Functional Expression and Characterization of a Panel of Cobalt and Iron-Dependent Nitrile Hydratases
Abstract
1. Introduction
2. Results and Discussion
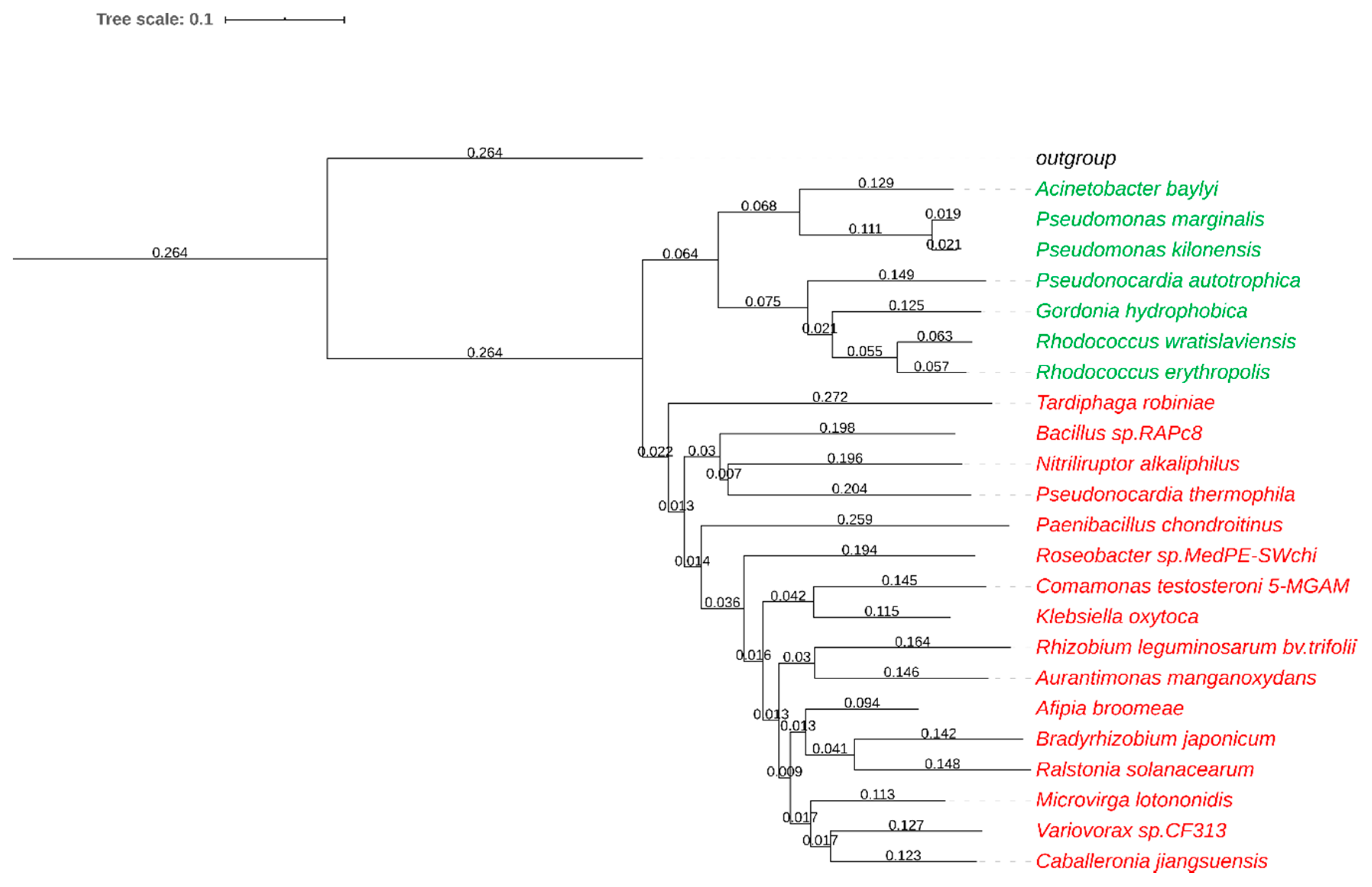
2.1. Identification and Production of Nitrile Hydratases
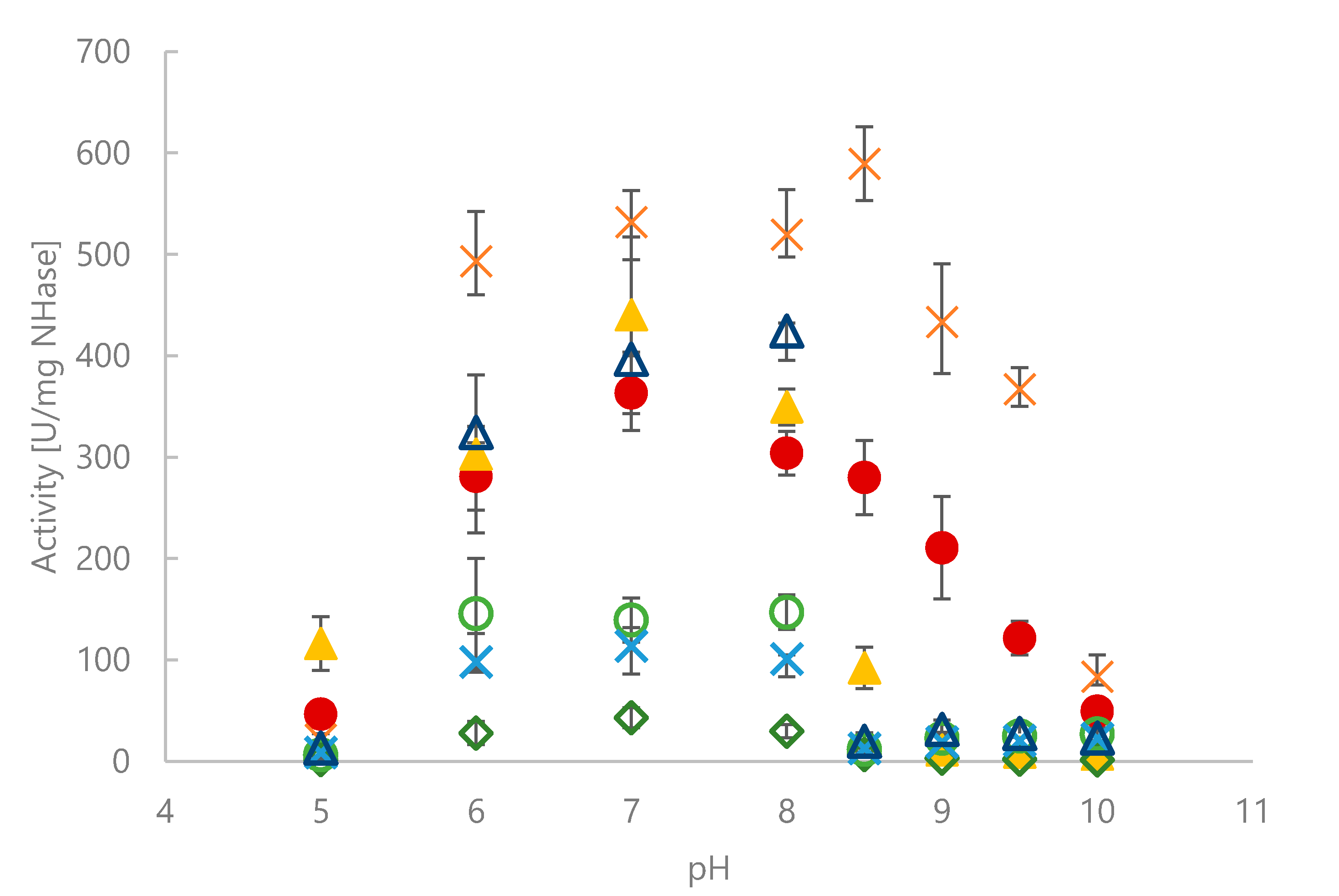
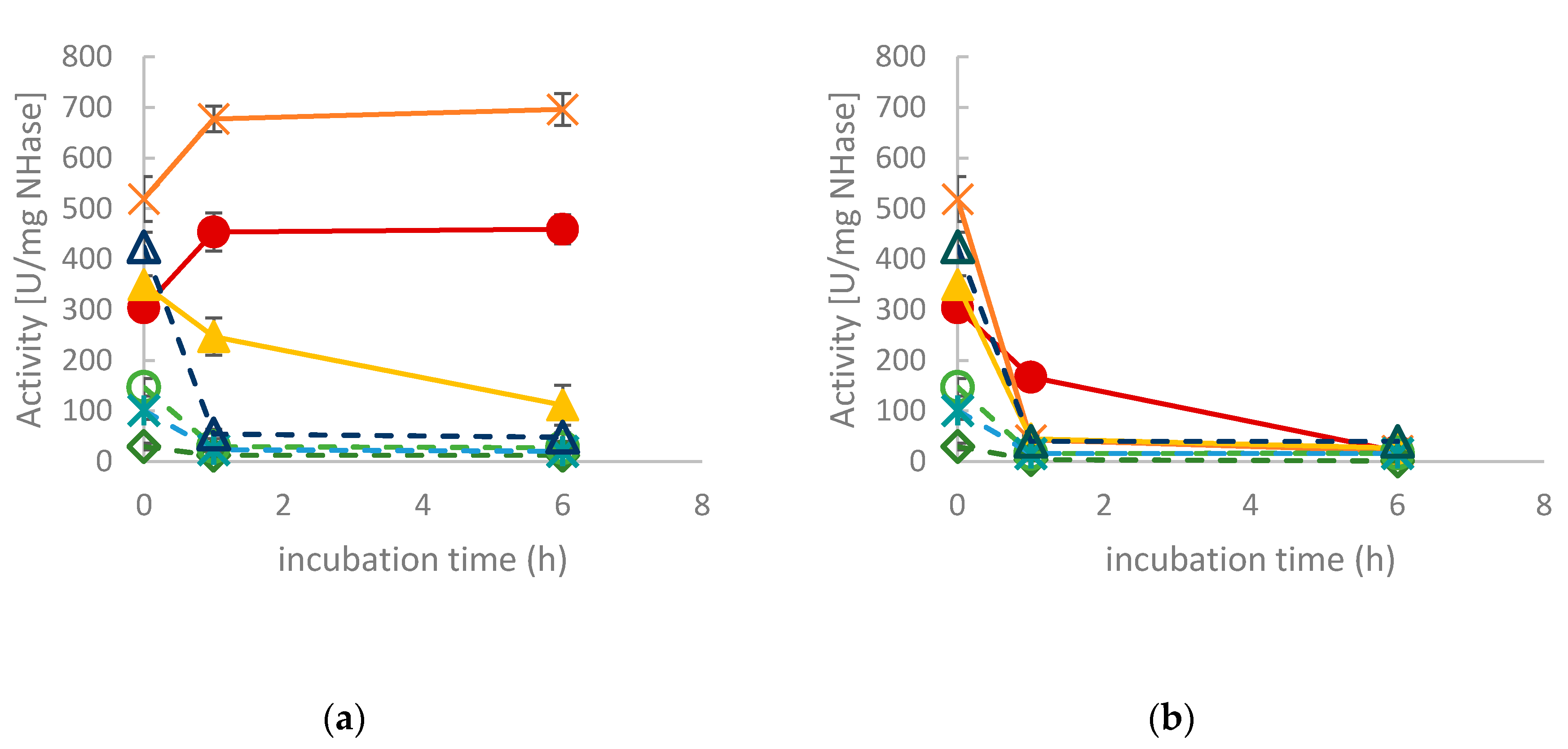
2.2. Activity for Methacrylonitrile and Characterization of pH Optimum and Stability
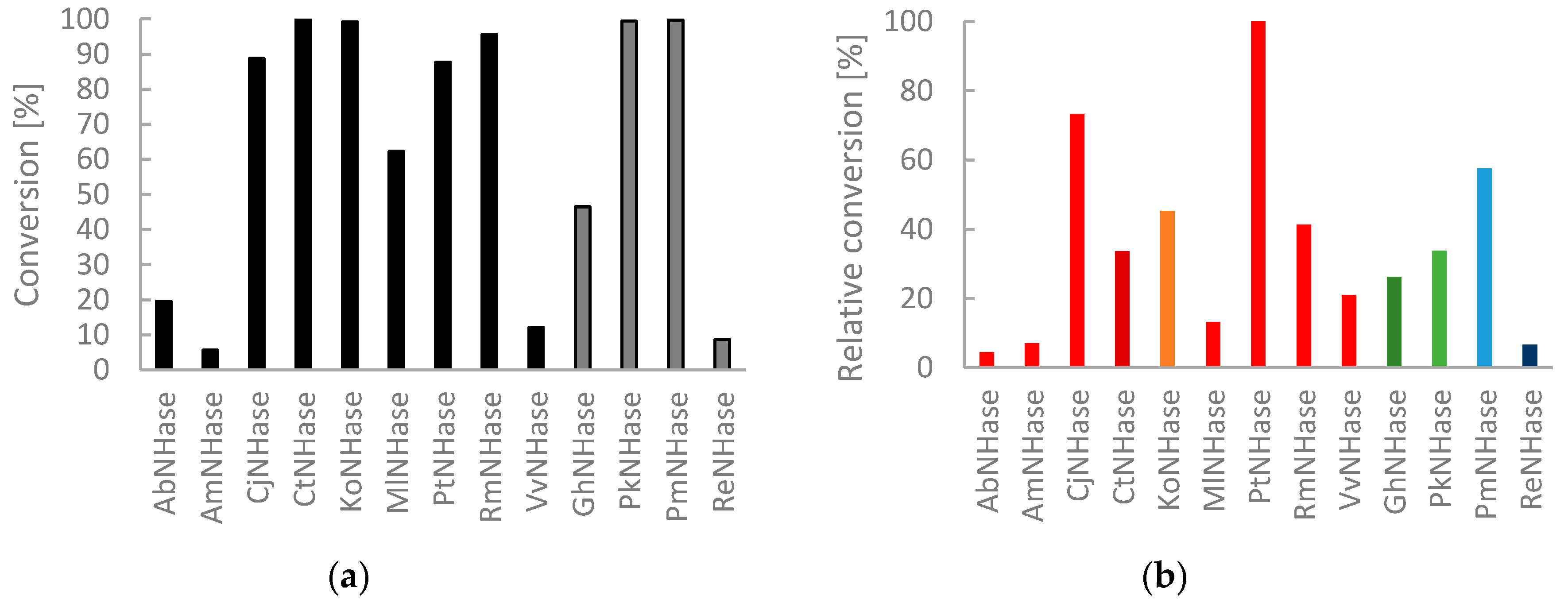
2.3. Exploration of Substrate Scope
3. Materials and Methods
3.1. General
3.2. Sequence Identification and Cloning
3.3. Protein Expression
3.4. Spectrophotometric MAN Assay
3.5. Synthesis
3.6. Chromatographic Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Prasad, S.; Bhalla, T.C. Nitrile hydratases (NHases): At the interface of academia and industry. Biotechnol. Adv. 2010, 28, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Mashweu, A.R.; Chhiba-Govindjee, V.P.; Bode, M.L.; Brady, D. Substrate Profiling of the Cobalt Nitrile Hydratase from Rhodococcus rhodochrous ATCC BAA 870. Molecules 2020, 25, 238. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.-C.; Zheng, Y.-G.; Shen, Y.-C. Acrylamide, Microbial Production by Nitrile Hydratase. In Encyclopedia of Industrial Biotechnology; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2010; pp. 1–39. [Google Scholar]
