Discovery of a Natural Product That Binds to the Mycobacterium tuberculosis Protein Rv1466 Using Native Mass Spectrometry
Abstract
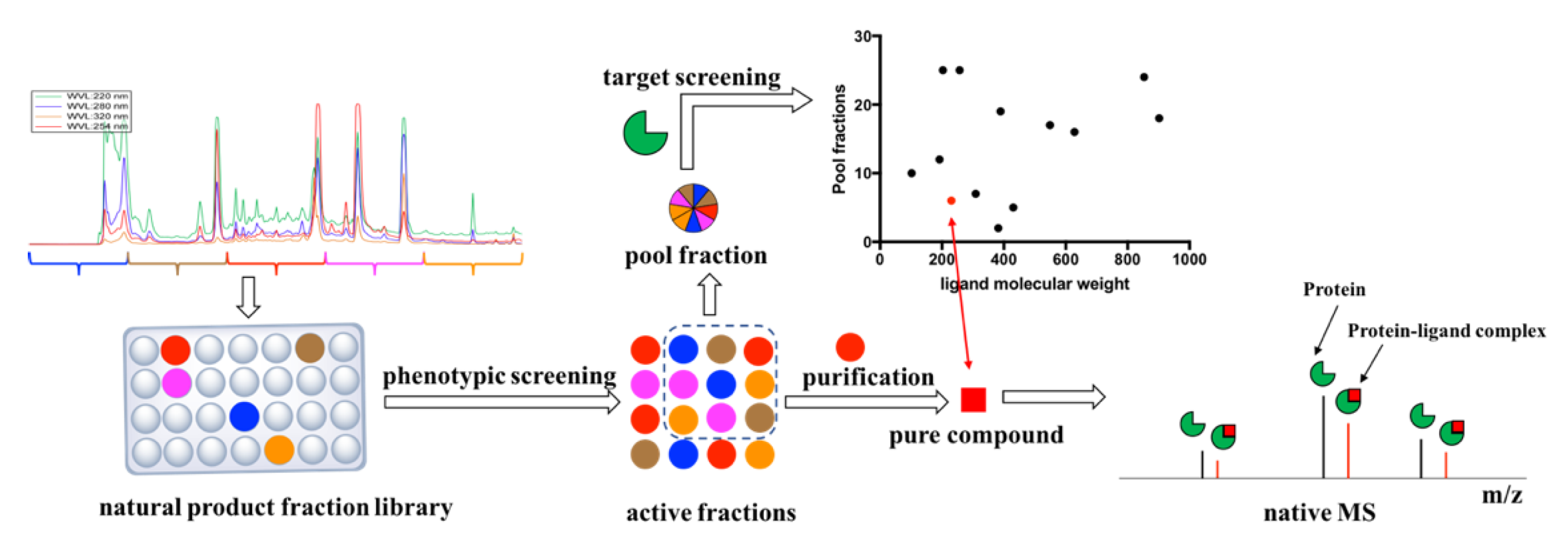
1. Introduction
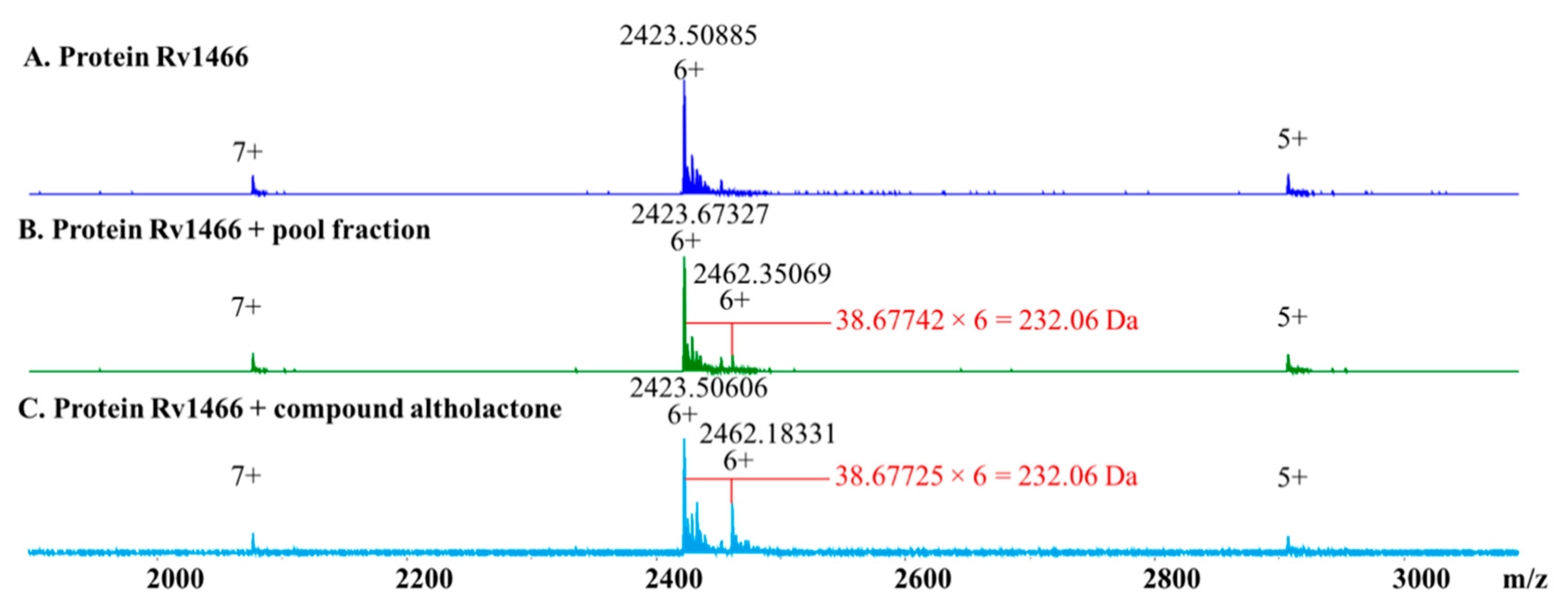
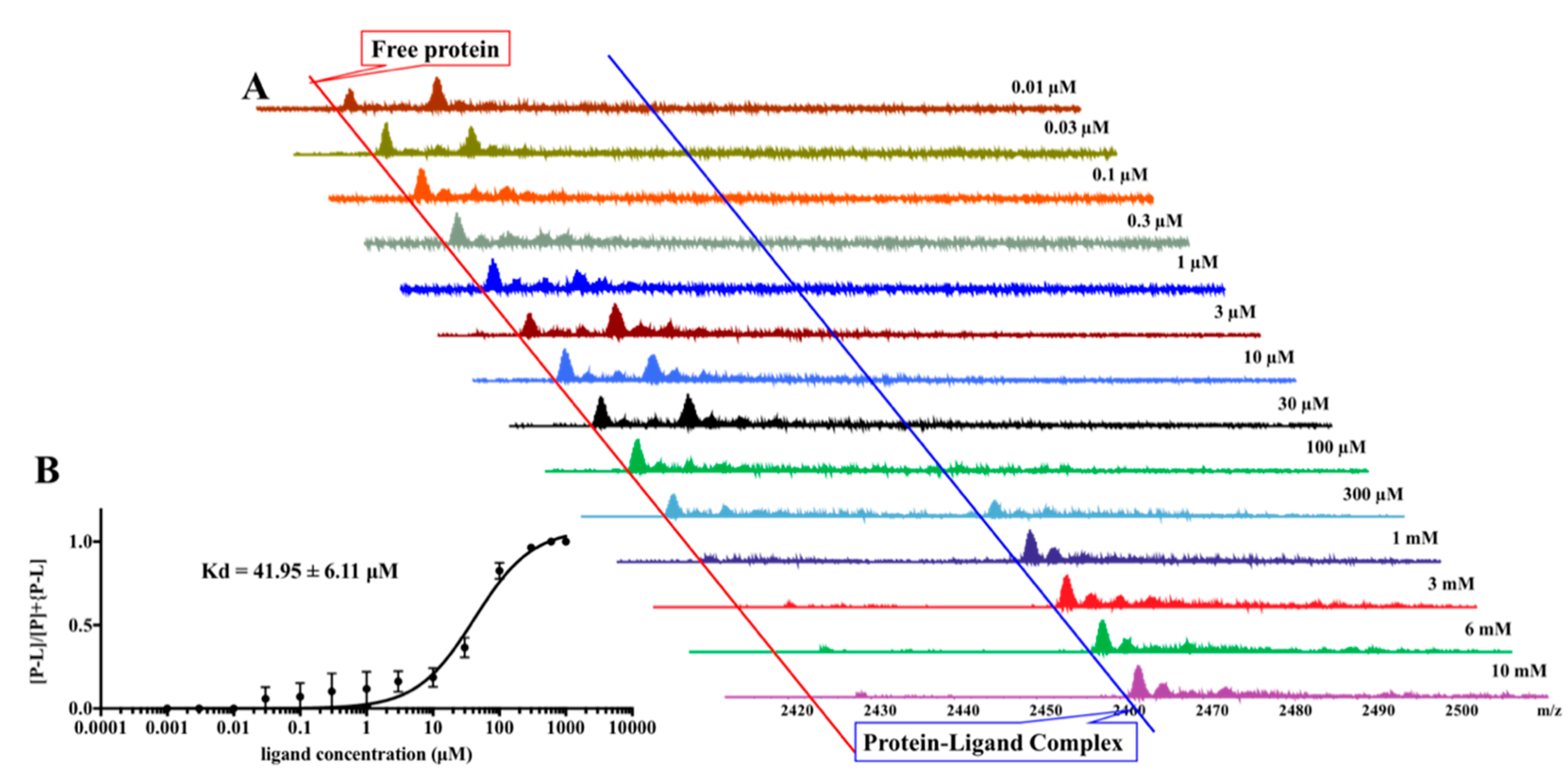
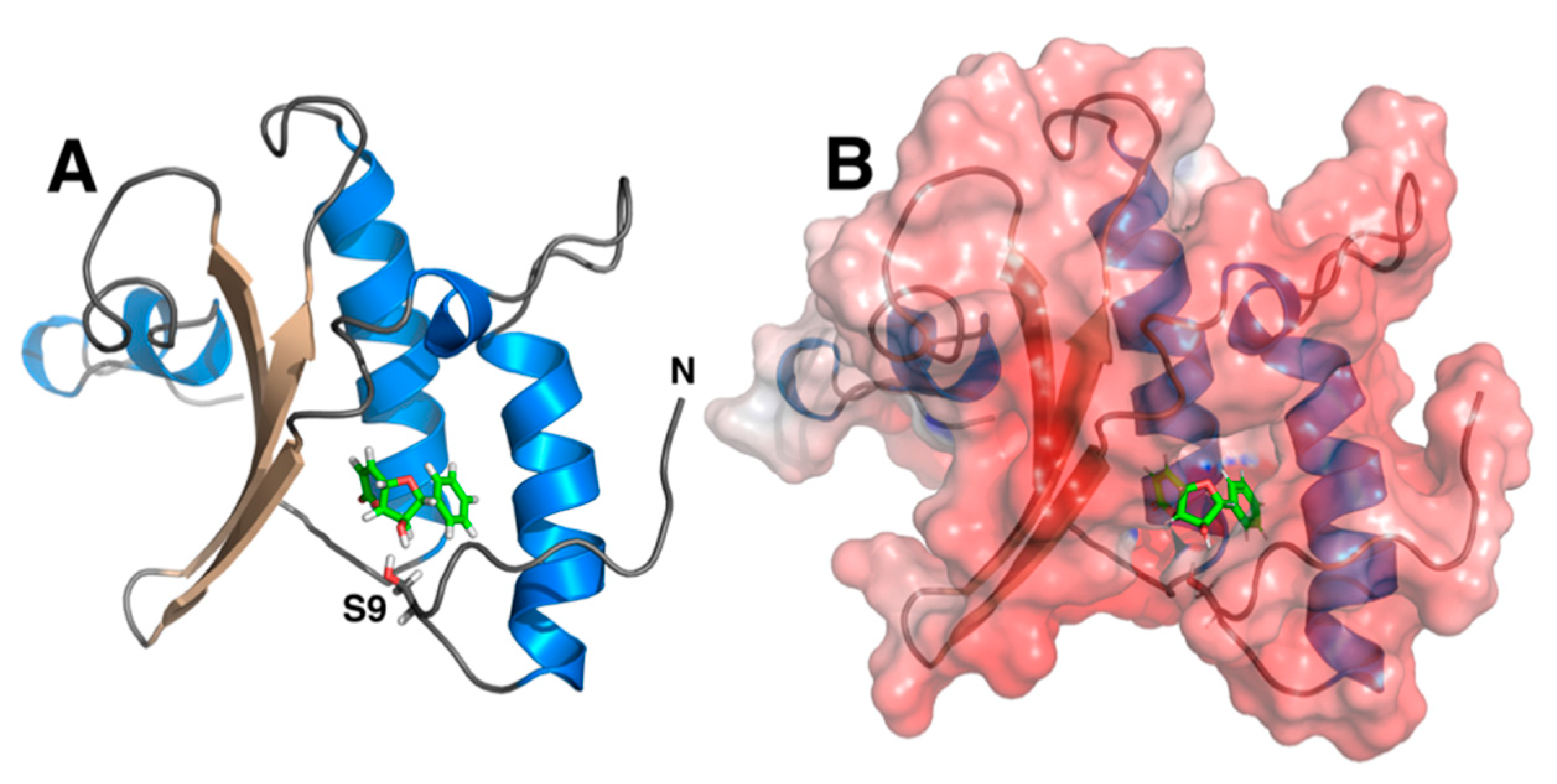
2. Results
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Natural Product Fraction Library
4.3. Phenotypic Screening against M. tuberculosis H37Rv
4.4. Re-Extraction and Purification
4.5. Altholactone
4.6. Biological Assays
4.7. Target Screening
4.8. Pseudo-Kd Determination
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2019; World Health Organization: Geneva, Switzerland, 2019; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Blaser, M.J.; Kirschner, D. The equilibria that allow bacterial persistence in human hosts. Nature 2007, 449, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.G.; Barry, C.E.; Flynn, J.L. Tuberculosis: What we don’t know can, and does, hurt us. Science 2010, 328, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Zhang, J.Y.; Song, Z.J.; Liu, M.M.; Hu, J.S.; Hou, C.J.; Zhu, G.L.; Jiang, L.; Xia, X.K.; Quinn, R.J.; et al. Genome- and MS-based mining of antibacterial chlorinated chromones and xanthones from the phytopathogenic fungus Bipolaris sorokiniana strain 11134. Appl. Microbiol. Biot. 2019, 103, 5167–5181. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Abdel-Mageed, W.M.; Ren, B.; He, W.N.; Huang, P.; Li, X.L.; Bolla, K.; Guo, H.; Chen, C.X.; Song, F.H.; et al. Endophytic Streptomyces sp Y3111 from traditional Chinese medicine produced antitubercular pluramycins. Appl. Microbiol. Biot. 2014, 98, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; El-Hossary, E.M.; Oelschlaeger, T.A.; Donia, M.S.; Quinn, R.J.; Abdelmohsen, U.R. Potential of marine natural products against drug-resistant bacterial infections. Lancet Infect. Dis. 2019, 19, 237–245. