Extraction, Antioxidant Capacity, 5-Lipoxygenase Inhibition, and Phytochemical Composition of Propolis from Eastern Canada
Abstract
1. Introduction
2. Results and Discussion
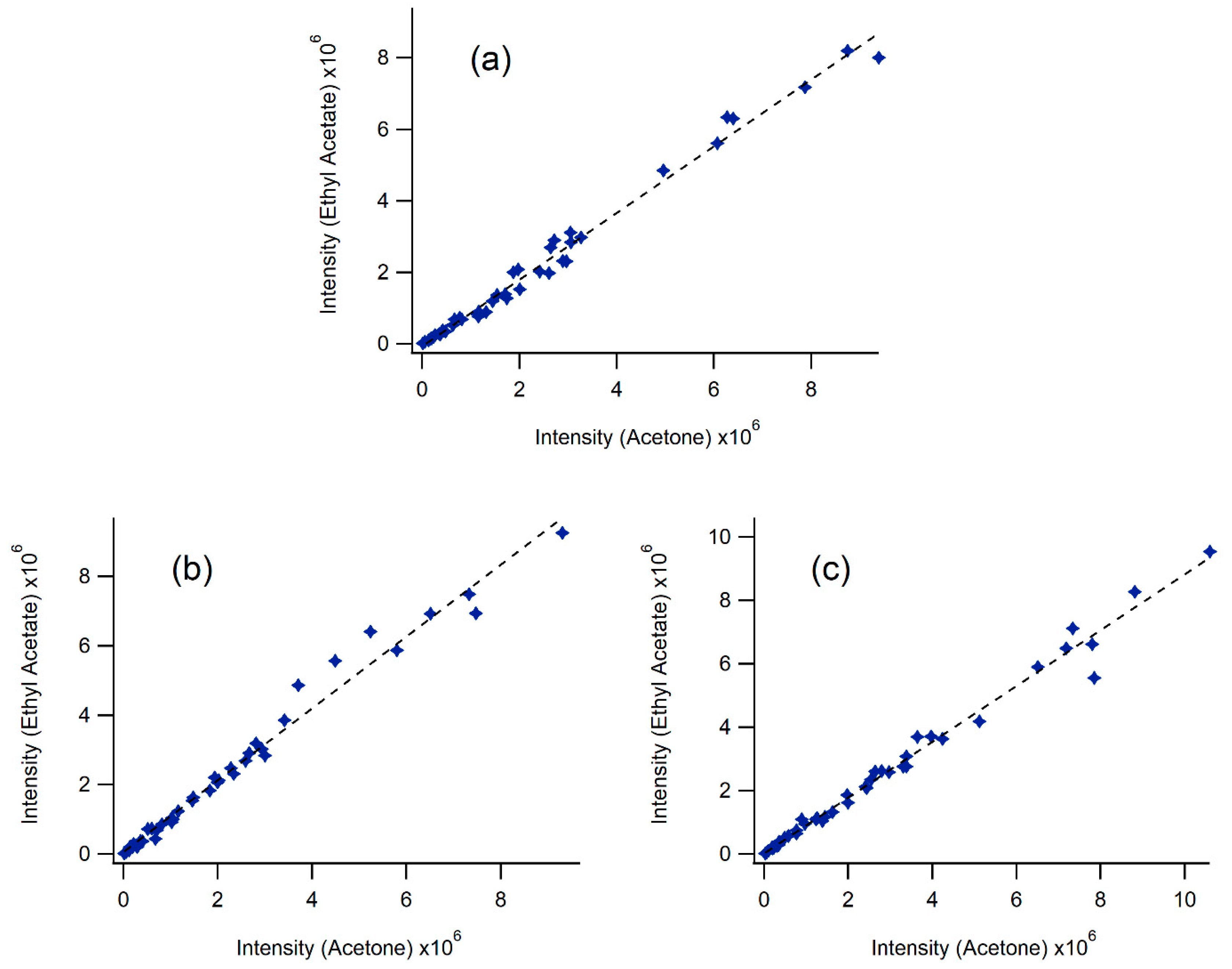
2.1. Extractions
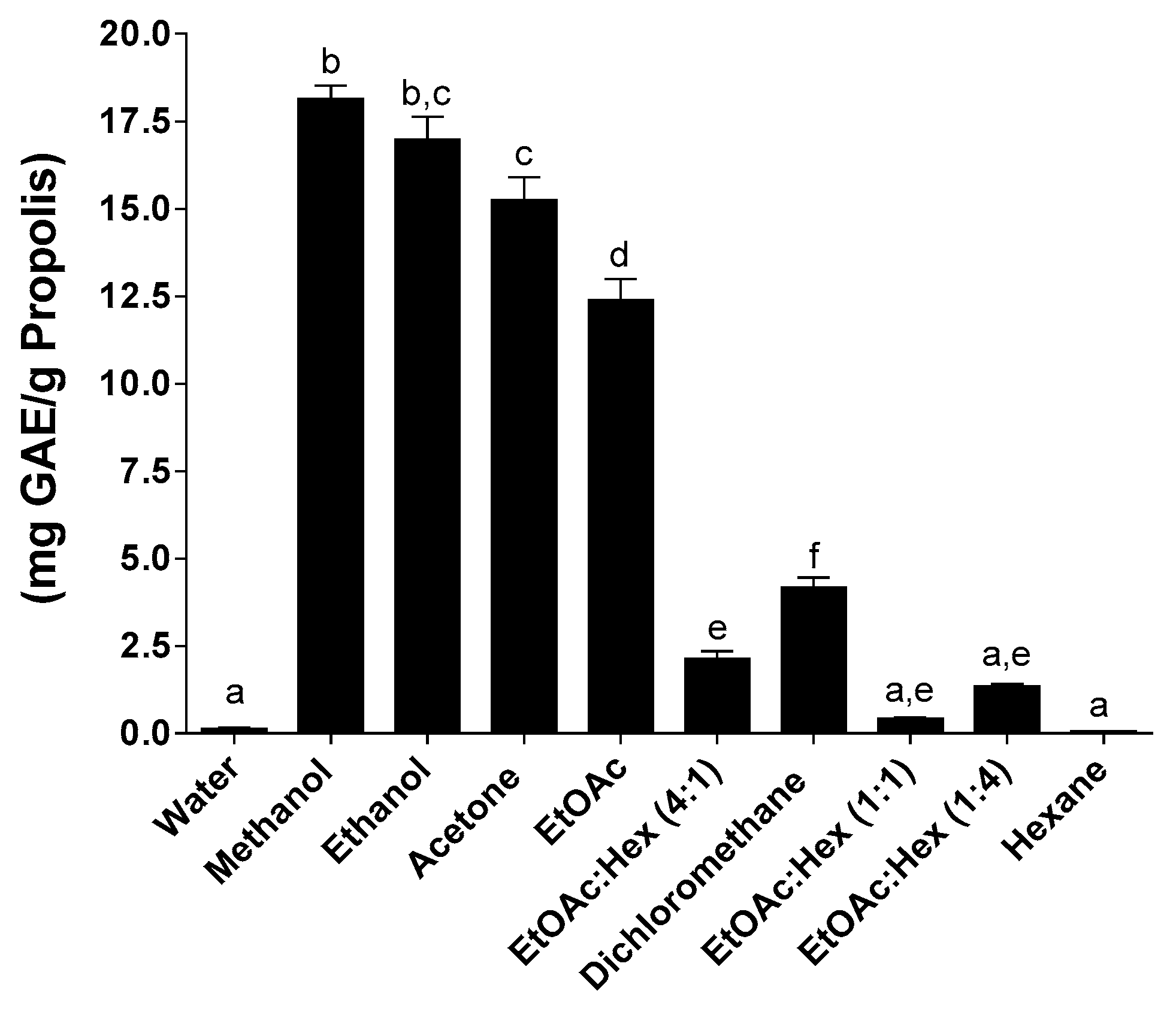
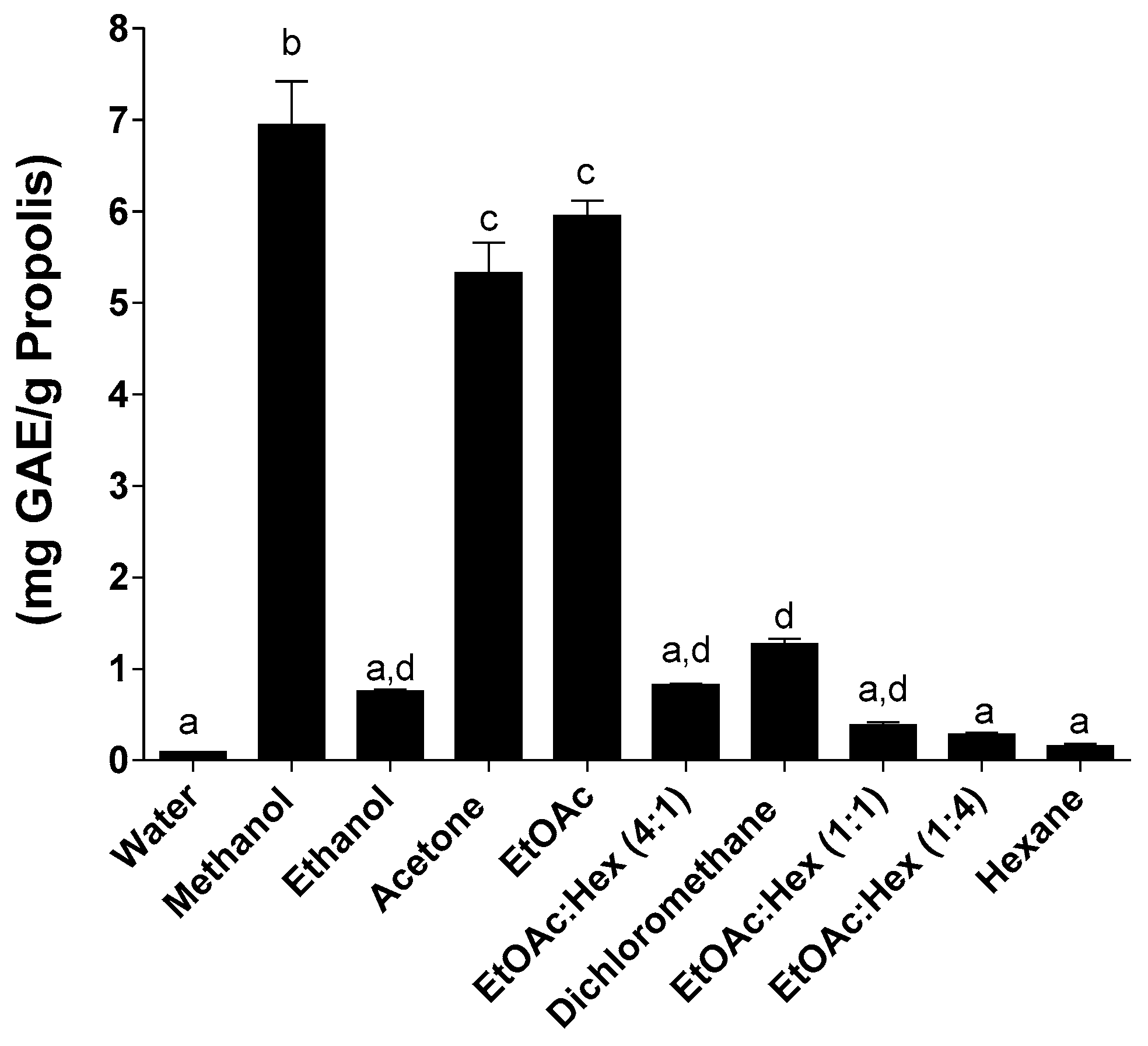
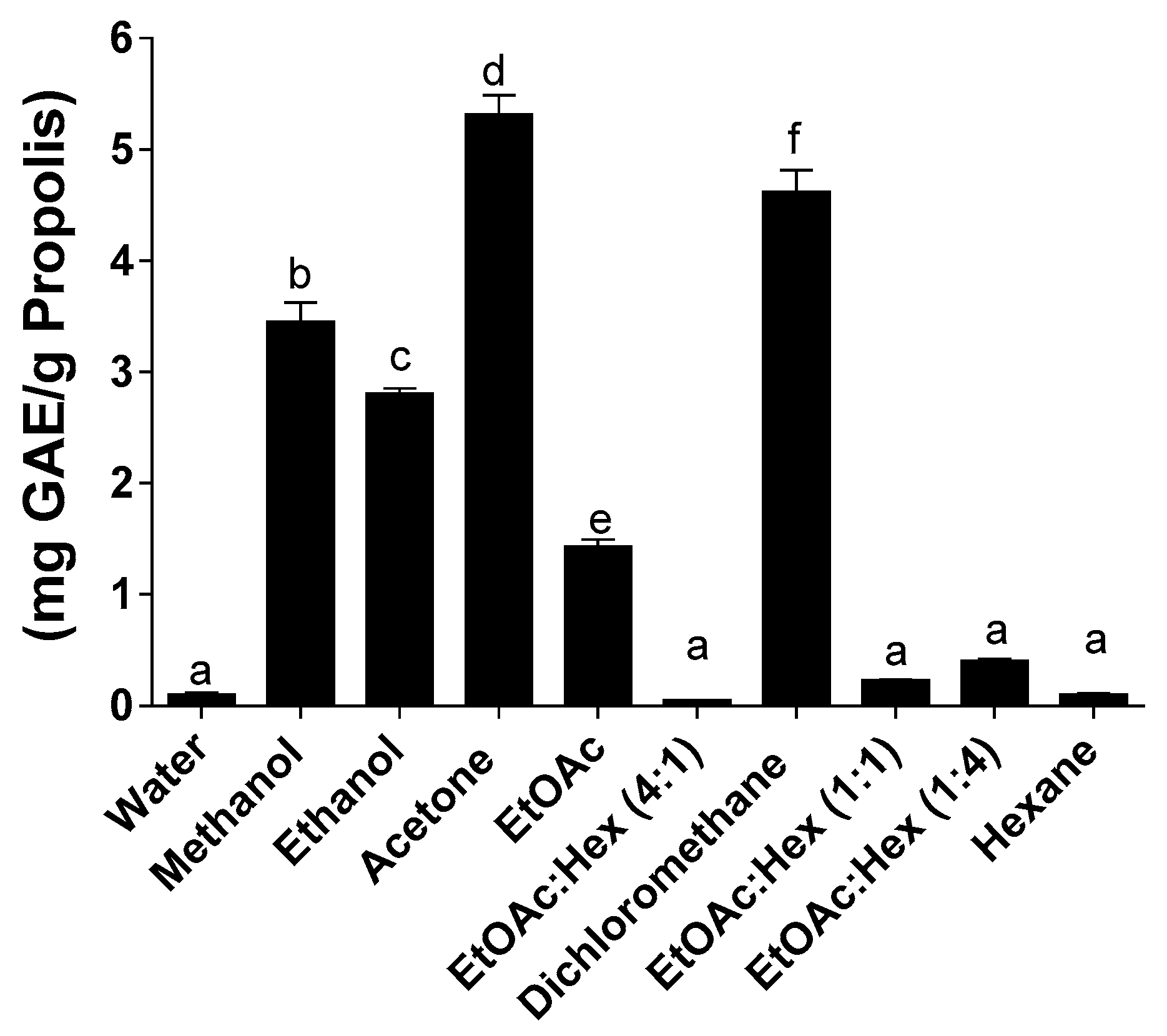
2.2. Total Phenolic Content
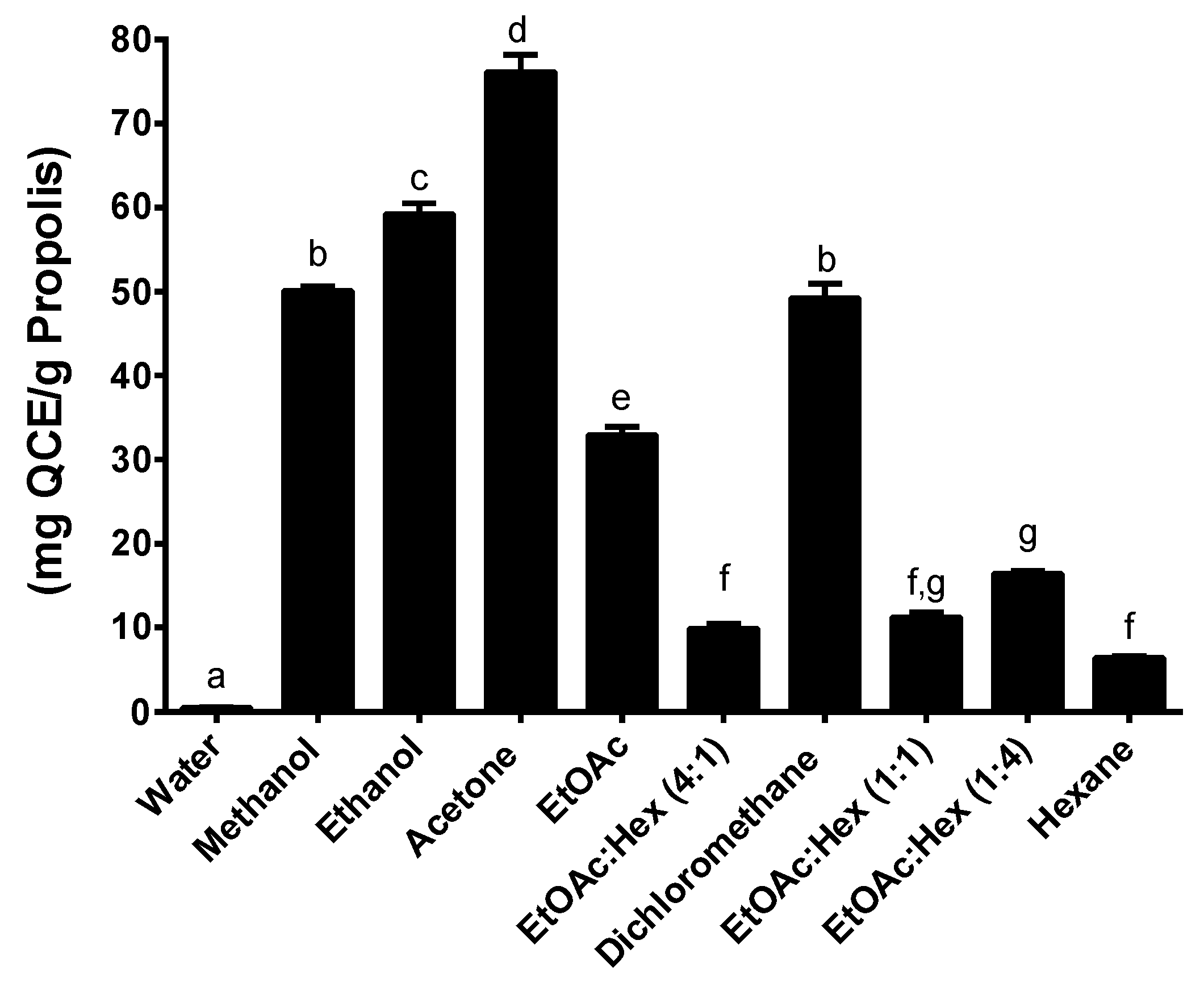
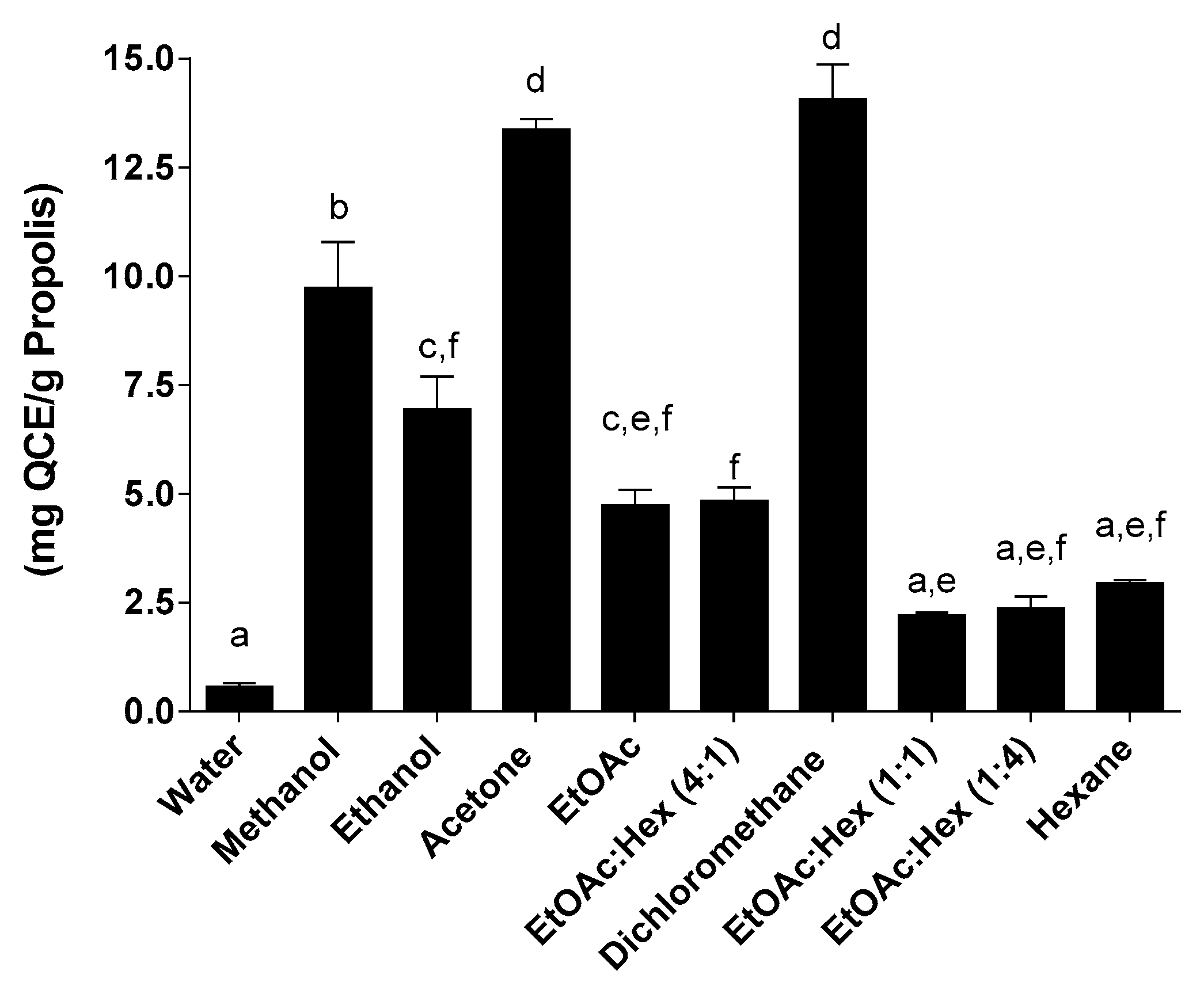
2.3. Total Flavonoid Content
2.4. Antioxidant Activity
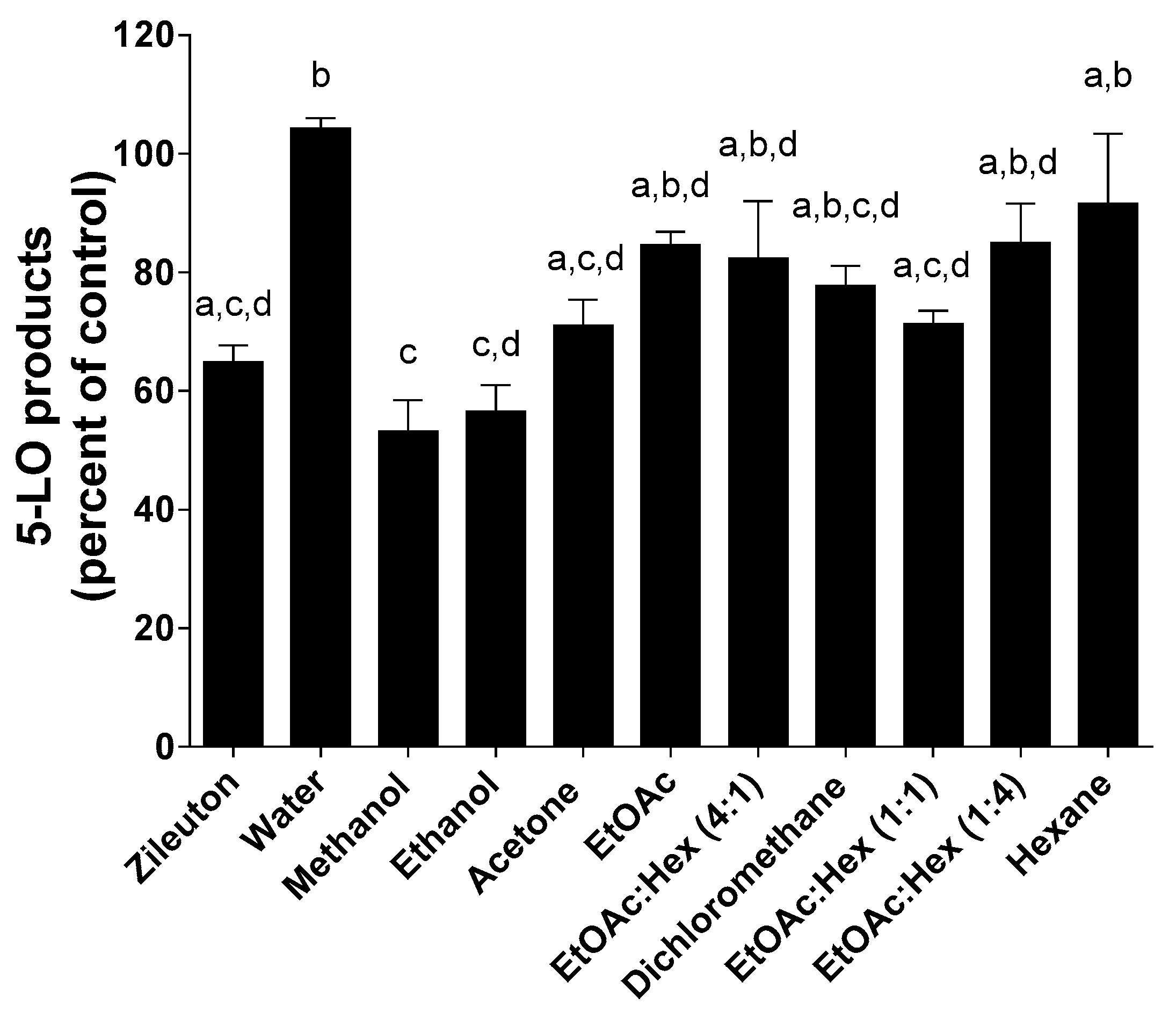
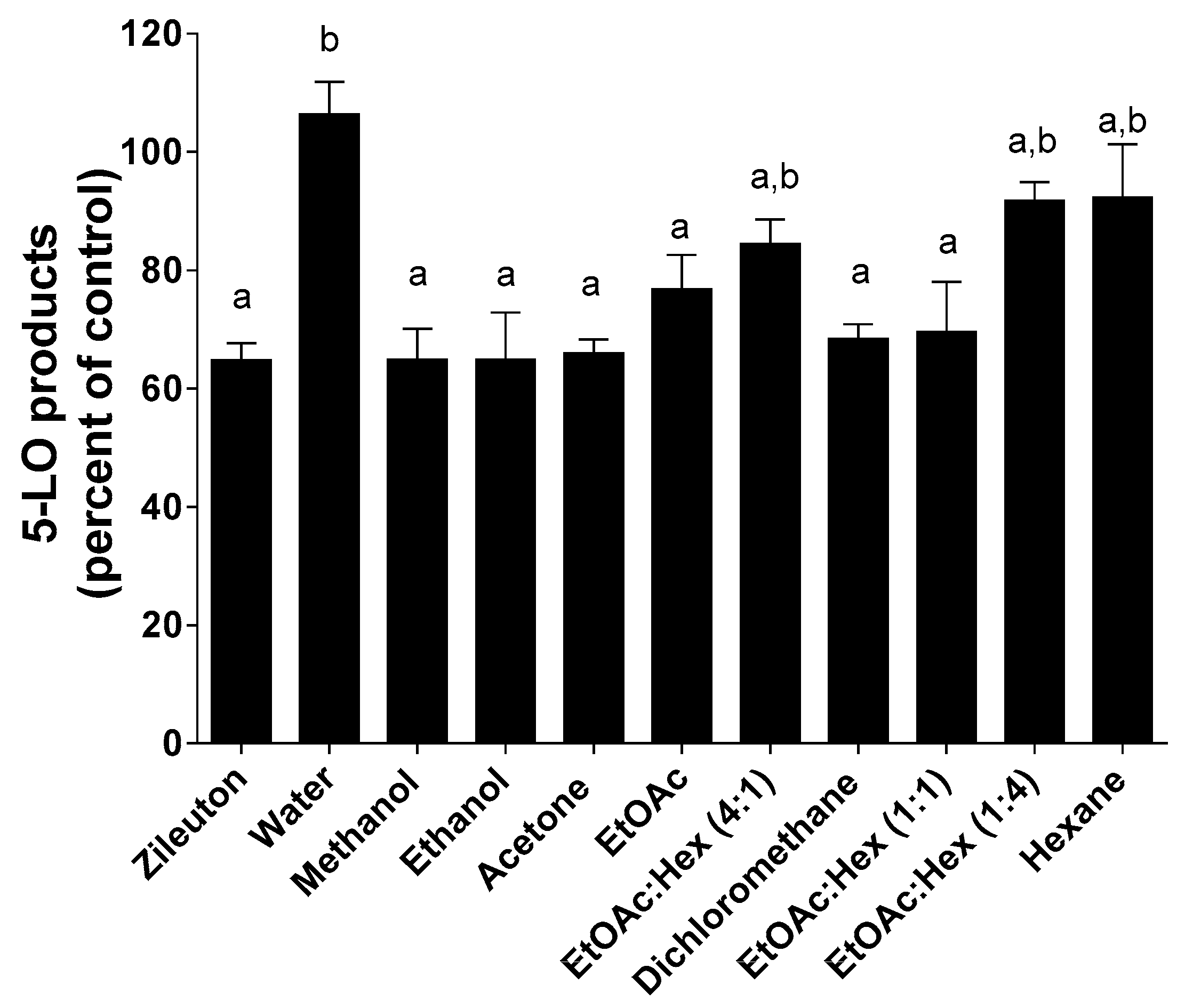
2.5. 5-LO Product Biosynthesis Assays in HEK293 Cells
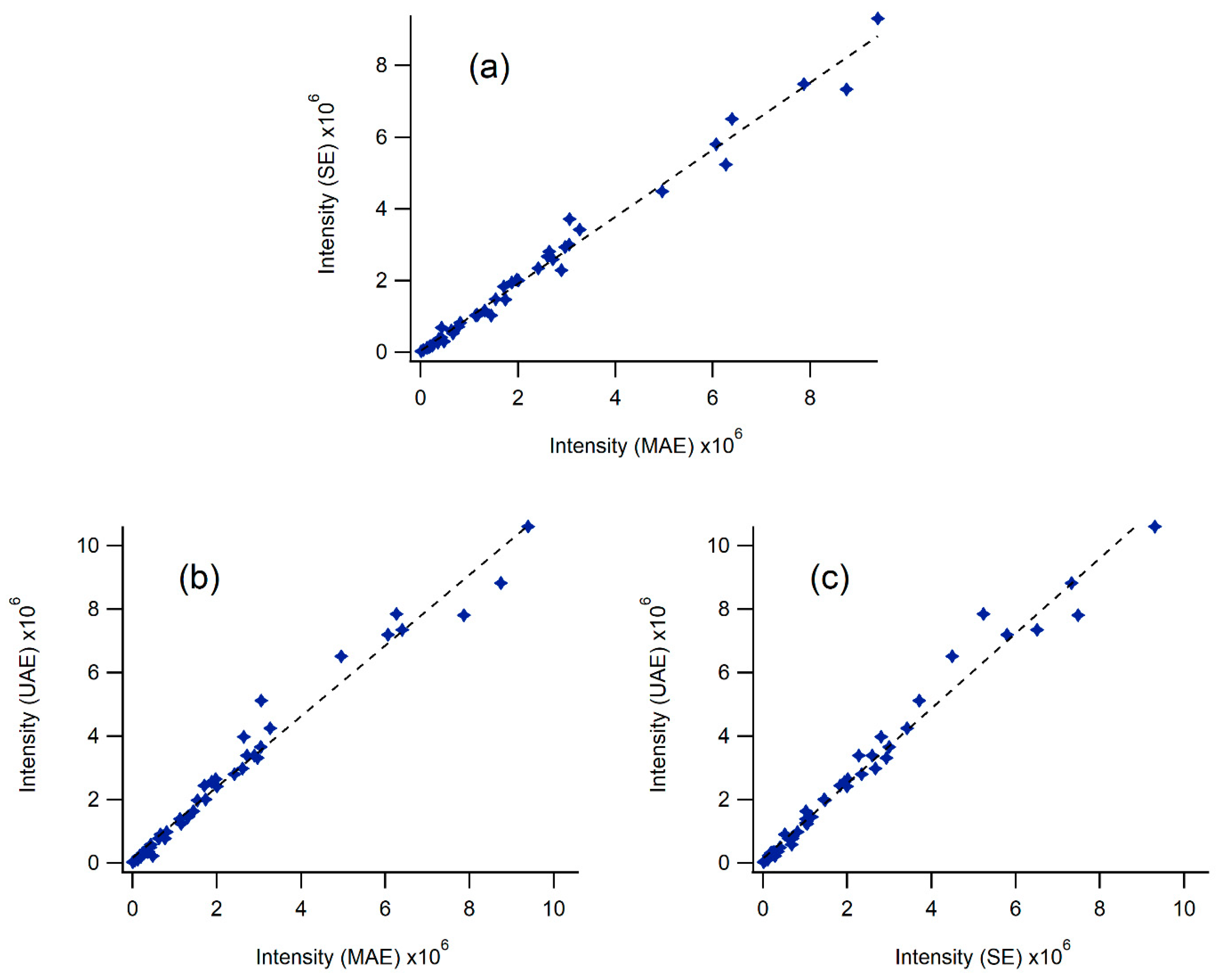
2.6. Identification of Chemical Compounds in Propolis from Eastern Canada Obtained Following SE, MAE and UAE
2.7. Semi-Quantitative Analysis of 46 Compounds in Propolis by HPLC-MS Analysis
3. Materials and Methods
3.1. Materials
3.2. Soxhlet Extraction (SE)
3.3. Ultrasound-Assisted Extraction (UAE)
3.4. Microwave-Assisted Extraction (MAE)
3.5. Total Phenolic Content (TPC)
3.6. Total Flavonoid Content (TFC)
3.7. Scavenging of Free Radicals
3.8. 5-LO Product Biosynthesis Assays in HEK293 Cells
3.9. Identification of Chemical Profiles by LC-MS/MS Analysis
3.10. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ahn, M.R.; Kumazawa, S.; Hamasaka, T.; Bang, K.S.; Nakayama, T. Antioxidant activity and constituents of propolis collected in various regions of Korea. J. Agric. Food Chem. 2004, 52, 7286–7292. [Google Scholar] [CrossRef] [PubMed]
- Trusheva, B.; Popova, M.; Bankova, V.; Somova, S.; Marcucci, M.C.; Miorin, P.L.; Pasin, F.D.R.; Tsvetkova, I. Bioactive constituents of Brazilian red propolis. Evid. Based Complementary Altern. Med. 2006, 3, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Silici, S.; Kutluca, S. Chemical composition and antibacterial activity of propolis collected by three different races of honeybees in the same region. J. Ethnopharmacol. 2005, 99, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, C.P.; Wang, K.; Li, G.Q.; Hu, F.L. Recent advances in the chemical composition of propolis. Molecules 2014, 19, 19610–19632. [Google Scholar] [CrossRef] [PubMed]
- Wilson-Rich, N. Genetic, Individual, and Group Facilitation of Disease Resistance in Honeybees (Apis mellifera) and Two Species of Paper Wasps (P. dominolus and P. fuscatus); Tufts University: Medford, MA, USA, 2011. [Google Scholar]