- Cheng, Z.; Xia, Y.; Zhou, Z. Recent Advances and Promises in Nitrile Hydratase: From Mechanism to Industrial Applications. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Supreetha, K.; Rao, S.N.; Srividya, D.; Anil, H.S.; Kiran, S. Advances in cloning, structural and bioremediation aspects of nitrile hydratases. Mol. Biol. Rep. 2019, 46, 4661–4673. [Google Scholar] [CrossRef]
- Wu, S.; Fallon, R.D.; Payne, M.S. Over-production of stereoselective nitrile hydratase from Pseudomonas putida 5B in Escherichia coli: Activity requires a novel downstream protein. Appl. Microbiol. Biotechnol. 1997, 48, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Yukl, E.T.; Wilmot, C.M. Cofactor biosynthesis through protein post-translational modification. Curr. Opin. Chem. Biol. 2012, 16, 54–59. [Google Scholar] [CrossRef]
- Guo, F.-M.; Wu, J.-P.; Yang, L.-R.; Xu, G. Overexpression of a nitrile hydratase from Klebsiella oxytoca KCTC 1686 in Escherichia coli and its biochemical characterization. Biotechnol. Bioprocess Eng. 2015, 20, 995–1004. [Google Scholar] [CrossRef]
- Cameron, R.A.; Sayed, M.; Cowan, D.A. Molecular analysis of the nitrile catabolism operon of the thermophile Bacillus pallidus RAPc8. Biochim. Biophys. Acta 2005, 1725, 35–46. [Google Scholar] [CrossRef]
- Miyanaga, A.; Fushinobu, S.; Ito, K.; Wakagi, T. Crystal Structure of Cobalt-Containing Nitrile Hydratase. Biochem. Biophys. Res. Commun. 2001, 288, 1169–1174. [Google Scholar] [CrossRef]