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug. Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Marques, A.; Martins, I.S.; Kastner, T.; Plutzar, C.; Theurl, M.C.; Eisenmenger, N.; Huijbregts, M.A.J.; Wood, R.; Stadler, K.; Bruckner, M.; et al. Increasing impacts of land use on biodiversity and carbon sequestration driven by population and economic growth. Nat. Ecol. Evol. 2019, 3, 628–637. [Google Scholar] [CrossRef]
- Daskin, J.H.; Pringle, R.M. Warfare and wildlife declines in Africa’s protected areas. Nature 2018, 553, 328–332. [Google Scholar] [CrossRef]
- Parmesan, C. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 637–669. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; Mace, G.M.; Tilman, D.; Wardle, D.A. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Zhang, H.L.; Wang, C.Y.; Yao, S.C.L.; Yao, S.Q. Target identification of natural products and bioactive compounds using affinity-based probes. Nat. Prod. Rep. 2016, 33, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M. Recent advances in target identification of bioactive natural products. Biosci. Biotechnol. Biochem. 2019, 83, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Schenone, M.; Dancik, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef]
- Carver, T.E.; Bordeau, B.; Cummings, M.D.; Petrella, E.C.; Pucci, M.J.; Zawadzke, L.E.; Dougherty, B.A.; Tredup, J.A.; Bryson, J.W.; Yanchunas, J.J.; et al. Decrypting the biochemical function of an essential gene from Streptococcus pneumoniae using thermofluor technology. J. Biol. Chem. 2005, 280, 11704–11712. [Google Scholar] [CrossRef]
- Pedro, L.; Quinn, R.J. Native mass spectrometry in fragment-based drug discovery. Molecules 2016, 21, 984. [Google Scholar] [CrossRef]