- Bankova, V. Chemical diversity of propolis and the problem of standardization. J. Ethnopharmacol. 2005, 100, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Celińska-Janowicz, K.; Zaręba, I.; Lazarek, U.; Teul, J.; Tomczyk, M.; Pałka, J.; Miltyk, W. Constituents of propolis: Chrysin, caffeic acid, p-coumarin and ferulic acid iinduce PRODH/POX-dependent apoptosis in human tongue squamous cell carcinoma cell (CAL-27). Front. Pharm. 2018, 9, 336. [Google Scholar] [CrossRef]
- Khodabakhshi, D.; Eskandarinia, A.; Kefayat, A.; Rafienia, M.; Navid, S.; Karbasi, S.; Moshtaghian, J. In vitro and in vivo performance of a propolis-coated polyurethane wound dressing with high porosity and antibacterial efficacy. Colloids Surf B Biointerfaces 2019, 178, 177–184. [Google Scholar] [CrossRef]
- Thamnopoulos, I.I.; Michailidis, G.F.; Fletouris, D.J.; Badeka, A.; Kontominas, M.G.; Angelidis, A.S. Inhibitory activity of propolis against Listeria monocytogenes in milk stored under refrigeration. Food Microbiol. 2018, 73, 168–176. [Google Scholar] [CrossRef]
- Yildirim, A.; Duran, G.G.; Duran, N.; Jenedi, K.; Bolgul, B.S.; Miraloglu, M.; Muz, M. Antiviral activity of Hatay propolis against replication of herpes simplex virus type 1 and 2. Med. Sci. Monit. 2016, 9, 422–430. [Google Scholar] [CrossRef]
- Yangi, B.; Cengiz Ustuner, M.; Dincer, M.; Ozbayer, C.; Tekin, N.; Ustuner, D.; Colak, E.; Kolac, U.K.; Entok, E. Propolis protects endotoxin induced acute lung and liver inflammation through attenuating inflammatory responses and oxidative stress. J. Med. Food 2018, 21, 1096–1105. [Google Scholar] [CrossRef]
- Khurshid, Z.; Naseem, M.; Zafar, M.S.; Najeeb, S.; Zohaib, S. Propolis: A natural biomaterial for dental and oral health care. J. Dent. Res. Dent. Clin. Dent. Prospect. 2017, 11, 265–274. [Google Scholar]
- Henshaw, F.R.; Bolton, T.; Nube, V.; Hood, A.; Veldhoen, D.; Pfrunder, L.; McKew, G.L.; Macleod, C.; McLennan, S.V.; Twigg, S.M. Topical application of the beehive protectant propolis is well tolerated and improves human diabetic foot ulcer healing in a prospective feasibility study. J. Diabetes Its Complicat. 2014, 28, 850–857. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rzepecka-Stojko, A.; Stojko, J.; Kurek-Gorecka, A.; Gorecki, M.; Kabala-Dzok, A.; Kubina, R.; Mozdzierz, A.; Buszman, E. Polyphenols from bee pollen: Structure, absorption, metabolism and biological activity. Molecules 2015, 20, 21732–21749. [Google Scholar] [CrossRef] [PubMed]
- Bonamigo, T.; Campos, J.F.; Oliveira, A.S.; Torquato, H.F.V.; Balestieri, J.B.P.; Cardoso, C.A.L.; Paredes-Gamero, E.J.; de Picoli Souza, K.; Dos Santos, E.L. Antioxidant and cytotoxic activity of propolis of Plebeia droryana and Apis mellifera (Hymenoptera, Apidae) from the brazilian Cerrado biome. PLoS ONE 2017, 12, e0183983. [Google Scholar] [CrossRef] [PubMed]
