Bioactive Potential of Extracts of Labrenzia aggregata Strain USBA 371, a Halophilic Bacterium Isolated from a Terrestrial Source
Abstract
1. Introduction
2. Results and Discussion
2.1. Metabolic Profiling of Crude Extracts from Chloroform Extractions
2.2. Cytotoxic Activity Analysis of Major Compounds in Chloroform Extracts
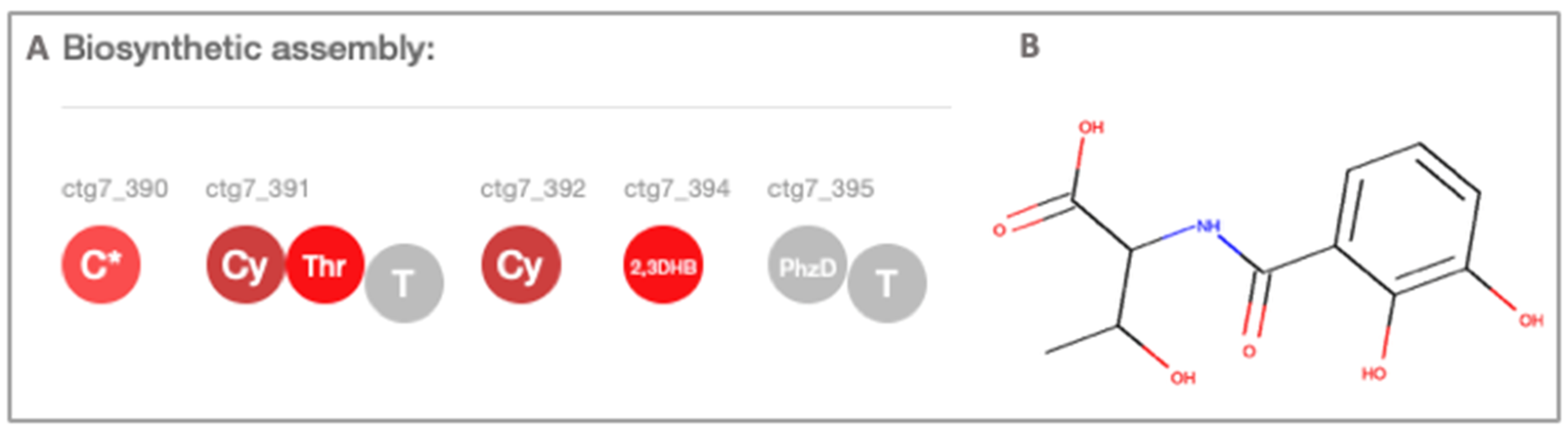
2.3. Secondary Metabolite Profiling, Genome Mining and Proteomics
3. Materials and Methods
3.1. Accession Numbers
3.2. Culturing Labrenzia aggregata USBA 371 to Obtain Metabolites in Crude Extracts from Chloroform Extraction
3.3. Metabolic Profiling of the Crude Extract from Labrenzia aggregata USBA 371
3.4. Isolation of Compounds from Crude Extract of L. aggregata USBA 371
3.5. Cytotoxic Activity Assay of Major Compounds Isolated after Chloroform Extraction of Supernatants
3.6. Proteomic Analysis
3.7. Genome Sequencing, Assembly and Annotation
4. Conclusion and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sinha, S.; Nge, C.E.; Leong, C.Y.; Ng, V.; Crasta, S.; Alfatah, M.; Goh, F.; Low, K.N.; Zhang, H.; Arumugam, P.; et al. Genomics-driven discovery of a biosynthetic gene cluster required for the synthesis of BII-Rafflesfungin from the fungus Phoma sp. F3723. BMC Genom. 2019, 20, 374. [Google Scholar] [CrossRef] [PubMed]
- Quadri, I.; Hassani, I.I.; L’Haridon, S.; Chalopin, M.; Hacène, H.; Jebbar, M. Characterization and antimicrobial potential of extremely halophilic archaea isolated from hypersaline environments of the Algerian Sahara. Microbiol. Res. 2016, 186, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, G.; Bu, T.; Zhang, Y.; Wang, Y.; Liu, M.; Lin, X. Phylogenetic analysis and screening of antimicrobial and cytotoxic activities of moderately halophilic bacteria isolated from the Weihai Solar Saltern (China). World J. Microbiol. Biotechnol. 2010, 26, 879–888. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.-N.; Fu, C.-M.; Wang, G.-Y. Phylogenetic Analysis and Screening of Antimicrobial and Antiproliferative Activities of Culturable Bacteria Associated with the Ascidian Styela clava from the Yellow Sea, China. BioMed Res. Int. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Mazguene, S.; Rossi, M.; Gogliettino, M.; Palmieri, G.; Cocca, E.; Mirino, S.; Imadalou-Idres, N.; Benallaoua, S. Isolation and characterization from solar salterns of North Algeria of a haloarchaeon producing a new halocin. Extremophiles 2018, 22, 259–270. [Google Scholar] [CrossRef]
- Tang, X.-X.; Liu, S.-Z.; Yan, X.; Tang, B.-W.; Fang, M.-J.; Wang, X.-M.; Wu, Z.; Qiu, Y.-K. Two New Cytotoxic Compounds from a Deep-Sea Penicillum citreonigrum XT20-134. Mar. Drugs 2019, 17, 509. [Google Scholar] [CrossRef]
- Karthikeyan, P.; Bhat, S.G.; Chandrasekaran, M. Halocin SH10 production by an extreme haloarchaeon Natrinema sp. BTSH10 isolated from salt pans of South India. Saudi J. Biol. Sci. 2013, 20, 205–212. [Google Scholar] [CrossRef][Green Version]
