Bioassay Guided Isolation and Docking Studies of a Potential β-Lactamase Inhibitor from Clutia myricoides
Abstract
1. Introduction
2. Results and Discussion
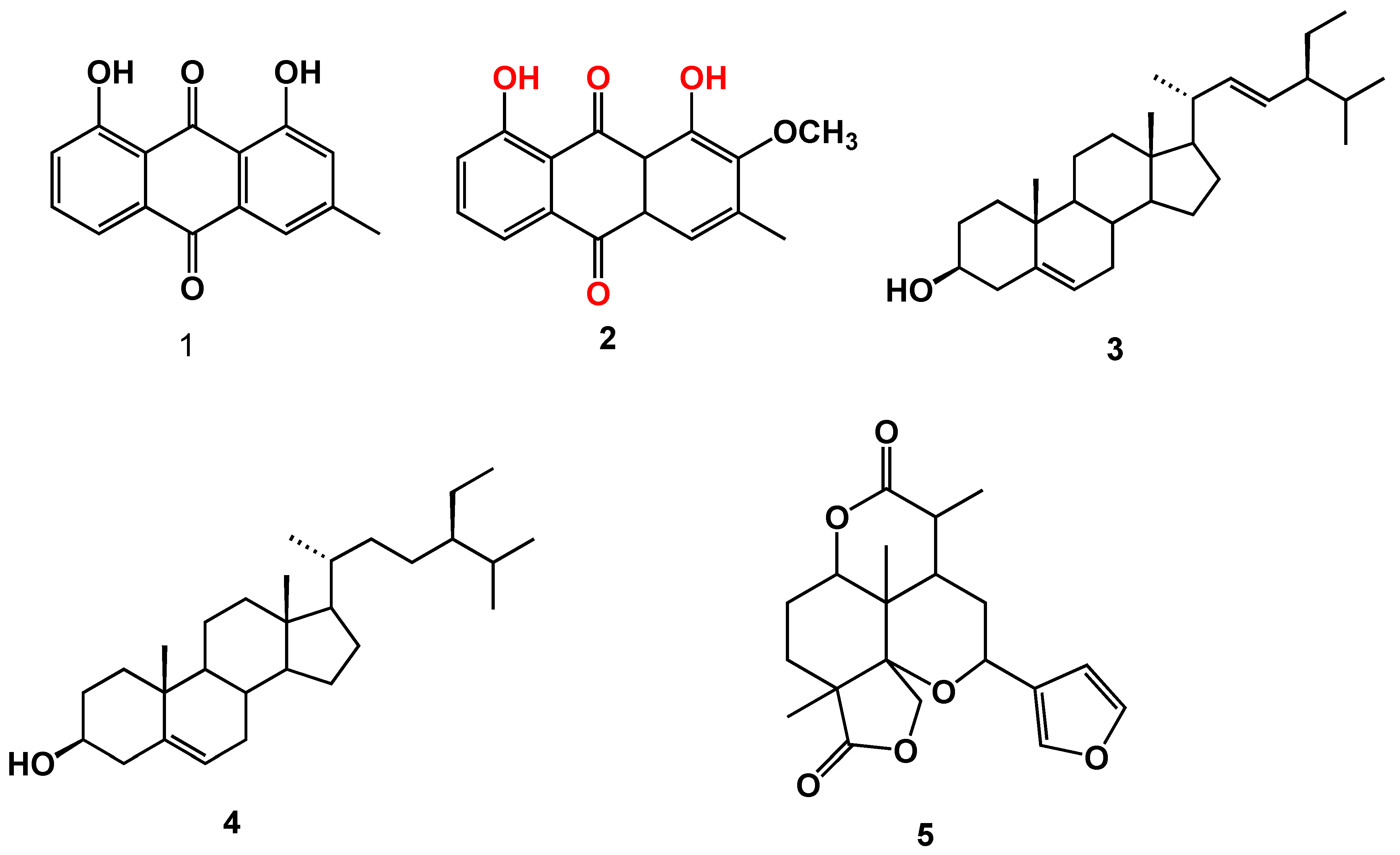
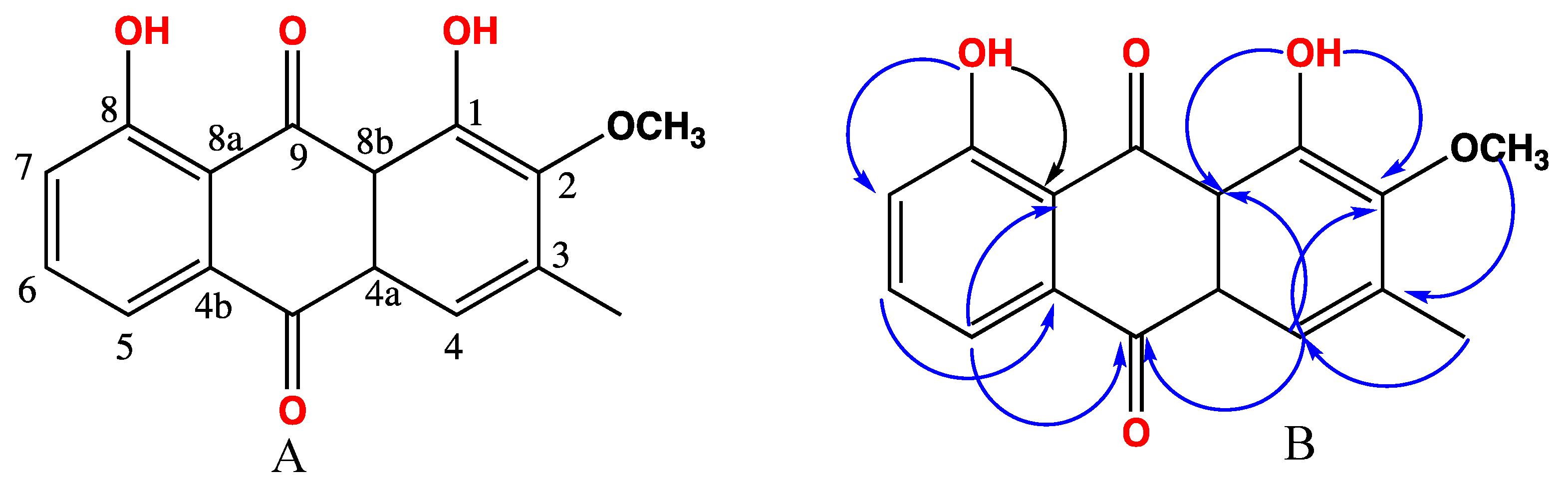
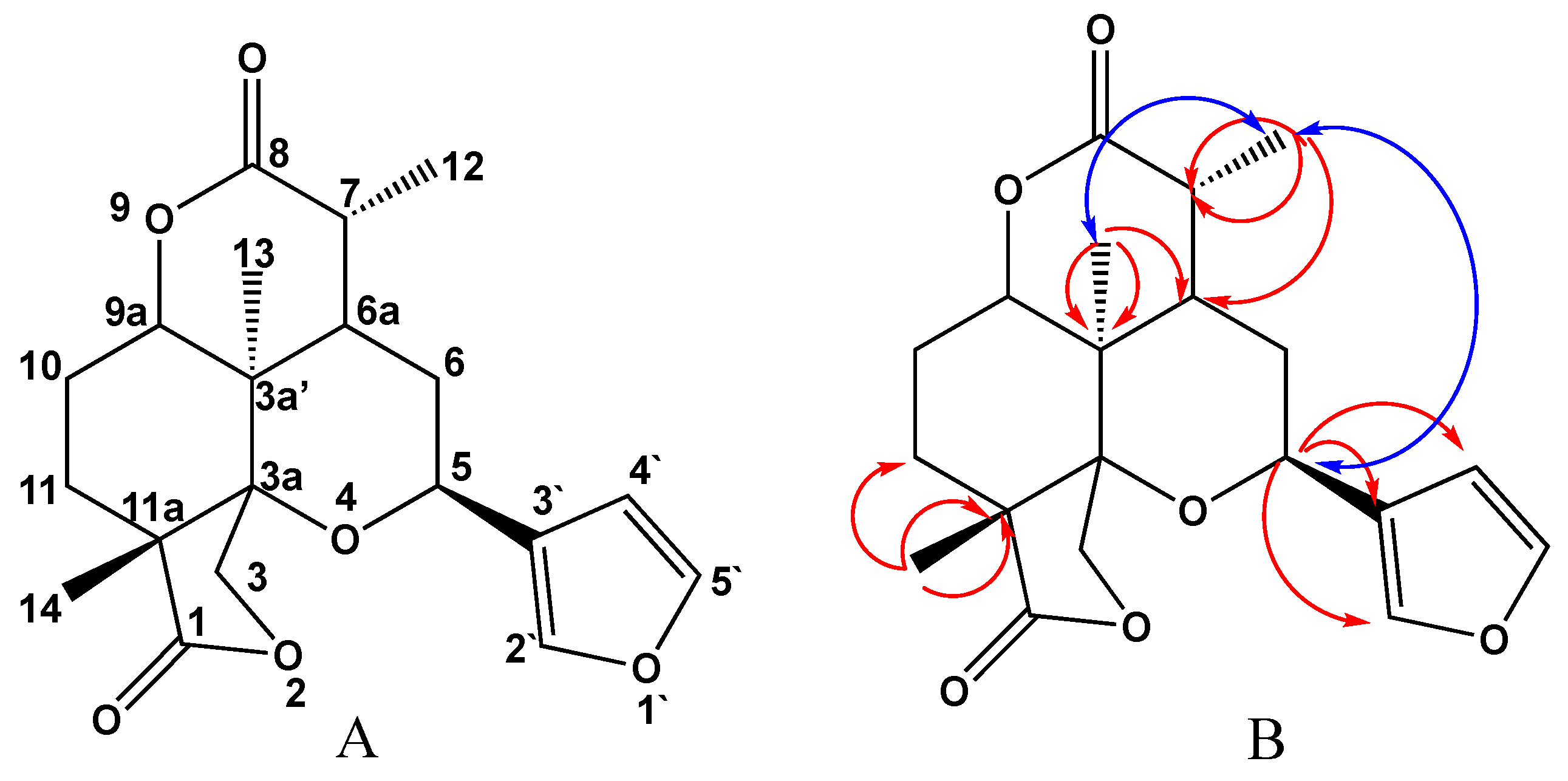
2.1. Chemical Investigation of the Chloroform Fraction
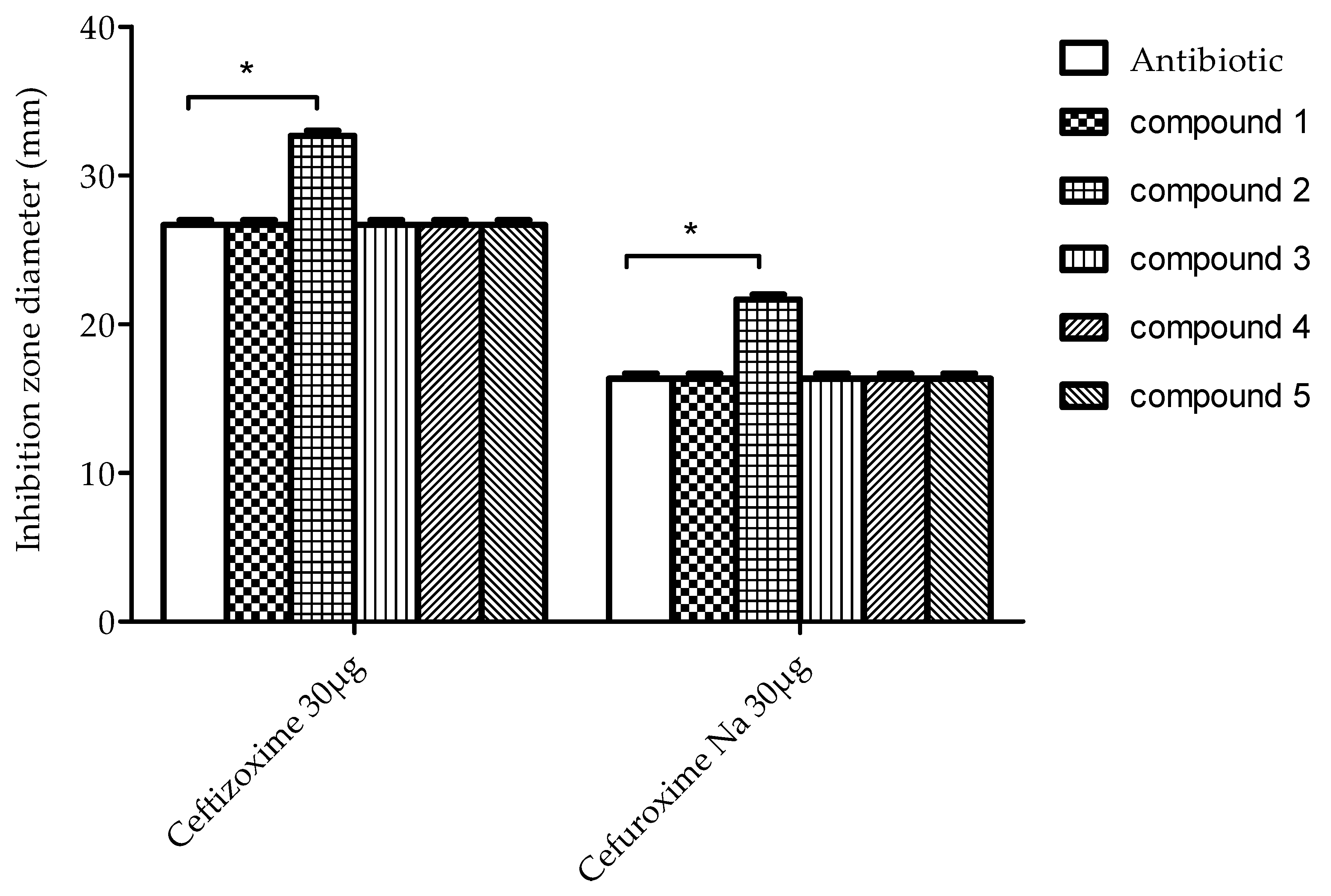
2.2. Testing for Antibacterial Activity
2.3. Characterization of Possible ESBL Inhibitory Activities
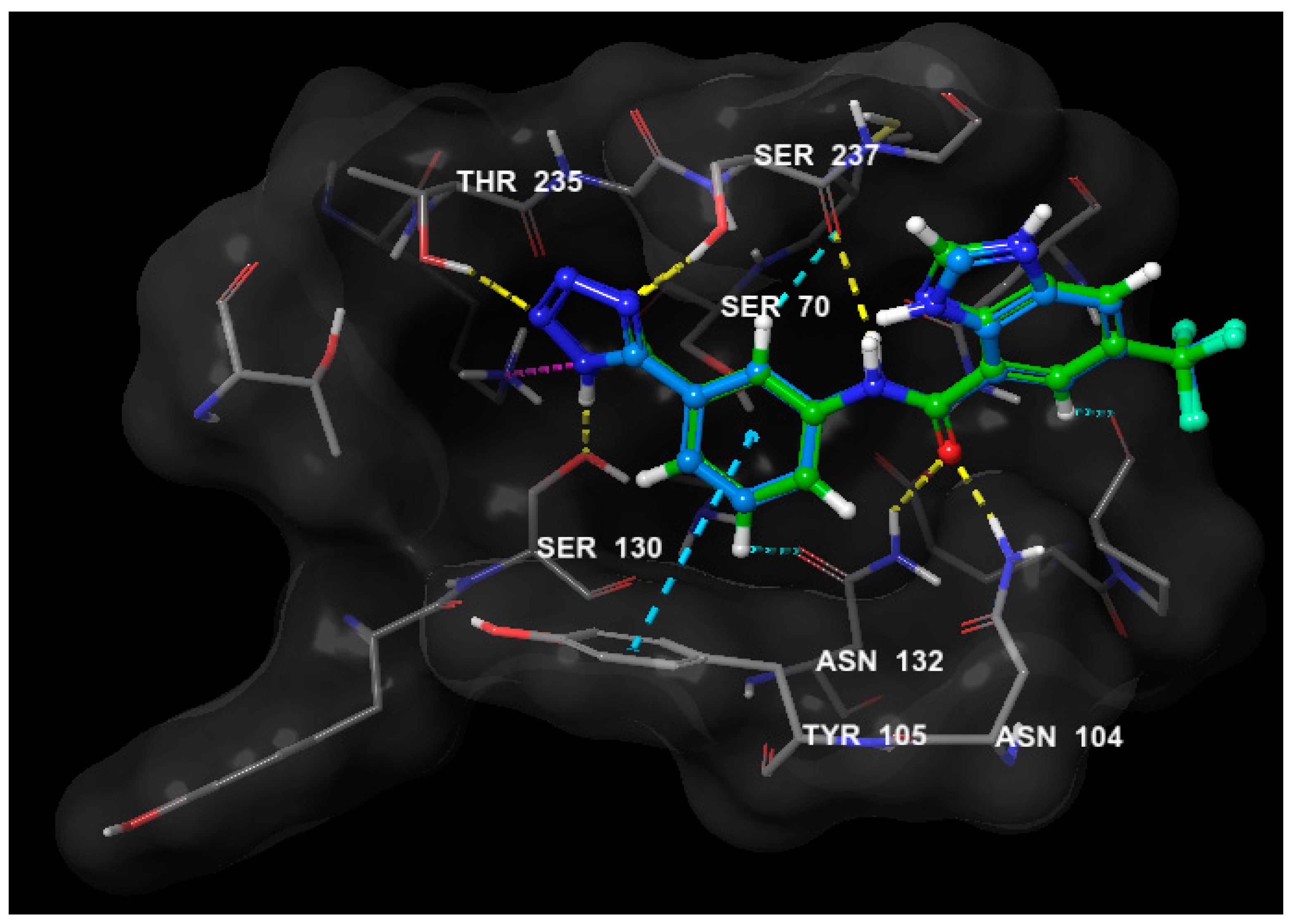
2.4. Molecular Modeling Study
3. Materials and Methods
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Spectral Data
3.5. Bacterial Strain
3.6. Screening for Antibacterial and Anti-ESBL Activity
3.7. Molecular Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lee, K.; Chong, Y.; Shin, H.; Kim, Y.; Yong, D.; Yum, J. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobactet species. Clin. Microbiol. Infect. 2001, 7, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Aggarwal, A. Occurrence of the CTX-M, SHV and the TEM Genes among the extended spectrum β-Lactamase producing isolates of enterobacteriaceae in a tertiary care hospital of North India. J. Clin. Diagn. Res. 2013, 7, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Hujer, A.M.; Hujer, K.M.; Helfand, M.S.; Anderson, V.E.; Bonomo, R.A. Amino acid substitutions at Ambler position Gly238 in the SHV-1 β-lactamase: Exploring sequence requirements for resistance to penicillins and cephalosporins. Antimicrob. Agents Chemother. 