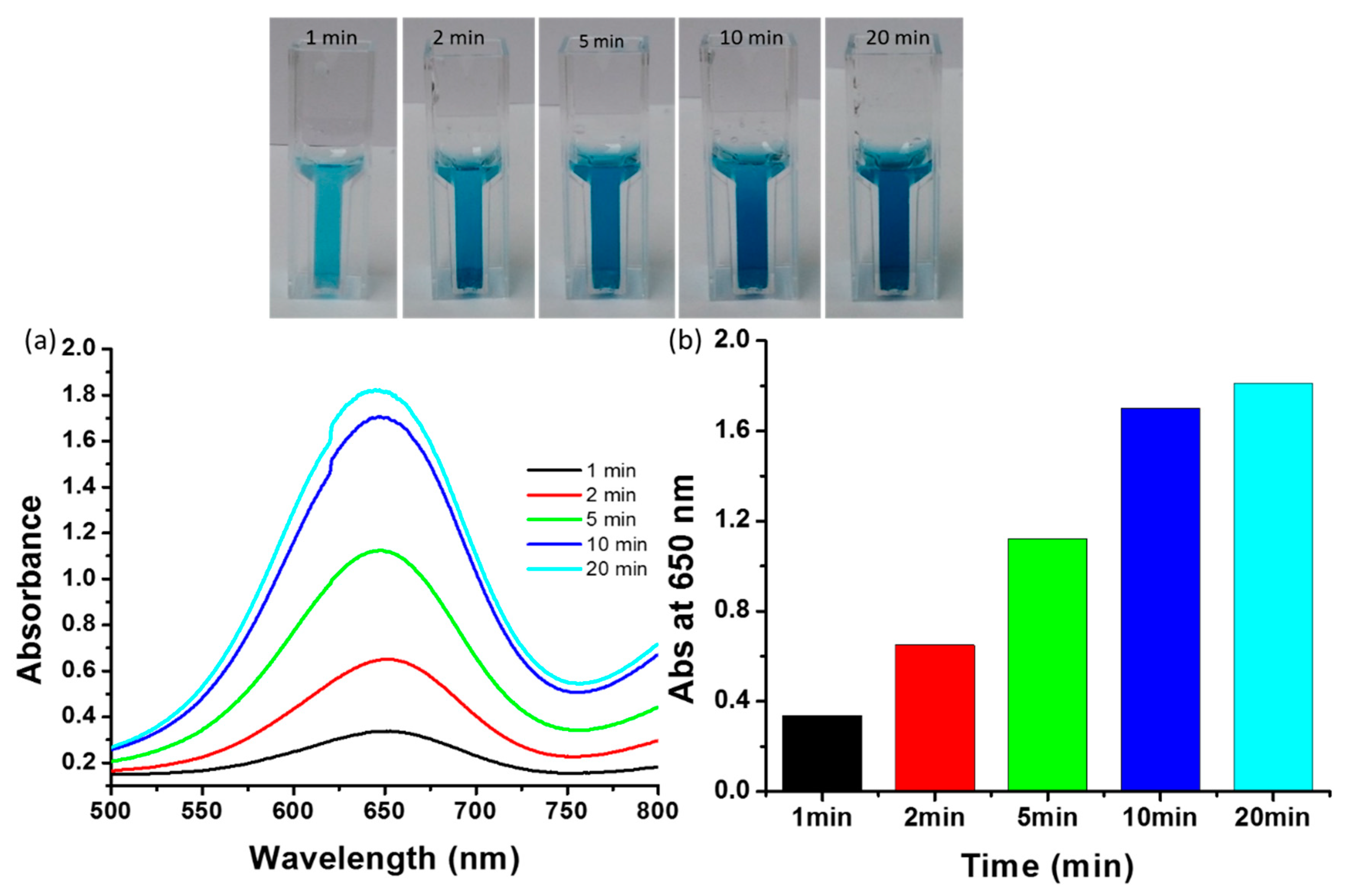
2.9. Influence of Incubation Time on Peroxidase Mimetic Activity
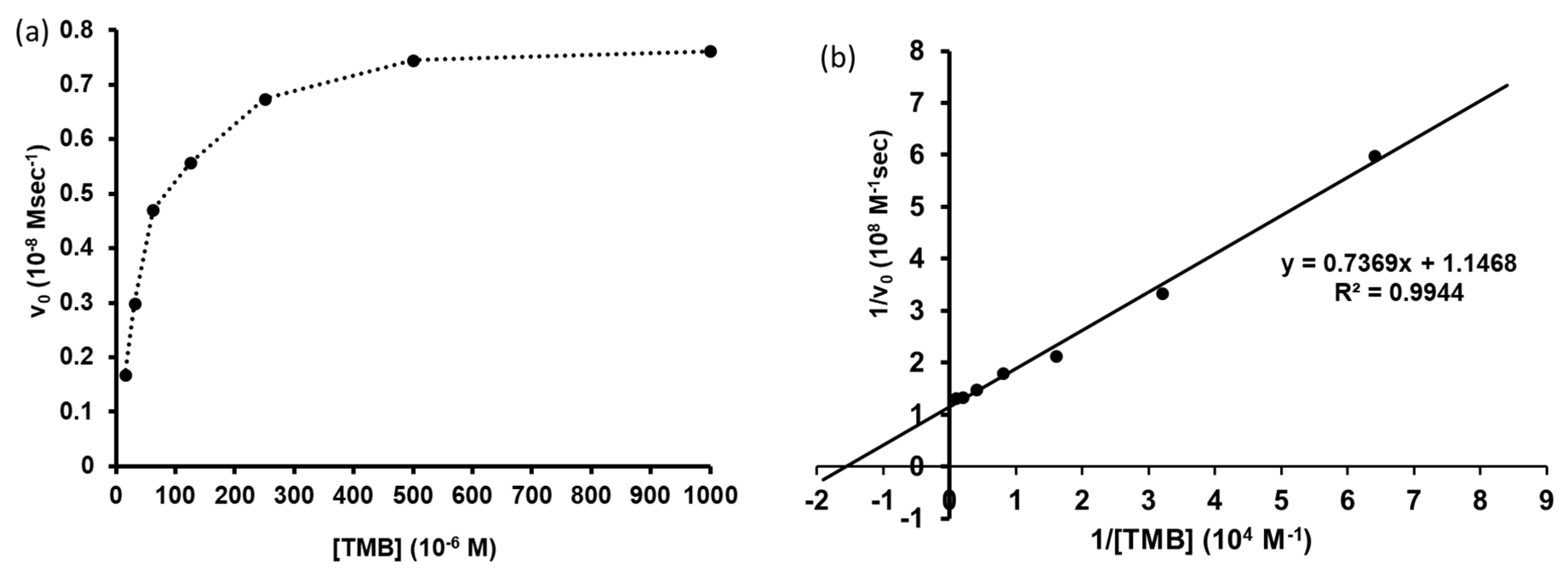
2.10. Determination of Catalytic Activity for TMB Oxidation
3. Materials and Methods
3.1. Materials
3.2. E. annuus Leaf Extract Preparation
3.3. Biosynthesis of ZnO-Pd Nanosheets
3.4. Characterization of Nanosheets
3.5. Peroxidase Mimetic Activity
3.6. Determination of Catalytic Activity for TMB Oxidation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tripathi, R.M.; Gupta, R.K.; Bhadwal, A.S.; Singh, P.; Shrivastav, A.; Shrivastav, B. Fungal biomolecules assisted biosynthesis of Au–Ag alloy nanoparticles and evaluation of their catalytic property. IET Nanobiotechnol. 2015, 9, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Bhadwal, A.S.; Kumar, N.; Shrivastav, A.; Shrivastav, B.R.; Singh, M.P.; Zafar, F.; Tripathi, R.M. Catalytic degradation of methylene blue by biosynthesised copper nanoflowers using F. benghalensis leaf extract. IET Nanobiotechnol. 2016, 10, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, N.; Tripathi, R.M.; Zafar, F.; Singh, M.P. Catalytic degradation of dichlorvos using biosynthesized zero valent iron nanoparticles. IEEE Trans. Nanobiosci. 2017, 16, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Chung, S.J. Reclamation of hexavalent chromium using catalytic activity of highly recyclable biogenic Pd (0) nanoparticles. Sci. Rep. 2020, 10, 640. [Google Scholar] [CrossRef]
- Tripathi, R.; Bhadwal, A.S.; Gupta, R.K.; Singh, P.; Shrivastav, A.; Shrivastav, B. ZnO nanoflowers: Novel biogenic synthesis and enhanced photocatalytic activity. J. Photochem. Photobiol. B 2014, 141, 288–295. [Google Scholar] [CrossRef]
- Tripathi, R.M.; Shrivastav, B.R.; Shrivastav, A. Antibacterial and catalytic activity of biogenic gold nanoparticles synthesised by Trichoderma harzianum. IET Nanobiotechnol. 2018, 12, 509–513. [Google Scholar] [CrossRef]
- Tripathi, R.M.; Pudake, R.N.; Shrivastav, B.; Shrivastav, A. Antibacterial activity of poly (vinyl alcohol)—biogenic silver nanocomposite film for food packaging material. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018, 9, 025020. [Google Scholar] [CrossRef]