- Pei, X.; Zhang, H.; Meng, L.; Xu, G.; Yang, L.; Wu, J. Efficient cloning and expression of a thermostable nitrile hydratase in Escherichia coli using an auto-induction fed-batch strategy. Process Biochem. 2013, 48, 1921–1927. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Yang, Z.; Wang, A.; Yang, L.; Wu, J. Identification and functional analysis of the activator gene involved in the biosynthesis of Co-type nitrile hydratase from Aurantimonas manganoxydans. J. Biotechnol. 2017, 251, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Yamaki, T.; Oikawa, T.; Ito, K.; Nakamura, T. Cloning and sequencing of a nitrile hydratase gene from Pseudonocardia thermophila JCM3095. J. Ferment. Bioeng. 1997, 83, 474–477. [Google Scholar] [CrossRef]
- Van Pelt, S.; Zhang, M.; Otten, L.G.; Holt, J.; Sorokin, D.Y.; van Rantwijk, F.; Black, G.W.; Perry, J.J.; Sheldon, R.A. Probing the enantioselectivity of a diverse group of purified cobalt-centred nitrile hydratases. Org. Biomol. Chem. 2011, 9. [Google Scholar] [CrossRef]
- Petrillo, K.L.; Wu, S.; Hann, E.C.; Cooling, F.B.; Ben-Bassat, A.; Gavagan, J.E.; DiCosimo, R.; Payne, M.S. Over-expression in Escherichia coli of a thermally stable and regio-selective nitrile hydratase from Comamonas testosteroni 5-MGAM-4D. Appl. Microbiol. Biotechnol. 2005, 67, 664–670. [Google Scholar] [CrossRef]