2002, 46, 3971–3977. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wright, G.D. Bacterial resistance to antibiotics: Enzymatic degradation and modification. Adv. Drug Deliv. Rev. 2005, 57, 1451–1470. [Google Scholar] [CrossRef]
- Chen, J.; Shang, X.; Hu, F.; Lao, X.; Gao, X.; Zheng, H.; Yao, W. β-Lactamase inhibitors: An update. Mini Rev. Med. Chem. 2013, 13, 1846–1861. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Gangoué-Piéboji, J.; Baurin, S.; Frère, J.M.; Ngassam, P.; Ngameni, B.; Azebaze, A.; Pegnyemb, D.E.; Watchueng, J.; Goffin, C.; Galleni, M. Screening of some medicinal plants from Cameroon for β-lactamase inhibitory activity. Phytother. Res. 2007, 21, 284–287. [Google Scholar] [CrossRef]
- Nascimento, G.G.; Locatelli, J.; Freitas, P.C.; Silva, G.L. Antibacterial activity of plant extracts and phytochemicals on antibiotic-resistant bacteria. Braz. J. Microbiol. 2000, 31, 247–256. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Hu, Z.-Q.; Okubo, S.; Hara, Y.; Shimamura, T. Mechanism of synergy between epigallocatechin gallate and β-lactams against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 1737–1742. [Google Scholar] [CrossRef]
- Aqil, F.; Khan, M.S.A.; Owais, M.; Ahmad, I. Effect of certain bioactive plant extracts on clinical isolates of β-lactamase producing methicillin resistant Staphylococcus aureus. J. Basic Microbiol. 2005, 45, 106–114. [Google Scholar] [CrossRef]