- Werz, O. 5-lipoxygenase: Cellular biology and molecular pharmacology. Curr. Drug Targets-Inflamm. Allergy 2007, 1, 23–44. [Google Scholar] [CrossRef]
- Peters-Golden, M.; Henderson Jr, W.R. Leukotrienes. N. Engl. J. Med. 2007, 357, 1841–1854. [Google Scholar] [CrossRef]
- Boudreau, L.H.; Maillet, J.; LeBlanc, L.M.; Jean-Francois, J.; Touaibia, M.; Flamand, N.; Surette, M.E. Caffeic acid phenethyl ester and its amide analogue are potent inhibitors of leukotriene biosynthesis in human polymorphonuclear leukocytes. PLoS ONE 2012, 7, e31833. [Google Scholar] [CrossRef]
- Bittencourt, M.L.F.; Ribeiro, P.R.; Franco, R.L.P.; Hilhorst, H.W.M.; de Castro, R.D.; Fernandez, L.G. Metabolite profiling, antioxidant and antibacterial activities of Brazilian propolis: Use of correlation and multivariate analyses to identify potential bioactive compounds. Food Res. Int. 2015, 76, 449–457. [Google Scholar] [CrossRef]
- Gomez-Caravaca, A.M.; Gomez-Romero, M.; Arraez-Roman, D.; Segura-Carretero, A.; Fernandez-Gutierrez, A. Advances in the analysis of phenolic compounds in products derived from bees. J. Pharm. Biomed. Anal. 2006, 41, 1220–1234. [Google Scholar] [CrossRef]
- Viuda-Martos, M.; Ruiz-Navajas, Y.; Fernandez-Lopez, J.; Perez-Alvarez, J.A. Functional properties of honey, propolis and royal jelly. J. Food Sci. 2008, 73, R117–R124. [Google Scholar] [CrossRef]
- Cao, J.; Peng, L.Q.; Du, L.J.; Zhang, Q.D.; Xu, J.J. Ultrasound –assisted ionic liquid-based micellar extraction combined with microcrystalline cellulose as sorbent in dispersive microextraction for the determination of phenolic compounds in propolis. Anal. Chim. Acta 2017, 963, 24–32. [Google Scholar] [CrossRef]
- Darendeliogliu, E.; Aykutoglu, G.; Tartik, M.; Baydas, G. Turkish propolis protects human endothelial cells in vitro from homocysteine-induced apoptosis. Acta Histochem. 2016, 118, 369–376. [Google Scholar] [CrossRef]
- Machado, B.A.S.; De Abreu Barreto, G.; Costa, A.S.; Costa, S.S.; Silva, R.P.D.; Da Silva, D.F. Determination of parameters for the supercritical extraction of antioxidant compounds from green propolis using carbon dioxide and ethanol as co-solvent. PLoS ONE 2015, 10, e0134489. [Google Scholar] [CrossRef]
- Trusheva, B.; Trunkova, D.; Bankova, V. Different extraction methods of biologically active components from propolis: A preliminary study. Chem. Cent. J. 2007, 1, 13. [Google Scholar] [CrossRef]
- Reis, J.H.dO.; Barreto, G.dA.; Cerquiera, J.C.; Anjos, J.P.D.; Andrade, L.N.; Padilha, F.F.; Machado, B.A.S. Evaluation of the antioxidant profile and cytotoxic activity of red propolis extracts from different regions of northeastern Brazil obtained by conventional and ultrasound-assisted extraction. PLoS ONE 2019, 14, e0219063. [Google Scholar] [CrossRef] [PubMed]