- Tripathi, R.; Chung, S.J. Biogenic nanomaterials: Synthesis, characterization, growth mechanism, and biomedical applications. J. Microbiol. Methods 2019, 157, 65–80. [Google Scholar] [CrossRef]
- Tripathi, R.; Park, S.H.; Kim, G.; Kim, D.-H.; Ahn, D.; Kim, Y.M.; Kwon, S.J.; Yoon, S.-Y.; Kang, H.J.; Chung, S.J. Metal-induced redshift of optical spectra of gold nanoparticles: An instant, sensitive, and selective visual detection of lead ions. Int. Biodeterior. Biodegrad. 2019, 144, 104740. [Google Scholar] [CrossRef]
- Mehrotra, N.; Tripathi, R.M. Short interfering RNA therapeutics: Nanocarriers, prospects and limitations. IET Nanobiotechnol. 2015, 9, 386–395. [Google Scholar] [CrossRef]
- Tripathi, R.; Shrivastav, A.; Shrivastav, B. Biogenic gold nanoparticles: As a potential candidate for brain tumor directed drug delivery. Artif. Cells Nanomed. Biotechnol. 2015, 43, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.M.; Ranac, D.; Shrivastav, A.; Singh, R.P.; Shrivastav, B.R. Biogenic synthesis of silver nanoparticles using saraca indica leaf extract and evaluation of their antibacterial activity. Nano Biomed. Eng. 2013, 5, 50–56. [Google Scholar] [CrossRef]
- Tripathi, R.; Rao, R.P.; Tsuzuki, T. Green synthesis of sulfur nanoparticles and evaluation of their catalytic detoxification of hexavalent chromium in water. RSC Adv. 2018, 8, 36345–36352. [Google Scholar] [CrossRef]
- Mahajan, R.; Bhadwal, A.S.; Kumar, N.; Madhusudanan, M.; Pudake, R.N.; Tripathi, R.M. Green synthesis of highly stable carbon nanodots and their photocatalytic performance. IET Nanobiotechnol. 2016, 11, 360–364. [Google Scholar] [CrossRef]
- Tripathi, R.; Kumar, N.; Bhadwal, A.S.; Gupta, R.K.; Shrivastav, B.; Shrivastav, A. Facile and rapid biomimetic approach for synthesis of HAp nanofibers and evaluation of their photocatalytic activity. Mater. Lett. 2015, 140, 64–67. [Google Scholar] [CrossRef]
- Singh, V.K.; Yadav, P.K.; Chandra, S.; Bano, D.; Talat, M.; Hasan, S.H. Peroxidase mimetic activity of fluorescent NS-carbon quantum dots and their application in colorimetric detection of H2O2 and glutathione in human blood serum. J. Mater. Chem. B 2018, 6, 5256–5268. [Google Scholar] [CrossRef]