- Asili, J.; Emami, S.A.; Eynolghozat, R.; Noghab, Z.S.; Bazzaz, B.S.F.; Sahebkar, A. Chemical Composition and In Vitro Efficacy of Essential Oil of Seven Artemisia Species Against ESBL Producing Multidrug-Resistant Escherichia coli. J. Essent. Oil Bear. Plants 2015, 18, 124–145. [Google Scholar] [CrossRef]
- Lin, R.D.; Chin, Y.P.; Lee, M.H. Antimicrobial activity of antibiotics in combination with natural flavonoids against clinical extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae. Phytother. Res. 2005, 19, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Parveen, M.; Ahmad, F.; Malla, A.M.; Azaz, S.; Alam, M.; Basudan, O.A.; Silva, M.R.; Silva, P.S.P. Acetylcholinesterase and cytotoxic activity of chemical constituents of Clutia lanceolata leaves and its molecular docking study. Nat. Prod. Bioprospect. 2016, 6, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Mossa, J.S.; Cassady, J.M.; Kozlowski, J.F.; Zennie, T.M.; Antoun, M.D.; Pellechia, M.G.; McKenzie, A.T.; Byrn, S.R. Novel 6, 7-seco-6, 11-cyclolabdane diterpenes from Cluytia richardiana. Tetrahedron Lett. 1988, 29, 3627–3630. [Google Scholar] [CrossRef]
- Muhammad, I.; Mossa, J.S.; Al-Yahya, M.A.; Mirza, H.H.; El-Feraly, F.S.; Mcphail, A.T. Modified labdane diterpenoids from Cluytia richardiana. Phytochemistry 1994, 37, 1377–1381. [Google Scholar] [CrossRef]