- Murakami, T.; Nojiri, M.; Nakayama, H.; Dohmae, N.; Takio, K.; Odaka, M.; Endo, I.; Nagamune, T.; Yohda, M. Post-translational modification is essential for catalytic activity of nitrile hydratase. Protein Sci. 2000, 9, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yang, S.; Huang, G. Design, synthesis and biological activity of pyrazinamide derivatives for anti-Mycobacterium tuberculosis. J. Enzyme Inhib. Med. Chem. 2017, 32, 1183–1186. [Google Scholar] [CrossRef]
- Mauger, J.; Nagasawa, T.; Yamada, H. Nitrile hydratase-catalyzed production of isonicotinamide, picolinamide and pyrazinamide from 4-cyanopyridine, 2-cyanopyridine and cyanopyrazine in Rhodococcus rhodochrous J1. J. Biotechnol. 1988, 8, 87–95. [Google Scholar] [CrossRef]
- Chhiba, V.; Bode, M.L.; Mathiba, K.; Kwezi, W.; Brady, D. Enantioselective biocatalytic hydrolysis of β-aminonitriles to β-amino-amides using Rhodococcus rhodochrous ATCC BAA-870. J. Mol. Catal. B Enzym. 2012, 76, 68–74. [Google Scholar] [CrossRef]
- Preiml, M.; Hillmayer, K.; Klempier, N. A new approach to beta-amino acids: Biotransformation of N-protected beta-amino nitriles. Tetrahedron Lett. 2003, 44, 5057–5059. [Google Scholar] [CrossRef]
- Preiml, M.; Hönig, H.; Klempier, N. Biotransformation of β-amino nitriles: The role of the N-protecting group. J. Mol. Catal. B Enzym. 2004, 29, 115–121. [Google Scholar] [CrossRef]