- Scepankova, H.; Martins, M.; Estevinho, L.; Delgadillo, I.; Saraiva, J.A. Enhancement of bioactivity of natural extracts by non-thermal high hydrostatic pressure extraction. Plant Foods Hum. Nutr. 2018, 73, 253–267. [Google Scholar] [CrossRef]
- Devequi-Nunes, D.; Machado, B.A.S.; Barreto, G.dA.; Reboucas Silva, J.; da Silva, D.F.; da Rocha, J.L.C. Chemical characterization and biological activity of six different extracts of propolis through conventional methods and supercritical extraction. PLoS ONE 2018, 13, e0207676. [Google Scholar] [CrossRef]
- Andrade, J.K.S.; Denaddai, M.; Andrade, G.R.S.; da Cuhna Nascimento, C.; Barbosa, P.F.; Jesus, M.S. Development and characterization of microcapsules containing spray dried powder obtained from Brazilian brown, green and red propolis. Food Res. Int. 2018, 109, 278–287. [Google Scholar] [CrossRef]
- Sadhana, N.; Lohidasan, S.; Mahadik, K.R. Marker-based standardization and investigation of nutraceutical potential of Indian propolis. J. Integr. Med. 2017, 15, 483–494. [Google Scholar] [CrossRef]
- Radmark, O.; Werz, O.; Steinhilber, D.; Samuelsson, B. 5-Lipoxygenase: Regulation of expression and enzyme activity. Trends Biochem. Sci. 2007, 32, 332–341. [Google Scholar] [CrossRef]
- Doiron, J.; Boudreau, L.H.; Picot, N.; Villebonet, B.; Surette, M.E.; Touaibia, M. Synthesis and 5-Lipoxygenase Inhibitory Activity of New Cinnamoyl and Caffeoylclusters. Bioorg. Med. Chem. Lett. 2009, 19, 1118–1121. [Google Scholar] [CrossRef]
- Sawaya, A.C.H.F.; Tomazela, D.M.; Cunha, I.B.S.; Bankova, V.S.; Marcucci, M.C.; Custodio, A.R.; Eberlin, M.N. Electrospray ionization mass spectrometry fingerprinting of propolis. Analyst 2004, 129, 739–744. [Google Scholar] [CrossRef]
- Trudic, B.; Andelkovic, B.; Orlovic, S.; Tesevic, V.; Pilipovic, A.; Cvetkovic, M.; Stankovic, J. HPLC/MS—TOF analysis of surface resins from three poplar clones grown in Serbia. South-East Eur. For. 2016, 7, 129–133. [Google Scholar] [CrossRef]
- Costa, A.G., Jr.; Yoshida, N.C.; Garcez, W.S.; Perdomo, R.T.; Matos, M.C.; Garcez, F.R. Metabolomics approach expands the classification of propolis samples from midwest Brazil. J. Nat. Prod. 2020, 83, 333–343. [Google Scholar] [CrossRef]
- Shi, H.; Yang, H.; Zhang, X.; Yu, L. Identification and quantification of phytochemical composition and anti-inflammatory and radical scavenging properties of methanolic extracts of Chinese propolis. J. Agric. Food Chem. 2012, 60, 12403–12410. [Google Scholar] [CrossRef]