- Camacho, M.; Redondo-Gómez, S.; Rodrìguez-Llorente, I.; Rohde, M.; Spröer, C.; Schumann, P.; Klenk, H.P.; del Carmen Montero-Calasanz, M. Labrenzia salina sp. Nov., isolated from the rhizosphere of the halophyte Arthrocnemum macrostachyum. Int. J. Syst. Evol. Microbiol. 2016, 66, 5173–5180. [Google Scholar] [CrossRef]
- Rodrigues, G.N.; Lago-Lestón, A.; Costa, R.; Keller-Costa, T. Draft genome sequence of Labrenzia sp. strain EL143, a coral-associated alphaproteobacterium with versatile symbiotic living capability and strong halogen degradation potential. Genome Announc. 2018, 6, 1–2. [Google Scholar] [CrossRef]
- Biebl, H.; Pukall, R.; Lünsdorf, H.; Schulz, S.; Allgaier, M.; Tindall, B.J.; Wagner-Döbler, I. Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., and of Stappia alba as Labrenzia alba comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1095–1107. [Google Scholar] [CrossRef]
- Kačar, D.; Schleissner, C.; Cañedo, L.M.; Rodríguez, P.; de la Calle, F.; Galán, B.; García, J.L. Genome of Labrenzia sp. PHM005 Reveals a Complete and Active Trans-AT PKS Gene Cluster for the Biosynthesis of Labrenzin. Front. Microbiol. 2019, 10, 2561. [Google Scholar] [CrossRef] [PubMed]
- Schleissner, C.; Cañedo, L.M.; Rodríguez, P.; Crespo, C.; Zúñiga, P.; Peñalver, A.; De La Calle, F.; Cuevas, C. Bacterial Production of a Pederin Analogue by a Free-Living Marine Alphaproteobacterium. J. Nat. Prod. 2017, 80, 2170–2173. [Google Scholar] [CrossRef] [PubMed]
- Raj Sharma, A.; Zhou, T.; Harunari, E.; Oku, N.; Trianto, A.; Igarashi, Y. Labrenzbactin from a coral-associated bacterium Labrenzia sp. J. Antibiot. (Tokyo) 2019, 72, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, J.A.; Dávila-Céspedes, A.; Kehraus, S.; Crüsemann, M.; Köse, M.; Müller, C.E.; König, G.M. Cyclopropane-containing fatty acids from the marine bacterium Labrenzia sp. 011 with antimicrobial and GPR84 activity. Mar. Drugs 2018, 16, 369. [Google Scholar] [CrossRef]
- Díaz-Cárdenas, C.; Cantillo, A.; Rojas, L.Y.; Sandoval, T.; Fiorentino, S.; Robles, J.; Ramos, F.A.; Zambrano, M.M.; Baena, S. Microbial diversity of saline environments: Searching for cytotoxic activities. AMB Express 2017, 7, 223. [Google Scholar] [CrossRef]
- Borthwick, A.D.; Da Costa, N.C. 2,5-diketopiperazines in food and beverages: Taste and bioactivity. Crit. Rev. Food Sci. Nutr. 2017, 57, 718–742. [Google Scholar] [CrossRef]
- Lautru, S.; Gondry, M.; Genet, R.; Pernodet, J.L. The albonoursin gene cluster of S. noursei: Biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem. Biol. 2002, 9, 1355–1364. [Google Scholar] [CrossRef]
- Yao, T.; Liu, J.; Liu, Z.; Li, T.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Gu, Q.; Li, W. Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery. Nat. Commun. 2018, 9, 4091. [Google Scholar] [CrossRef]