- Vu, H.; Quinn, R.J. Direct screening of natural product extracts using mass spectrometry. J. Biomol. Screen. 2008, 13, 265–275. [Google Scholar] [CrossRef]
- Camp, D.; Davis, R.A.; Campitelli, M.; Ebdon, J.; Quinn, R.J. Drug-like properties: Guiding principles for the design of natural product libraries. J. Nat. Prod. 2012, 75, 72–81. [Google Scholar] [CrossRef]
- Loder, J.W.; Nearn, R.H. Altholactone, a novel tetrahydrofuro[3,2b]pyran-5-one from a polyalthia species (Annonaceae). Heterocycles 1977, 7, 113–118. [Google Scholar] [CrossRef]
- Lekphrom, R.; Kanokmedhakul, S.; Kanokmedhakul, K. Bioactive styryllactones and alkaloid from flowers of Goniothalamus laoticus. J. Ethnopharmacol. 2009, 125, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Macabeo, A.P.G.; Lopez, A.D.A.; Schmidt, S.; Heilmann, J.; Dahse, H.M.; Alejandro, G.J.D.; Franzblau, S.G. Antitubercular and cytotoxic constituents from Goniothalamus gitingensis. Rec Nat. Prod. 2014, 8, 41–45. [Google Scholar]
- Xie, Y.; Feng, Y.; Di Capua, A.; Mak, T.; Buchko, G.W.; Myler, P.J.; Liu, M.; Quinn, R.J. A phenotarget approach for identifying an alkaloid interacting with the tuberculosis protein Rv1466. Mar. Drugs 2020, 18, 149. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.; Pedro, L.; Mak, T.; McCormick, B.; Rowley, J.; Liu, M.M.; Di Capua, A.; Williams-Noonan, B.; Pham, N.B.; Pouwer, R.; et al. Fragment-based screening of a natural product library against 62 potential malaria drug targets employing native mass spectrometry. ACS Infect. Dis. 2018, 4, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Feng, Y.J.; Vu, H.; McCormick, B.; Rowley, J.; Pedro, L.; Crowther, G.J.; Van Voorhis, W.C.; Forster, P.I.; Quinn, R.J. Bioaffinity mass spectrometry screening. J. Biomol. Screen. 2016, 21, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Van Pelt, C.K.; Wilson, D.B. Quantitative determination of noncovalent binding interactions using automated nanoelectrospray mass spectrometry. Anal. Chem. 2003, 75, 3010–3018. [Google Scholar] [CrossRef]