- Yuan, Y.; Zheng, S.; Zeng, L.; Deng, Z.; Zhang, B.; Li, H. The phenolic compounds, metabolites, and antioxidant activity of propolis extracted by ultrasound-assisted method. J. Food Sci. 2019, 84, 3850–3865. [Google Scholar] [CrossRef]
- Greenaway, W.; May, J.; Scaysbrook, T.; Whatley, F.R. Identification by gas chromatography-mass spectrometry of 150 compounds in propolis. Z. Nat. C 1991, 46, 111–121. [Google Scholar] [CrossRef]
- Romero, M.; Freire, J.; Pastene, E.; Garcia, A.; Aranda, M.; Gonzalez, C. Propolis polyphenolic compounds affect the viability and structure of Heliicobacter pyroli in vitro. Braz. J. Pharmacogn. 2019, 29, 325–332. [Google Scholar] [CrossRef]
- Zingue, S.; Nde, C.B.M.; Michel, T.; Ndinteh, D.T.; Tchatchou, J.; Adamou, M.; Fernandez, X.; Fohouo, F.N.T.; Clyne, C.; Njamen, D. Ethanol-extracted Cameroonian propolis exerts estrogenic effects and alleviates hot flushes in ovariectomized Wistar rats. BMC Complementary Altern. Med. 2017, 17, 65–82. [Google Scholar] [CrossRef]
- deGroot, A.C.; Popova, M.P.; Bankova, V.S. An update on the constituents of poplar-type propolis. 2014. [Google Scholar]
- Bakdash, A.; Almohammadi, O.H.; Taha, N.A.; Abu-Rumman, A.; Kumar, S. Chemical composition of propolis from the Baha region in Saudi Arabia. Czech J. Food Sci. 2018, 36, 1–9. [Google Scholar]
- Allain, E.P.; Boudreau, L.H.; Flamand, N.; Surette, M.E. The Intracellular Localisation and Phosphorylation Profile of the Human 5-Lipoxygenase Δ13 Isoform Differs from That of Its Full Length Counterpart. PLoS ONE 2015, 10, e0132607. [Google Scholar] [CrossRef]
Sample Availability: Samples of all extracts are available from the authors. |


| Solvents | m (g/5 g of Dry Propolis) | |||
|---|---|---|---|---|
| SE | MAE | UAE | ||
| Water | Mean | 0.047 | 0.044 | 0.065 |
| CI | 0.013 to 0.081 | 0.034 to 0.055 | 0.033 to 0.096 | |
| Methanol | Mean | 1.774 | 1.362 | 1.352 |
| CI | 1.553 to 1.995 | 1.245 to 1.478 | 1.283 to 1.420 | |
| Ethanol | Mean | 1.936 | 0.876 | 0.737 |
| CI | 1.853 to 2.018 | 0.785 to 0.968 | 0.535 to 0.938 | |
| Acetone | Mean | 1.614 | 0.825 | 1.466 |
| CI | 1.251 to 1.977 | 0.680 to 0.971 | 1.299 to 1.632 | |
| EtOAc | Mean | 2.283 | 1.079 | 0.768 |
| CI | 1.960 to 2.605 | 0.790 to 1.368 | 0.329 to 1.207 | |
| Dichloromethane | Mean | 2.08 | 0.937 | 1.028 |
| CI | 1.493 to 2.680 | 0.619 to 1.255 | 0.933 to 1.122 | |
| EtOAc:Hex (1:1) | Mean | 0.761 | 0.425 | 0.153 |
| CI | 0.612 to 0.910 | 0.336 to 0.515 | 0.144 to 0.162 | |
| EtOAc:Hex (4:1) | Mean | 0.781 | 0.641 | 0.335 |
| CI | 0.555 to 1.007 | 0.541 to 0.741 | 0.226 to 0.443 | |
| EtOAc:Hex (1:4) | Mean | 1.091 | 0.483 | 0.31 |