- Hnasko, R.M. Bioconjugation of Antibodies to Horseradish Peroxidase (HRP). In ELISA; Springer: Berlin, Germany, 2015; pp. 43–50. [Google Scholar]
- Singh, N.; Singh, J. An enzymatic method for removal of phenol from industrial effluent. Prep. Biochem. Biotechnol. 2002, 32, 127–133. [Google Scholar] [CrossRef]
- Zhang, Z.; Lai, J.; Wu, K.; Huang, X.; Guo, S.; Zhang, L.; Liu, J. Peroxidase-catalyzed chemiluminescence system and its application in immunoassay. Talanta 2018, 180, 260–270. [Google Scholar] [CrossRef]
- Radhapyari, K.; Kotoky, P.; Khan, R. Detection of anticancer drug tamoxifen using biosensor based on polyaniline probe modified with horseradish peroxidase. Mater. Sci. Eng. C 2013, 33, 583–587. [Google Scholar] [CrossRef]
- Wang, T.; Tian, J.; Li, T.; Li, J. Enzyme properties and thermal stability of horseradish peroxidase (HRP-DL). In Proceedings of the 2016 International Conference on Engineering and Technology Innovations, Wuhan, China, 26–27 March 2016. [Google Scholar]
- Song, Y.; Wang, X.; Zhao, C.; Qu, K.; Ren, J.; Qu, X. Label-free colorimetric detection of single nucleotide polymorphism by using single-walled carbon nanotube intrinsic peroxidase-like activity. Chem. Eur. J. 2010, 16, 3617–3621. [Google Scholar] [CrossRef]
- Shi, W.; Wang, Q.; Long, Y.; Cheng, Z.; Chen, S.; Zheng, H.; Huang, Y. Carbon nanodots as peroxidase mimetics and their applications to glucose detection. Chem. Commun. 2011, 47, 6695–6697. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Qu, K.; Zhao, C.; Ren, J.; Qu, X. Graphene oxide: Intrinsic peroxidase catalytic activity and its application to glucose detection. Adv. Mater. 2010, 22, 2206–2210. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, S.; Bao, J.; Ju, H. Nanostructured FeS as a mimic peroxidase for biocatalysis and biosensing. Chem. Eur. J. 2009, 15, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wu, X.; Liu, J.; Hu, X.; Zhang, K.; Hou, S.; Zhou, W.; Xie, S. Design of AgM bimetallic alloy nanostructures (M = Au, Pd, Pt) with tunable morphology and peroxidase-like activity. Chem. Mater. 2010, 22, 2988–2994. [Google Scholar] [CrossRef]