- Pedro, L.; Van Voorhis, W.C.; Quinn, R.J. Optimization of electrospray ionization by statistical design of experiments and response surface methodology: Protein-ligand equilibrium dissociation constant determinations. J. Am. Soc. Mass Spectr. 2016, 27, 1520–1530. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, 665–667. [Google Scholar] [CrossRef]
- Laureanti, J.A.; Brandi, J.; Offor, E.; Engel, D.; Rallo, R.; Ginovska, B.; Martinez, X.; Baaden, M.; Baker, N.A. Visualizing biomolecular electrostatics in virtual reality with UnityMol-APBS. Protein Sci. 2019, 29, 237–246. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update Autodock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Al Momani, F.; Alkofahi, A.S.; Mhaidat, N.M. Altholactone displays promising antimicrobial activity. Molecules 2011, 16, 4560–4566. [Google Scholar] [CrossRef] [PubMed]
- Euanorasetr, J.; Junhom, M.; Tantimavanich, S.; Vorasin, O.; Munyoo, B.; Tuchinda, P.; Panbangred, W. Halogenated benzoate derivatives of altholactone with improved anti-fungal activity. J. Asian Nat. Prod. Res. 2016, 18, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.W.; Masood, M.; Rasul, A.; Wei, W.; Wang, Y.; Ali, M.; Mustaqeem, M.; Li, J.; Li, X.M. Altholactone Inhibits NF-kappa B and STAT3 activation and induces reactive oxygen species-mediated apoptosis in prostate cancer DU145 cells. Molecules 2017, 22, 240. [Google Scholar] [CrossRef]
- Zhao, B.; Li, X.M. Altholactone induces reactive oxygen species-mediated apoptosis in bladder cancer T24 cells through mitochondrial dysfunction, MAPK-p38 activation and Akt suppression. Oncol. Rep. 2014, 31, 2769–2775. [Google Scholar] [CrossRef] [PubMed]