| CI | 0.868 to 1.313 | 0.258 to 0.709 | 0.252 to 0.368 | |
| Hexane | Mean | 0.599 | 0.324 | 0.53 |
| CI | 0.275 to 0.923 | 0.244 to 0.403 | 0.397 to 0.664 | |
| Solvents | IC50 (µg/mL) | |||
|---|---|---|---|---|
| SE | MAE | UAE | ||
| Water | Mean | 451.8 | 359.1 | 301.4 |
| CI | 432.4 to 472.0 | 354.9 to 363.4 | 282.8 to 321.1 | |
| Methanol | Mean | 76.01 | 63.22 | 79.12 |
| CI | 72.13 to 80.10 | 61.54 to 64.94 | 74.60 to 83.92 | |
| Ethanol | Mean | 59.03 | 70.03 | 74.38 |
| CI | 57.97 to 60.10 | 58.90 to 83.26 | 68.91 to 80.27 | |
| Acetone | Mean | 124.9 | 88.54 | 48.11 |
| CI | 120.8 to 129.2 | 84.13 to 93.18 | 44.97 to 51.48 | |
| EtOAc | Mean | 118.3 | 120.6 | 97.24 |
| CI | 114.7 to 122.1 | 110.4 to 131.8 | 87.76 to 107.8 | |
| Dichloromethane | Mean | 231.7 | 102.3 | 186.8 |
| CI | 213.6 to 251.4 | 80.95 to 129.3 | 174.0 to 200.6 | |
| EtOAc:Hex (1:1) | Mean | ~1290 | 246.9 | 188.2 |
| CI | (Very wide) | 210.2 to 289.9 | 176.5 to 200.7 | |
| EtOAc:Hex (4:1) | Mean | 590.6 | 140.5 | 187.5 |
| CI | 482.2 to 723.4 | 131.6 to 149.9 | 170.1 to 206.5 | |
| EtOAc:Hex (1:4) | Mean | 378.0 | 480.8 | 770.4 |
| CI | 356.0 to 401.4 | 468.6 to 493.4 | 731.0 to 812.0 | |
| Hexane | Mean | NI | NI | NI |
| Ascorbic acid | Mean | 10.91 | ||
| CI | 9.55 to 12.47 | |||
| Caffeic acid | Mean | 8.80 | ||
| CI | 8.16 to 9.48 | |||
| Quercetin | Mean | 6.68 | ||
| CI | 5.68 to 7.84 | |||
| [M − H]− (m/z) | MS/MS Fragments (m/z) | |||||||
|---|---|---|---|---|---|---|---|---|
| tR (min) | Compound | CAS No. | Formula | Calculated | Experimental | Mass Accuracy (ppm) | Q-Exactive | LTQ-XL |
| 13.1 | p-Hydroxybenzaldehyde | 123-8-0 | C7H6O2 | 121.0284 | 121.0278 | 5.0 | n.d. | n.d. |
| 13.1 | Caffeic acid | 331-39-5 | C9H8O4 | 179.0339 | 179.0337 | 1.1 | 135.0426 | 135.2 |
| 14.7 | p-Coumaric acid | 501-98-4 | C9H8O3 | 163.0390 | 163.0384 | 3.7 | 119.0477 | 119.2 |
| 15.2 | Ferulic acid | 1135-24-6 | C10H10O4 | 193.0495 | 193.0489 | 3.1 | 134.0348, 178.0246, 149.0582 | 149.2, 134.2, 178.1 |
| 15.9 | Cinnamic acid | 140-10-3 | C9H8O2 | 147.0441 | 147.0436 | 3.4 | 103.0541 | 103.2 |
| 16.2 | Benzoic acid | 65-85-0 | C7H6O2 | 121.0284 | 121.0278 | 5.0 | n.d. | n.d. |
| 17.6 | Quercetin | 117-39-5 | C15H10O7 | 301.0343 | 301.0332 | 3.7 | 151.0011, 286.0462, 178.9960 | 179.1, 151.1, 273.2 |
| 18.0 | Ethyl caffeate | 102-37-4 | C11H12O4 | 207.0652 | 207.0645 | 3.4 | 179.0325, 135.0426, 161.0219 | 179.2, 133.2, 161.2 |
| 18.8 | Apigenin | 520-36-5 | C15H10O5 | 269.0444 | 269.0437 | 2.6 | n.d. | 225.1, 149.1, 218.2 |
| 19.0 | Pinobanksin | 548-82-3 | C15H12O5 | 271.0601 | 271.0593 | 3.0 | 253.0486, 225.0536, 215.0691 | 253.2, 225.2, 215.2 |