- Jv, Y.; Li, B.; Cao, R. Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection. Chem. Commun. 2010, 46, 8017–8019. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Deng, L.; Li, J.; Guo, S.; Wang, E.; Dong, S. Hemin– graphene hybrid nanosheets with intrinsic peroxidase-like activity for label-free colorimetric detection of single-nucleotide polymorphism. ACS Nano 2011, 5, 1282–1290. [Google Scholar] [CrossRef]
- Nazaruk, J.; Kalemba, D. Chemical composition of the essential oils from the roots of erigeron acris L. and erigeron annuus (L.) Pers. Molecules 2009, 14, 2458–2465. [Google Scholar] [CrossRef]
- Tripathi, R.; Shrivastav, A.; Shrivastav, B. Biofabrication of gold nanoparticles using leaf of Ficus benghalensis and their characterization. Int. J. Pharm. Biol. Sci. 2012, 3, 551–558. [Google Scholar]
- Liu, C.-P.; Chen, K.-C.; Su, C.-F.; Yu, P.-Y.; Lee, P.-W. Revealing the active site of gold nanoparticles for the peroxidase-like activity: The determination of surface accessibility. Catalysts 2019, 9, 517. [Google Scholar] [CrossRef]
- Suwanboon, S. Structural and optical properties of nanocrystalline ZnO powder from sol-gel method. Sci. Asia 2008, 34, 31–34. [Google Scholar] [CrossRef]
- Vempati, S.; Mitra, J.; Dawson, P. One-step synthesis of ZnO nanosheets: A blue-white fluorophore. Nanoscale Res. Lett. 2012, 7, 470. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Yoon, S.-Y.; Ahn, D.; Chung, S.J. Facile synthesis of triangular and hexagonal anionic gold nanoparticles and evaluation of their cytotoxicity. Nanomaterials 2019, 9, 1774. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.-L.; Cohen, A.; Petrutik, N.; Shlomovich, A.; Burstein, L.; Pang, S.-P.; Gozin, M. Highly insensitive and thermostable energetic coordination nanomaterials based on functionalized graphene oxides. J. Mater. Chem. A 2016, 4, 9941–9948. [Google Scholar] [CrossRef]
- Shankar, S.S.; Rai, A.; Ahmad, A.; Sastry, M. Rapid synthesis of Au, Ag, and bimetallic Au core–Ag shell nanoparticles using Neem (Azadirachta indica) leaf broth. J. Colloid Interface Sci. 2004, 275, 496–502. [Google Scholar] [CrossRef]
- Liao, G.; Fang, J.; Li, Q.; Li, S.; Xu, Z.; Fang, B. Ag-Based nanocomposites: Synthesis and applications in catalysis. Nanoscale 2019, 11, 7062–7096. [Google Scholar] [CrossRef]