- Muhammad, I.; Mossa, J.S.; Mirza, H.H.; El-Feraly, F.S. A new modified 6, 7-secolabdane diterpenoid from Clutia richardiana. Phytochemistry 1999, 50, 1225–1227. [Google Scholar] [CrossRef]
- Muhammad, I.; Mossa, J.S.; Al-Yahya, M.A.; Mirza, H.H.; El-Feraly, F.S.; McPhail, A.T. New modified labdane diterpenoids from Cluytia richardiana. J. Nat. Prod. 1994, 57, 248–255. [Google Scholar] [CrossRef]
- Mossa, J.S.; Cassady, J.M.; Antoun, M.D.; Byrn, S.R.; McKenzie, A.T.; Kozlowski, J.F.; Main, P. Saudin, a hypoglycemic diterpenoid, with a novel 6, 7-seco-labdane carbon skeleton, from Cluytia richardiana. J. Org. Chem. 1985, 50, 916–918. [Google Scholar] [CrossRef]
- Mossa, J.S.; Muhammad, I.; Al-Yahya, M.A.; Mirza, H.H.; El-Feraly, F.S.; McPhail, A.T. Five new modified 6, 7-secolabdane diterpenoids from Cluytia richardiana. J. Nat. Prod. 1996, 59, 224–231. [Google Scholar] [CrossRef]
- Waigh, R.D.; Zerihun, B.; Melvin, R. Three diterpenes with a secolabdane skeleton from Clutia abyssinica. Phytochemistry 1990, 29, 2935–2938. [Google Scholar] [CrossRef]
- Waigh, R.D.; Zerihun, B.M.; Maitland, D.J. Ten 5-methylcoumarins from Clutia abyssinica. Phytochemistry 1991, 30, 333–335. [Google Scholar] [CrossRef]
- Abdallah, H.M.; Asfour, H.Z.; El-Halawany, A.M.; Elfaky, M.A. Saudi plants as a source of potential β-lactamase inhibitors. Pak. J. Pharm. Sci. 2018, 31. [Google Scholar]