- Mareya, T.M.; Coady, D.T.M.; O’Reilly, D.C.; Kinsella, D.M.; Coffey, D.L.; Lennon, D.C.M. Process Optimisation Studies and Aminonitrile Substrate Evaluation of Rhodococcus erythropolis SET1, A Nitrile Hydrolyzing Bacterium. ChemistryOpen 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.; Cramp, R.; Pereira, R.; Graham, D.; Almatawah, Q. Biochemistry and biotechnology of mesophilic and thermophilic nitrile metabolizing enzymes. Extremophiles 1998, 2, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Frederick, J.; Hennessy, F.; Horn, U.; de la Torre Cortés, P.; van den Broek, M.; Strych, U.; Willson, R.; Hefer, C.A.; Daran, J.-M.G.; Sewell, T.; et al. The complete genome sequence of the nitrile biocatalyst Rhodococcus rhodochrous ATCC BAA-870. BMC Genomics 2020, 21, 3. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 3a and 3b are available from the authors. |


| NHase | Estimated NHase Content in CFE [mg/mL] | Activity[µmol min−1 mg−1]1 | Specific Activity [µmol min−1 mg−1]2 |
|---|---|---|---|
| Fe-type AcNHase | <0.1 | 3 | 0 |
| Fe-type GhNHase | 3.4 | 18 | 55 |
| Fe-type PkNHase | 2.9 | 182 | 615 |
| Fe-type PmNHase | 1.9 | 52 | 313 |
| Fe-type ReNHase | 2.9 | 248 | 605 |
| Co-type AbNHase | 1.6 | 14 | 53 |
| Co-type AmNHase | 0.8 | 3 | 37 |
| Co-type BjNHase | 0.1 | 2 | 14 |
| Co-type BrNHase | 0.1 | 4 | 20 |
| Co-type CjNHase | 2.9 | 23 | 88 |
| Co-type CtNHase | 3.4 | 118 | 336 |
| Co-type KoNHase | 2.9 | 165 | 613 |
| Co-type MlNHase | 3.9 | 30 | 98 |
| Co-type NaNHase | 1.0 | 62 | 505 |
| Co-type PcNHase | 1.7 | 1 | 5 |