- Tommonaro, G.; Abbamondi, G.R.; Iodice, C.; Tait, K.; De Rosa, S. Diketopiperazines Produced by the Halophilic Archaeon, Haloterrigena hispanica, Activate AHL Bioreporters. Microb. Ecol. 2012, 63, 490–495. [Google Scholar] [CrossRef]
- Nishanth Kumar, S.; Mohandas, C.; Siji, J.V.; Rajasekharan, K.N.; Nambisan, B. Identification of antimicrobial compound, diketopiperazines, from a Bacillus sp. N strain associated with a rhabditid entomopathogenic nematode against major plant pathogenic fungi. J. Appl. Microbiol. 2012, 113, 914–924. [Google Scholar] [CrossRef]
- Fdhila, F.; Vázquez, V.; Sánchez, J.L.; Riguera, R. dd-diketopiperazines: Antibiotics active against Vibrio anguillarum isolated from marine bacteria associated with cultures of Pecten maximus. J. Nat. Prod. 2003, 66, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Ku, S.K.; Choi, H.; Bae, J.S. Three diketopiperazines from marine-derived bacteria inhibit LPS-induced endothelial inflammatory responses. Bioorganic Med. Chem. Lett. 2016, 26, 1873–1876. [Google Scholar] [CrossRef] [PubMed]
- Alshaibani, M.M.; Zin, N.; Jalil, J.; Sidik, N.M.; Ahmad, S.J.; Kamal, N.; Edrada-Ebel, R. Isolation, Purification, and Characterization of Five Active Diketopiperazine Derivatives from Endophytic Streptomyces SUK 25 with Antimicrobial and Cytotoxic Activities. J. Microbiol. Biotechnol. 2017, 27, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- van der Merwe, E.; Huang, D.; Peterson, D.; Kilian, G.; Milne, P.J.; Van de Venter, M.; Frost, C. The synthesis and anticancer activity of selected diketopiperazines. Peptides 2008, 29, 1305–1311. [Google Scholar] [CrossRef]
- Gomes, N.G.M.; Pereira, R.B.; Andrade, P.B.; Valentão, P. Double the chemistry, double the fun: Structural diversity and biological activity of marine-derived diketopiperazine dimers. Mar. Drugs 2019, 17, 551. [Google Scholar] [CrossRef]
- Martins, M.B.; Carvalho, I. Diketopiperazines: Biological activity and synthesis. Tetrahedron 2007, 63, 9923–9932. [Google Scholar] [CrossRef]
- Yonezawa, K.; Yamada, K.; Kouno, I. New Diketopiperazine Derivatives Isolated from Sea Urchin-Derived Bacillus sp. Chem. Pharm. Bull. 2011, 59, 106–108. [Google Scholar] [CrossRef]
- Stierle, A.C.; Cardellina, J.H.; Strobel, G.A. Maculosin, a host-specific phytotoxin for spotted knapweed from Alternaria alternata. Proc. Natl. Acad. Sci. USA 1988, 85, 8008–8011. [Google Scholar] [CrossRef]
- Azab, A.; Nassar, A.; Azab, A.N. Anti-inflammatory activity of natural products. Molecules 2016, 21, 1321. [Google Scholar] [CrossRef]
- Deharo, E.; Ginsburg, H. Analysis of additivity and synergism in the anti-plasmodial effect of purified compounds from plant extracts. Malar. J. 2011, 10, S5. [Google Scholar] [CrossRef]