- Albar, H.A.; Alsofuani, T.A.; Khorshed, F.A. Phytochemical Analysis and Biological Screening of Leaf Extracts from Clutia myricoides. In Proceedings of the Oryx Conference for the Life Sciences, Riyadh, Saudi Arabia, 10–12 April 2007. [Google Scholar]
- El-Halawany, A.M.; Chung, M.H.; Nakamura, N.; Ma, C.-M.; Nishihara, T.; Hattori, M. Estrogenic and anti-estrogenic activities of Cassia tora phenolic constituents. Chem. Pharm. Bull. 2007, 55, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Zamzami, T.A.; Abdallah, H.M.; Shehata, I.A.; Mohamed, G.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Koshak, A.E.; Ibrahim, S.R. Macrochaetosides A and B, new rare sesquiterpene glycosides from Echinops macrochaetus and their cytotoxic activity. Phytochem. Lett. 2019, 30, 88–92. [Google Scholar] [CrossRef]
- El-Halawany, A.M.; Osman, S.M.; Abdallah, H.M. Cytotoxic constituents from Vicia monantha subsp. monantha seeds. Nat. Prod. Res. 2019, 33, 1783–1786. [Google Scholar] [CrossRef]
- Nair, M.G.D.; Mugunthu, R.; Kron, M.A.; Milev, Y.P. Anthraquinones and Process for the Preparation and Method of Use Thereof. US Patents 7658937 B2, 9 February 2010. [Google Scholar]
- Pemberton, O.A.; Zhang, X.; Nichols, D.A.; DeFrees, K.; Jaishankar, P.; Bonnet, R.; Adams, J.; Shaw, L.N.; Renslo, A.R.; Chen, Y. Antibacterial spectrum of a tetrazole-based reversible inhibitor of serine β-lactamases. Antimicrob. Agents Chemother. 2018, 62, e02563-17. [Google Scholar] [CrossRef]