| Co-type PtNHase | 1.2 | 41 | 243 |
| Co-type RlNHase 3 | 0.4 | 0 | 0 |
| Co-type RmNHase | 4.2 | 45 | 138 |
| Co-type RsNHase | 2.0 | 0 | 0 |
| Co-type TrNHase | 1.7 | 1 | 3 |
| Co-type VvNHase | 1.5 | 8 | 54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grill, B.; Glänzer, M.; Schwab, H.; Steiner, K.; Pienaar, D.; Brady, D.; Donsbach, K.; Winkler, M. Functional Expression and Characterization of a Panel of Cobalt and Iron-Dependent Nitrile Hydratases. Molecules 2020, 25, 2521. https://doi.org/10.3390/molecules25112521
Grill B, Glänzer M, Schwab H, Steiner K, Pienaar D, Brady D, Donsbach K, Winkler M. Functional Expression and Characterization of a Panel of Cobalt and Iron-Dependent Nitrile Hydratases. Molecules. 2020; 25(11):2521. https://doi.org/10.3390/molecules25112521
Chicago/Turabian StyleGrill, Birgit, Maximilian Glänzer, Helmut Schwab, Kerstin Steiner, Daniel Pienaar, Dean Brady, Kai Donsbach, and Margit Winkler. 2020. "Functional Expression and Characterization of a Panel of Cobalt and Iron-Dependent Nitrile Hydratases" Molecules 25, no. 11: 2521. https://doi.org/10.3390/molecules25112521
APA StyleGrill, B., Glänzer, M., Schwab, H., Steiner, K., Pienaar, D., Brady, D., Donsbach, K., & Winkler, M. (2020). Functional Expression and Characterization of a Panel of Cobalt and Iron-Dependent Nitrile Hydratases. Molecules, 25(11), 2521. https://doi.org/10.3390/molecules25112521