- Pan, Y.; Pang, Y.; Shi, Y.; Zheng, W.; Long, Y.; Huang, Y.; Zheng, H. One-pot synthesis of a composite consisting of the enzyme ficin and a zinc (II)-2-methylimidazole metal organic framework with enhanced peroxidase activity for colorimetric detection for glucose. Microchim. Acta 2019, 186, 213. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.; Liu, X.; Zhong, J.; Lin, T.; Guo, L.; Fu, F. Colorimetric assay of copper ions based on the inhibition of peroxidase-like activity of MoS2 nanosheets. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 185, 271–275. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Z.; Jiang, Y.; Chi, M.; Nie, G.; Lu, X.; Wang, C. Palladium nanoparticles modified electrospun CoFe2O4 nanotubes with enhanced peroxidase-like activity for colorimetric detection of hydrogen peroxide. RSC Adv. 2016, 6, 33636–33642. [Google Scholar] [CrossRef]
- Josephy, P.D.; Eling, T.; Mason, R.P. The horseradish peroxidase-catalyzed oxidation of 3, 5, 3′, 5′-tetramethylbenzidine. Free radical and charge-transfer complex intermediates. J. Biol. Chem. 1982, 257, 3669–3675. [Google Scholar]
- Jiang, D.; Ni, D.; Rosenkrans, Z.T.; Huang, P.; Yan, X.; Cai, W. Nanozyme: New horizons for responsive biomedical applications. Chem. Soc. Rev. 2019, 48, 3683–3704. [Google Scholar] [CrossRef]
Sample Availability: Samples of the ZnO-Pd nanosheets are available from the authors. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripathi, R.M.; Ahn, D.; Kim, Y.M.; Chung, S.J. Enzyme Mimetic Activity of ZnO-Pd Nanosheets Synthesized via a Green Route. Molecules 2020, 25, 2585. https://doi.org/10.3390/molecules25112585
Tripathi RM, Ahn D, Kim YM, Chung SJ. Enzyme Mimetic Activity of ZnO-Pd Nanosheets Synthesized via a Green Route. Molecules. 2020; 25(11):2585. https://doi.org/10.3390/molecules25112585
Chicago/Turabian StyleTripathi, Ravi Mani, Dohee Ahn, Yeong Mok Kim, and Sang J. Chung. 2020. "Enzyme Mimetic Activity of ZnO-Pd Nanosheets Synthesized via a Green Route" Molecules 25, no. 11: 2585. https://doi.org/10.3390/molecules25112585
APA StyleTripathi, R. M., Ahn, D., Kim, Y. M., & Chung, S. J. (2020). Enzyme Mimetic Activity of ZnO-Pd Nanosheets Synthesized via a Green Route. Molecules, 25(11), 2585. https://doi.org/10.3390/molecules25112585