- Cheesman, M.J.; Ilanko, A.; Blonk, B.; Cock, I.E. Developing new antimicrobial therapies: Are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn. Rev. 2017, 11, 57. [Google Scholar]
- Saeidi, S.; Boroujeni, N.A.; Ahmadi, H.; Hassanshahian, M. Antibacterial activity of some plant extracts against extended-spectrum beta-lactamase producing Escherichia coli isolates. Jundishapur J. Microbiol. 2015, 8. [Google Scholar] [CrossRef]
- Zuo, G.-Y.; Meng, F.-Y.; Han, J.; Hao, X.-Y.; Wang, G.-C.; Zhang, Y.-L.; Zhang, Q. In vitro activity of plant extracts and alkaloids against clinical isolates of extended-spectrum β-lactamase (ESBL)-producing strains. Molecules 2011, 16, 5453–5459. [Google Scholar] [CrossRef]
- Abreu, A.C.; McBain, A.J.; Simoes, M. Plants as sources of new antimicrobials and resistance-modifying agents. Nat. Prod. Rep. 2012, 29, 1007–1021. [Google Scholar] [CrossRef]
- Bouttier, S.; Fourniat, J.; Garofalo, C.; Gleye, C.; Laurens, A.; Hocquemiller, R. β-Lactamase Inhibitors from Anacardium occidentale. Pharm. Biol. 2002, 40, 231–234. [Google Scholar] [CrossRef]
- Vinod, N.; Shijina, R.; Dileep, K.; Sadasivan, C. Inhibition of beta-lactamase by 1, 4-naphthalenedione from the plant Holoptelea integrifolia. Appl. Biochem. Biotechnol. 2010, 160, 1752–1759. [Google Scholar] [CrossRef] [PubMed]
- Coudron, P.E.; Moland, E.S.; Sanders, C.C. Occurrence and detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae at a veterans medical center: Seek and you may find. J. Clin. Microbiol. 1997, 35, 2593–2597. [Google Scholar] [CrossRef]