| 19.1 | Kaempferol | 520-18-3 | C15H10O6 | 285.0394 | 285.0385 | 3.2 | 145.0269, 139.0374 | 255.2, 151.1, 229.2 |
| 19.2 | Isorhamnetin | 480-19-3 | C16H12O7 | 315.0499 | 315.0487 | 3.8 | 300.0253 | 300,2 |
| 19.3 | 3-Methylkaempferol | 1592-70-7 | C16H12O6 | 299.0550 | 299.0539 | 3.7 | 284.0308, 165.9882, 121.0270 | 284.2 |
| 20.2 | Rhamnetin | 90-19-7 | C16H12O7 | 315.0499 | 315.0486 | 4.1 | 165.0168, 121.0270, 300.0253 | 165.2, 193.1, 300.1 |
| 21.4 | Isosakuranetin | 480-43-3 | C16H14O5 | 285.0757 | 285.0748 | 3.2 | 164.0100, 243.0658, 270.0533 | 270.17, 243.17, 164.08 |
| 21.6 | Caffeic acid phenylethyl ester | 104594-70-9 | C17H16O4 | 283.0965 | 283.0955 | 3.5 | 179.0325, 135.0426, 161.0219 | 179.1, 135.2, 161.2 |
| 21.6 | Chrysin | 480-4-0 | C15H10O4 | 253.0495 | 253.0486 | 3.6 | 121.0270, 209.1525 | 209.2, 224.2, 181.2 |
| 22.0 | 4’-Methylkaempferol | 491-54-3 | C16H12O6 | 299.0550 | 299.0539 | 3.7 | 284.0308 | 284,2 |
| 22.0 | 7-Methylkaempferol | 569-92-6 | C16H12O6 | 299.0550 | 299.0539 | 3.7 | 284.0308, 256.0357, 121.0270 | 165.2, 271.1, 283.1 |
| 22.0 | Galangin | 548-83-4 | C15H10O5 | 269.0444 | 269.0437 | 2.6 | n.d. | 239.2, 197.2, 227.2 |
| 22.4 | Cinnamyl caffeate | 115610-79-2 | C18H16O4 | 295.0965 | 295.0952 | 4.4 | 178.0246, 137.0218, 134.0347 | 178.1, 134.2, 251.3 |
| [M − H]− (m/z) | [M −H]− ( m/z) | MS/MS Fragments (m/z) | ||||
|---|---|---|---|---|---|---|
| tR (min) | Experimental | Peak Intensity | Predicted Formula | Calculated | Mass Accuracy (ppm) | Q-Exactive |
| 14.1 | 237.0746 | 3.07 × 107 | C12H14O5 | 237.0757 | 4.6 | 145.0269, 119.0477, 163.0375 |
| 14.42 | 151.0374 | 9.22 × 107 | C8H8O3 | 151.0390 | 10.6 | 136.0140 |
| 15.39 | 295.0801 | 9.89 × 106 | C14H16O7 | 295.0812 | 3.7 | 161.0218, 59.0116, 135.0426 |
| 15.7 | 329.1004 | 5.3 × 107 | C18H18O6 | 329.1020 | 4.9 | 145.0269, 119.0477, 163.0375 |
| 15.8 | 359.1110 | 1.0 × 108 | C19H20O7 | 359.1125 | 4.2 | 145.0270, 163.0375, 119.0478 |
| 16.1 | 389.1213 | 4.0 × 107 | C20H22O8 | 389.1231 | 4.6 | 175.0375, 193.0482, 134.0348 |
| 16.3 | 177.0533 | 5.1 × 107 | C10H10O3 | 177.0546 | 7.3 | 162.0297 |
| 16.6 | 279.0854 | 8.8 × 107 | C14H16O6 | 279.0863 | 3.2 | 145.0269, 59.0116, 119.0477 |
| 16.6 | 317.1004 | 1.1 × 108 | C17H18O6 | 317.1020 | 5.0 | 121.0270, 222.9902, 250.9852 |
| 16.7 | 363.1058 | 3.9 × 107 | C18H20O8 | 363.1074 | 4.4 | 121.0270, 269. 0801, 164.0453 |
| 16.8 | 309.0959 | 2.5 × 107 | C15H18O7 | 309.0969 | 3.2 | 59.0116, 175.0375, 294.0724 |
| 16.9 | 251.0905 | 1.9 × 107 | C13H16O5 | 251.0914 | 3.6 | 161.0583, 145.0270, 133.0634 |
| 17.8 | 399.1057 | 1.1 × 107 | C21H20O8 | 399.1074 | 4.3 | 163.0375, 119.0477, 253.0697 |