- Prieto, K.; Cao, Y.; Mohamed, E.; Trillo-Tinoco, J.; Sierra, R.A.; Urueña, C.; Sandoval, T.A.; Fiorentino, S.; Rodriguez, P.C.; Barreto, A. Polyphenol-rich extract induces apoptosis with immunogenic markers in melanoma cells through the ER stress-associated kinase PERK. Cell Death Discov. 2019, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, T.A.; Urueña, C.P.; Llano, M.; Gómez-Cadena, A.; Hernández, J.F.; Sequeda, L.G.; Loaiza, A.E.; Barreto, A.; Li, S.; Fiorentino, S. Standardized Extract from Caesalpinia spinosa is Cytotoxic over Cancer Stem Cells and Enhance Anticancer Activity of Doxorubicin. Am. J. Chin. Med. 2016, 44, 1693–1717. [Google Scholar] [CrossRef] [PubMed]
- Adams, L.S.; Seeram, N.P.; Hardy, M.L.; Carpenter, C.; Heber, D. Analysis of the interactions of botanical extract combinations against the viability of prostate cancer cell lines. Evid.-Based Complement. Altern. Med. 2006, 3, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.J.; Liang, W.M.; Chen, C.J.; Tsang, H.; Chiou, J.S.; Liu, X.; Cheng, C.F.; Lin, T.H.; Liao, C.C.; Huang, S.M.; et al. Network analysis and mechanisms of action of Chinese herb-related natural compounds in lung cancer cells. Phytomedicine 2019, 58, 152893. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ma, Q.; Cui, H.; Liu, G.; Zhao, X.; Li, W.; Piao, G. How can synergism of traditional medicines benefit from network pharmacology? Molecules 2017, 22, 1135. [Google Scholar] [CrossRef]
- Lasso, P.; Llano Murcia, M.; Sandoval, T.A.; Urueña, C.; Barreto, A.; Fiorentino, S. Breast Tumor Cells Highly Resistant to Drugs Are Controlled Only by the Immune Response Induced in an Immunocompetent Mouse Model. Integr. Cancer Ther. 2019, 18, 1–16. [Google Scholar] [CrossRef]
- Nishanth Kumar, S.; Mohandas, C.; Nambisan, B.; Kumar, D.R.S.; Lankalapalli, R.S. Isolation of proline-based cyclic dipeptides from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. World J. Microbiol. Biotechnol. 2013, 29, 355–364. [Google Scholar] [CrossRef]
- Vázquez-Rivera, D.; González, O.; Guzmán-Rodríguez, J.; Díaz-Pérez, A.L.; Ochoa-Zarzosa, A.; López-Bucio, J.; Meza-Carmen, V.; Campos-García, J. Cytotoxicity of cyclodipeptides from pseudomonas aeruginosa PAO1 leads to apoptosis in human cancer cell lines. BioMed Res. Int. 2015, 2015, 9. [Google Scholar] [CrossRef]
- Kwak, M.-K.; Liu, R.; Kwon, J.-O.; Kim, M.-K.; Kim, A.H.; Kang, S.-O. Cyclic dipeptides from lactic acid bacteria inhibit proliferation of the influenza A virus. J. Microbiol. 2013, 51, 836–843. [Google Scholar] [CrossRef]
- Iimura, K.; Furukawa, T.; Yamamoto, T.; Negishi, L.; Suzuki, M.; Sakuda, S. The Mode of Action of Cyclo(L-Ala-L-Pro) in Inhibiting Aflatoxin Production of Aspergillus flavus. Toxins 2017, 9, 219. [Google Scholar] [CrossRef]