- Murray, P.; Baron, E.; Pfaller, M.; Tenover, F.; Yolke, R. Manual of Clinical Microbiology, 8th ed.; ASM Press: Washington, DC, USA, 1995. [Google Scholar]
- Chattopadhyay, D.; Dastidar, S.; Chakrabarty, A. Antimicrobial properties of methdilazine and its synergism with antibiotics and some chemotherapeutic agents. Arzneimittelforschung 1988, 38, 869–872. [Google Scholar]
Sample Availability: Not available. |


| No. | 2 | 5 | |||
|---|---|---|---|---|---|
| δH [Mult. J (Hz)] | δC | δH [Mult. J (Hz)] | δC | ||
| 1 | - | 159.7 | 1 | - | 178.0 |
| 2 | - | 166.4 | 3 | 4.18 d (9.4), 4.28 d (9.4) | 70.4 |
| 3 | - | 146.4 | 3a | - | 82.0 |
| 4 | 7.69 s | 121.5 | 3a′ | - | 35.2 |
| 4a | - | 129.0 | 5 | 5.11 d (1.7) | 92.0 |
| 4b | - | 133.1 | 6 | 1.25 m, 1.28 m | 29.7 |
| 5 | 7.83 dd (1.7,7.6) | 120.3 | 6a | 2.70 m | 38.4 |
| 6 | 7.70 d (7.6) | 137.4 | 7 | 3.02 m | 35.3 |
| 7 | 7.31 dd (1.7,7.6) | 124.9 | 8 | - | 173.0 |
| 8 | - | 162.6 | 9 | - | 82.0 |
| 8a | - | 115.7 | 9a | 4.37 dd (4.2,11.9) | 75.5 |
| 8b | - | 114.1 | 10 | 1.78 m, 1.84 m | 23.9 |
| 9 | - | 192.4 | 11 | 1.62 m, 2.42 m | 26.5 |
| 10 | - | 181.4 | 12 | 1.41 d (6.8) | 13.7 |
| OCH3 | 3.99 s | 52.8 | 13 | 1.20 s | 15.3 |
| CH3 | 2.44 s | 20.1 | 14 | 1.43 s | 23.0 |
| OH-1 | 12.37 | - | 2′ | 7.54 brs | 139.4 |
| OH-8 | 11.90 | - | 3′ | - | 121.0 |