| 18.1 | 343.1161 | 3.2 × 107 | C19H20O6 | 343.1176 | 4.4 | 147.0426, 164.0453 |
| 18.5 | 387.1420 | 4.2× 107 | C21H24O7 | 387.1438 | 4.6 | 119.0477, 145.0209, 163.0375 |
| 18.8 | 383.1106 | 7.2 × 107 | C21H20O7 | 383.1125 | 5.0 | 163.0375, 119.0477, 145.0269 |
| 18.9 | 301.0695 | 2.9 × 107 | C16H14O6 | 301.0707 | 4.0 | 165.9882, 109.9984, 194.9911 |
| 18.9 | 413.1212 | 2.8 × 107 | C22H22O8 | 413.1231 | 4.6 | 193.0481, 163.0375, 134.0348 |
| 19.7 | 191.0690 | 3.5 × 107 | C11H12O3 | 191.0703 | 6.8 | 145.0269, 119.0477, 163.0375 |
| 19.9 | 249.0749 | 3.0 × 107 | C13H14O5 | 249.0758 | 3.6 | 145.0269, 131.0477, 121.0269 |
| 21.2 | 485.1417 | 3.0 × 107 | C25H26O10 | 485.1442 | 5.2 | 193.0841, 134.0347, 175.0375 |
| 21.6 | 255.0643 | 1.2 × 107 | C15H12O4 | 255.0652 | 3.5 | 151.0011, 213.0533, 83.0114 |
| 22.5 | 253.0850 | 6.5 × 108 | C16H14O3 | 253.0859 | 3.6 | 121.0270, 145.0269, 162.0297 |
| 22.9 | 283.0954 | 6.0 × 107 | C17H16O4 | 283.0965 | 3.9 | 177.0168, 133.0269, 268.0721 |
| 23.1 | 267.1007 | 4.3 × 107 | C17H16O3 | 267.1016 | 3.4 | 119.0477, 163.0375, 145.0269 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sambou, M.; Jean-François, J.; Ndongou Moutombi, F.J.; Doiron, J.A.; Hébert, M.P.A.; Joy, A.P.; Mai-Thi, N.-N.; Barnett, D.A.; Surette, M.E.; Boudreau, L.H.; et al. Extraction, Antioxidant Capacity, 5-Lipoxygenase Inhibition, and Phytochemical Composition of Propolis from Eastern Canada. Molecules 2020, 25, 2397. https://doi.org/10.3390/molecules25102397
Sambou M, Jean-François J, Ndongou Moutombi FJ, Doiron JA, Hébert MPA, Joy AP, Mai-Thi N-N, Barnett DA, Surette ME, Boudreau LH, et al. Extraction, Antioxidant Capacity, 5-Lipoxygenase Inhibition, and Phytochemical Composition of Propolis from Eastern Canada. Molecules. 2020; 25(10):2397. https://doi.org/10.3390/molecules25102397
Chicago/Turabian StyleSambou, Mariama, Jacques Jean-François, Fanta J. Ndongou Moutombi, Jérémie A. Doiron, Mathieu P.A. Hébert, Andrew P. Joy, Ngoc-Nu Mai-Thi, David A. Barnett, Marc E. Surette, Luc H. Boudreau, and et al. 2020. "Extraction, Antioxidant Capacity, 5-Lipoxygenase Inhibition, and Phytochemical Composition of Propolis from Eastern Canada" Molecules 25, no. 10: 2397. https://doi.org/10.3390/molecules25102397
APA StyleSambou, M., Jean-François, J., Ndongou Moutombi, F. J., Doiron, J. A., Hébert, M. P. A., Joy, A. P., Mai-Thi, N.-N., Barnett, D. A., Surette, M. E., Boudreau, L. H., & Touaibia, M. (2020). Extraction, Antioxidant Capacity, 5-Lipoxygenase Inhibition, and Phytochemical Composition of Propolis from Eastern Canada. Molecules, 25(10), 2397. https://doi.org/10.3390/molecules25102397