- Holden, M.T.G.; Chhabra, S.R.; De Nys, R.; Stead, P.; Bainton, N.J.; Hill, P.J.; Manefield, M.; Kumar, N.; Labatte, M.; England, D.; et al. Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0--a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gondry, M.; Sauguet, L.; Belin, P.; Thai, R.; Amouroux, R.; Tellier, C.; Tuphile, K.; Jacquet, M.; Braud, S.; Courçon, M.; et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. 2009, 5, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Merwin, N.J.; Dejong, C.A.; Magarvey, N.A. Global analysis of prokaryotic tRNA-derived cyclodipeptide biosynthesis. BMC Genom. 2018, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Shaw-Reid, C.A.; Kelleher, N.L.; Losey, H.C.; Gehring, A.M.; Berg, C.; Walsh, C.T. Assembly line enzymology by multimodular nonribosomal peptide synthetases: The thioesterase domain of E. coil EntF catalyzes both elongation and cyclolactonization. Chem. Biol. 1999, 6, 385–400. [Google Scholar] [CrossRef]
- Mishra, A.K.; Choi, J.; Choi, S.J.; Baek, K.H. Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules 2017, 22, 1796. [Google Scholar] [CrossRef]
- Gil de la Fuente, A.; Godzien, J.; Fernández López, M.; Rupérez, F.J.; Barbas, C.; Otero, A. Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef]
- Hartmann, E.M.; Allain, F.; Gaillard, J.C.; Pible, O.; Armengaud, J. Taking the shortcut for high-throughput shotgun proteomic analysis of bacteria. Methods Mol. Biol. 2014, 1197, 275–285. [Google Scholar]
- Klein, G.; Mathé, C.; Biola-Clier, M.; Devineau, S.; Drouineau, E.; Hatem, E.; Marichal, L.; Alonso, B.; Gaillard, J.C.; Lagniel, G.; et al. RNA-binding proteins are a major target of silica nanoparticles in cell extracts. Nanotoxicology 2016, 10, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Huntemann, M.; Ivanova, N.N.; Mavromatis, K.; Tripp, H.J.; Paez-Espino, D.; Palaniappan, K.; Szeto, E.; Pillay, M.; Chen, I.-M.A.; Pati, A.; et al. The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4). Stand. Genom. Sci. 2015, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, K.; Chen, I.M.A.; Chu, K.; Ratner, A.; Seshadri, R.; Kyrpides, N.C.; Ivanova, N.N.; Mouncey, N.J. IMG-ABC v.5.0: An update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2020, 48, D422–D430. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1, 2 and mix 3a and 3b are available from the authors. |

| Compound | Molecular Formula | Molecular Weight (DB) g/mol | Mass Error (ppm) | Observed Ion | Description |
|---|---|---|---|---|---|
| Peptides | |||||
| Cyclo (l-leucyl-l-phenylalanyl) | C15H20N2O2 | 260.1525 | 3 | M + H | Cyclic dipeptides or diketopiperazines (DKPs) are a large class of natural products with biological activities. Dipeptide and tripeptides are incomplete breakdown product of protein digestion or protein catabolism. |
| Cyclo (l-leucyl-l-leucyl) | C12H22N2O2 | 226.1681 | 4 | M + H | |
| Cyclo (l-Pro-l-Tyr) | C14H16N2O3 | 260.151 | 1 | M + H | |
| Cyclo (l-Ala-l-Pro) | C8H12N2O2 | 168.07 | 1 | M + H | |
| Aspartyl-Lysine//Gly Val Ser | C10H19N3O5 | 261.1325 | 3 | M + Na | |
| Arg Gln Gln | C16H30N8O6 | 430.2288 | 8 | M + Na | |
| Lysyl-Asparagine | C10H20N4O4 | 260.1485 | 6 | M + H | |