| 4′ | 6.45 brs | 107.0 | |||
| 5′ | 7.40 brs | 144.8 | |||
| Code | Antibacterial/Anti ESBL Activity |
|---|---|
| 1 | −/− |
| 2 | −/+ * |
| 3 | −/− |
| 4 | −/− |
| 5 | −/− |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elfaky, M.A.; El-Halawany, A.M.; Koshak, A.E.; Alshali, K.Z.; El-Araby, M.E.; Khayat, M.T.; Abdallah, H.M. Bioassay Guided Isolation and Docking Studies of a Potential β-Lactamase Inhibitor from Clutia myricoides. Molecules 2020, 25, 2566. https://doi.org/10.3390/molecules25112566
Elfaky MA, El-Halawany AM, Koshak AE, Alshali KZ, El-Araby ME, Khayat MT, Abdallah HM. Bioassay Guided Isolation and Docking Studies of a Potential β-Lactamase Inhibitor from Clutia myricoides. Molecules. 2020; 25(11):2566. https://doi.org/10.3390/molecules25112566
Chicago/Turabian StyleElfaky, Mahmoud A., Ali M. El-Halawany, Abdulrahman E. Koshak, Khalid Z. Alshali, Moustafa E. El-Araby, Maan T. Khayat, and Hossam M. Abdallah. 2020. "Bioassay Guided Isolation and Docking Studies of a Potential β-Lactamase Inhibitor from Clutia myricoides" Molecules 25, no. 11: 2566. https://doi.org/10.3390/molecules25112566
APA StyleElfaky, M. A., El-Halawany, A. M., Koshak, A. E., Alshali, K. Z., El-Araby, M. E., Khayat, M. T., & Abdallah, H. M. (2020). Bioassay Guided Isolation and Docking Studies of a Potential β-Lactamase Inhibitor from Clutia myricoides. Molecules, 25(11), 2566. https://doi.org/10.3390/molecules25112566