- Forbes, L.; Ebsworth-Mojica, K.; DiDone, L.; Li, S.G.; Freundlich, J.S.; Connell, N.; Dunman, P.M.; Krysan, D.J. A high throughput screening assay for anti-Mycobacterial small molecules based on adenylate kinase release as a reporter of cell lysis. PLoS ONE 2015, 10, e0129234. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.; Miller, C.H.; Bellows, D.S.; O’Toole, R. Evaluation of the Mycobacterium smegmatis and BCG models for the discovery of Mycobacterium tuberculosis inhibitors. Tuberculosis 2010, 90, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Huet, G.; Daffe, M.; Saves, I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: Evidence for its implication in the pathogen’s survival. J. Bacteriol 2005, 187, 6137–6146. [Google Scholar] [CrossRef]
- Charan, M.; Singh, N.; Kumar, B.; Srivastava, K.; Siddiqi, M.I.; Habib, S. Sulfur mobilization for Fe-S cluster assembly by the essential SUF pathway in the Plasmodium falciparum apicoplast and its inhibition. Antimicrob. Agents Chemother. 2014, 58, 3389–3398. [Google Scholar] [CrossRef]
- Choby, J.E.; Mike, L.A.; Mashruwala, A.A.; Dutter, B.F.; Dunman, P.M.; Sulikowski, G.A.; Boyd, J.M.; Skaar, E.P. A small-molecule inhibitor of iron-sulfur cluster assembly uncovers a link between virulence regulation and metabolism in Staphylococcus aureus. Cell Chem. Biol. 2016, 23, 1351–1361. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef]
- Willemse, D.; Weber, B.; Masino, L.; Warren, R.M.; Adinolfi, S.; Pastore, A.; Williams, M.J. Rv1460, a SufR homologue, is a repressor of the suf operon in Mycobacterium tuberculosis. PLoS ONE 2018, 13, e0200145. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.A.; Widen, J.C.; Harki, D.A.; Brummond, K.M. Covalent modifiers: A chemical perspective on the reactivity of alpha, beta-unsaturated carbonyls with thiols via hetero-Michael addition reactions. J. Med. Chem. 2017, 60, 839–885. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Zhao, J.Y.; Yao, Y.; Wang, C.Y.; Zhang, J.B.; Shu, X.H.; Sun, X.L.; Li, Y.X.; Liu, K.X.; Yuan, H.; et al. Covalent binding design strategy: A prospective method for discovery of potent targeted anticancer agents. Eur. J. Med. Chem. 2017, 142, 493–505. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds altholactone and protein Rv1466 are available from the authors. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elnaas, A.R.; Grice, D.; Han, J.; Feng, Y.; Capua, A.D.; Mak, T.; Laureanti, J.A.; Buchko, G.W.; Myler, P.J.; Cook, G.; et al. Discovery of a Natural Product That Binds to the Mycobacterium tuberculosis Protein Rv1466 Using Native Mass Spectrometry. Molecules 2020, 25, 2384. https://doi.org/10.3390/molecules25102384
Elnaas AR, Grice D, Han J, Feng Y, Capua AD, Mak T, Laureanti JA, Buchko GW, Myler PJ, Cook G, et al. Discovery of a Natural Product That Binds to the Mycobacterium tuberculosis Protein Rv1466 Using Native Mass Spectrometry. Molecules. 2020; 25(10):2384. https://doi.org/10.3390/molecules25102384
Chicago/Turabian StyleElnaas, Ali R., Darren Grice, Jianying Han, Yunjiang Feng, Angela Di Capua, Tin Mak, Joseph A. Laureanti, Garry W. Buchko, Peter J. Myler, Gregory Cook, and et al. 2020. "Discovery of a Natural Product That Binds to the Mycobacterium tuberculosis Protein Rv1466 Using Native Mass Spectrometry" Molecules 25, no. 10: 2384. https://doi.org/10.3390/molecules25102384
APA StyleElnaas, A. R., Grice, D., Han, J., Feng, Y., Capua, A. D., Mak, T., Laureanti, J. A., Buchko, G. W., Myler, P. J., Cook, G., Quinn, R. J., & Liu, M. (2020). Discovery of a Natural Product That Binds to the Mycobacterium tuberculosis Protein Rv1466 Using Native Mass Spectrometry. Molecules, 25(10), 2384. https://doi.org/10.3390/molecules25102384