| Tyr Trp Ile | C26H32N4O5 | 480.2373 | 3 | M + H | |
| His Phe Trp | C26H28N6O4 | 488.2172 | 4 | M + H | |
| Trp Tyr Phe | C29H30N4O5 | 514.2216 | 4 | M + H | |
| Alkaloids | |||||
| Anthcolorin G | C33H47NO4 | 521.7305 | 1 | M + H | |
| Fatty Acyls | |||||
| Dodecanamide | C12H25NO | 199.1936 | 1 | M + H | Fatty acids comprise components of the dual-membrane envelope in bacteria. |
| Linoleic acid | C18H32O2 | 280.2402 | 4 | M + H | |
| α-Linolenic acid | C18H30O2 | 278.2246 | 4 | M + H | |
| N-palmitoyl isoleucine//Arachidoyl glycine | C22H43NO3 | 369.3243 | 5 | M + H | |
| Oleamide | C18H35NO | 281.2719 | 4 | M + H | |
| Palmitic acid | C16H32O2 | 256.2402 | 5 | M + H | |
| Oleic acid | C18H34O2 | 282.2559 | 3 | M + H | |
| N-Butyl arachidonoyl amine | C24H41NO | 359.3188 | 4 | M + H | |
| Tetradecenoyl-CoA | C35H60N7O17P3S | 975.2979 | 2 | M + Na | |
| Glycerolipids | |||||
| Monoacylglyceride (16:0) | C19H38O4 | 330.277 | 5 | M + H | Glycerolipids are complex lipids formed by the condensation of one, two, or three fatty acid molecules on glycerol. |
| Diglyceride (44:6) | C47H80O5 | 724.6006 | 6 | M + Na | |
| Glycerophospholipids | |||||
| Lysophosphatidylcholine (LPC) (16:1)//Phosphatidylethanolamine (PE) (19:1) | C24H48NO7P | 493.3168 | 4 | M + H | Glycerophospholipids comprise components of the dual-membrane envelope of Gram-negative bacteria with biological functions in protein binding, transport of proteins across inner membranes. |
| Lysophosphatidylcholine (O-18:0) | C26H52NO7P | 521.3481 | 4 | M + H | |
| Phosphatidylethanolamine (39:5) | C44H78NO8P | 779.5465 | 2 | M + H | |
| Phosphatidylethanolamine (41:6) | C46H80NO8P | 805.5622 | 2 | M + H | |
| Phosphatidylethanolamine (39:4) | C44H80NO8P | 781.5622 | 6 | M + H | |
| Phosphatidylethanolamine (41:5) | C46H82NO8P | 807.5778 | 1 | M + H | |
| Phosphatidylglycerol (31:3) | C37H67O10P | 702.4471 | 5 | M + H-H20 | |
| Cytotoxic Activity IC50 | ||
|---|---|---|
| Strain/Fraction | Cellular Line | |
| MCF-7 | 4T1 | |
| Strain USBA 371 | 4.5 ± 1.3 µg/mL | 5.5 ± 3.44 µg/mL |
| Compound 1 | 87.74 ± 2.32 | 57.09 ± 2.11 µM |
| Compound 2 | 890.53 ± 3.45 µM | 875.93 ± 5.14 µM |
| Compound mix 3a and 3b | 16.10 ± 1.66 µg/mL | 13.41 ± 1.64 µg/mL |
| Cell Line | Cytotoxic Activity of Fractions IC50 (µM) | Selectivity Index (SI) | |
|---|---|---|---|
| Compound 1 | |||
| Murine Breast Cancer | 4T1 H17 * | 40.38 ± 1.94 | 7.78 |
| TSA | 196.66 ± 4.18 | 1.59 | |
| Murine Melanoma | B16 | 80.87 ± 3.67 | 3.88 |
| Murine Colon | MCA 38 | 29.85 ± 1.55 | 10.52 |
| Human Uterus Sarcome | MES–SA/DX5P-Pgp (+) | 225.28 ± 1.23 | 1.39 |
| MES–SA/DX5P-Ppg (−) | 91.32 ± 5.64 | 3.44 | |
| Lung | 3LL | 51.71 ± 0.55 | 6.07 |
| Myeloid Acute Leukemia | K562 | ND | ND |
| U937 | ND | ND | |
| Murine Fibroblast | 3T3 | 314.30 ± 3.41 | ND |
| Biosynthetic Gene Cluster (BGC) Type * | Labrenzia Aggregata Strain USBA 371 | Labrenzia Aggregata CECT 4801 | Labrenzia Aggregata LZB033 | Labrenzia Marina DSM 17023 | Labrenzia Suaedae DSM 22153 | Labrenzia Alba CECT 5096 | Labrenzia Alba CECT 7551 | Labrenzia Alba VG12 | Labrenzia Alba CECT 5095 | Labrenzia Alba CECT 5094 |
|---|---|---|---|---|---|---|---|---|---|---|
| Betalactone | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| Bacteriocin | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| Ectoine, Hserlactone | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NAGGN | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| T1PKS | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| Terpene | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| NRPS | 2 | 4 | 6 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| TransAT-PKS, T3PKS-NRPS | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| Thiopeptide | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| NRPS-T1PKS | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| TOTAL BGC | 8 | 9 | 11 | 8 | 8 | 5 | 4 | 6 | 5 | 5 |
| Biosynthetic Gene Cluster (BGC) Type | From | To | Most Similar Known Cluster | Similarity | MIBiG BGC-ID |
|---|---|---|---|---|---|
| NRPS-T1PKS | 872,702 | 926,444 | Sporolide-nrps-t1pks | 4% | BGC0000150 |
| T1PKS | 963,299 | 1,010,906 | ND | ||
| Bacteriocin | 125,918 | 136,814 | ND | ||
| Betalactone | 82,927 | 105,536 | ND | ||
| Terpene | 492,697 | 513,548 | ND | ||
| NRPS | 405,475 | 443,932 | Turnerbactin | 30% | BGC0000451 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Cárdenas, C.; Rojas, L.Y.; Fiorentino, S.; Cala, M.P.; Díaz, J.I.; Ramos, F.A.; Armengaud, J.; Restrepo, S.; Baena, S. Bioactive Potential of Extracts of Labrenzia aggregata Strain USBA 371, a Halophilic Bacterium Isolated from a Terrestrial Source. Molecules 2020, 25, 2546. https://doi.org/10.3390/molecules25112546
Díaz-Cárdenas C, Rojas LY, Fiorentino S, Cala MP, Díaz JI, Ramos FA, Armengaud J, Restrepo S, Baena S. Bioactive Potential of Extracts of Labrenzia aggregata Strain USBA 371, a Halophilic Bacterium Isolated from a Terrestrial Source. Molecules. 2020; 25(11):2546. https://doi.org/10.3390/molecules25112546
Chicago/Turabian StyleDíaz-Cárdenas, Carolina, Laura Yinneth Rojas, Susana Fiorentino, Monica P. Cala, Jorge I Díaz, Freddy A. Ramos, Jean Armengaud, Silvia Restrepo, and Sandra Baena. 2020. "Bioactive Potential of Extracts of Labrenzia aggregata Strain USBA 371, a Halophilic Bacterium Isolated from a Terrestrial Source" Molecules 25, no. 11: 2546. https://doi.org/10.3390/molecules25112546
APA StyleDíaz-Cárdenas, C., Rojas, L. Y., Fiorentino, S., Cala, M. P., Díaz, J. I., Ramos, F. A., Armengaud, J., Restrepo, S., & Baena, S. (2020). Bioactive Potential of Extracts of Labrenzia aggregata Strain USBA 371, a Halophilic Bacterium Isolated from a Terrestrial Source. Molecules, 25(11), 2546. https://doi.org/10.3390/molecules25112546








