Aptamers Chemistry: Chemical Modifications and Conjugation Strategies
Abstract
1. Introduction
2. Aptamers and Selection Methods
3. Chemical Modifications of Aptamers and Their Impact on Pharmacological Properties
3.1. Modifications on Nucleic Acids Terminals
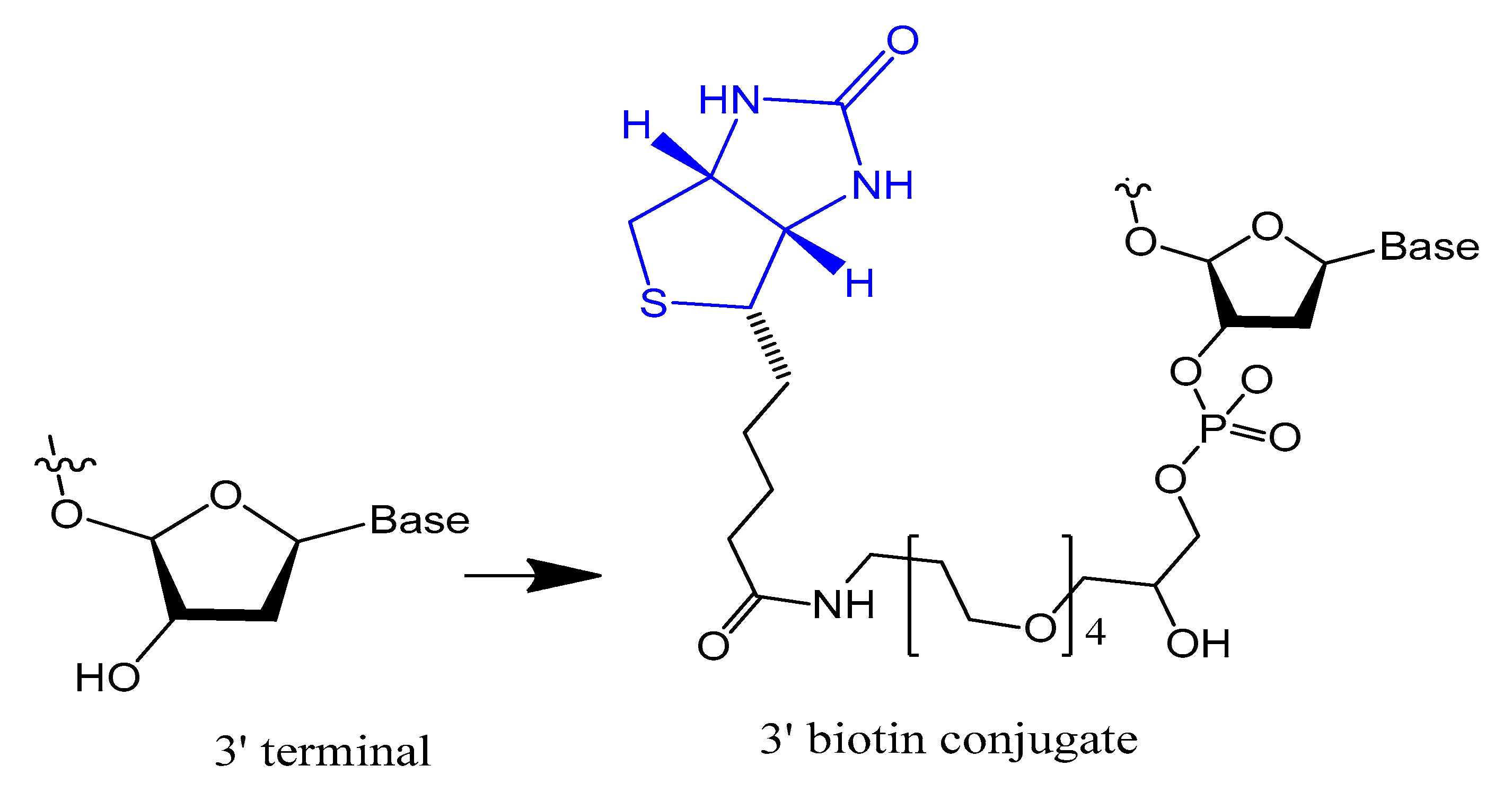
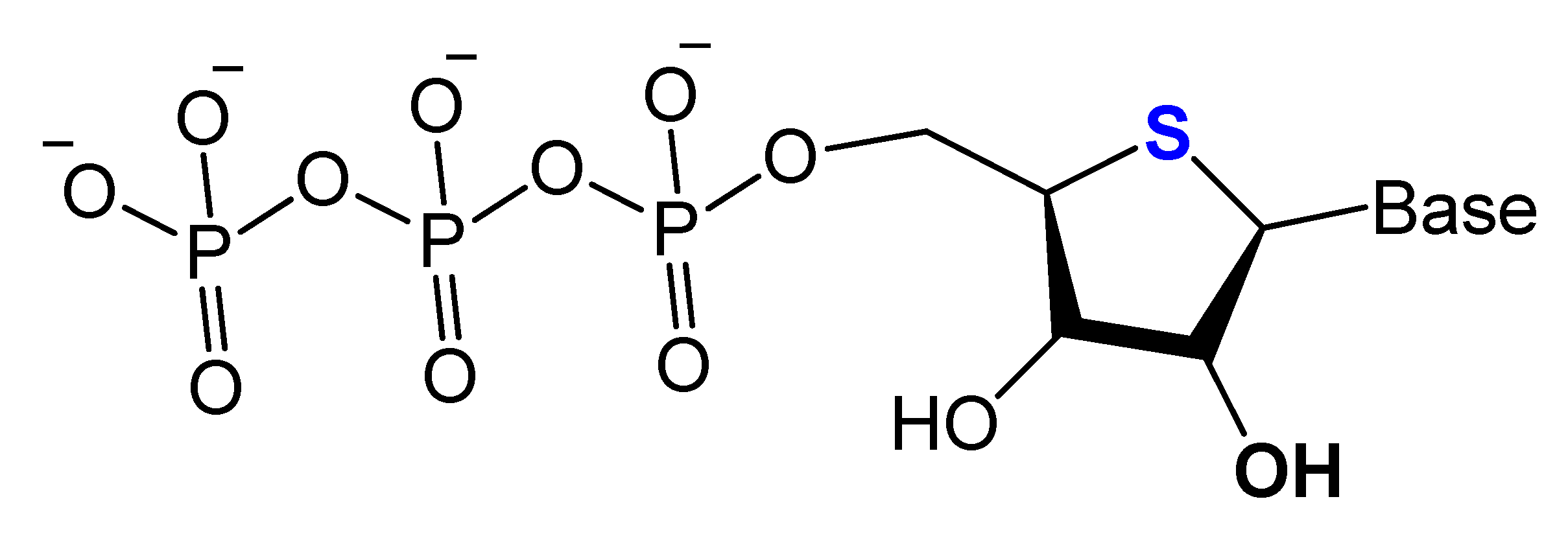
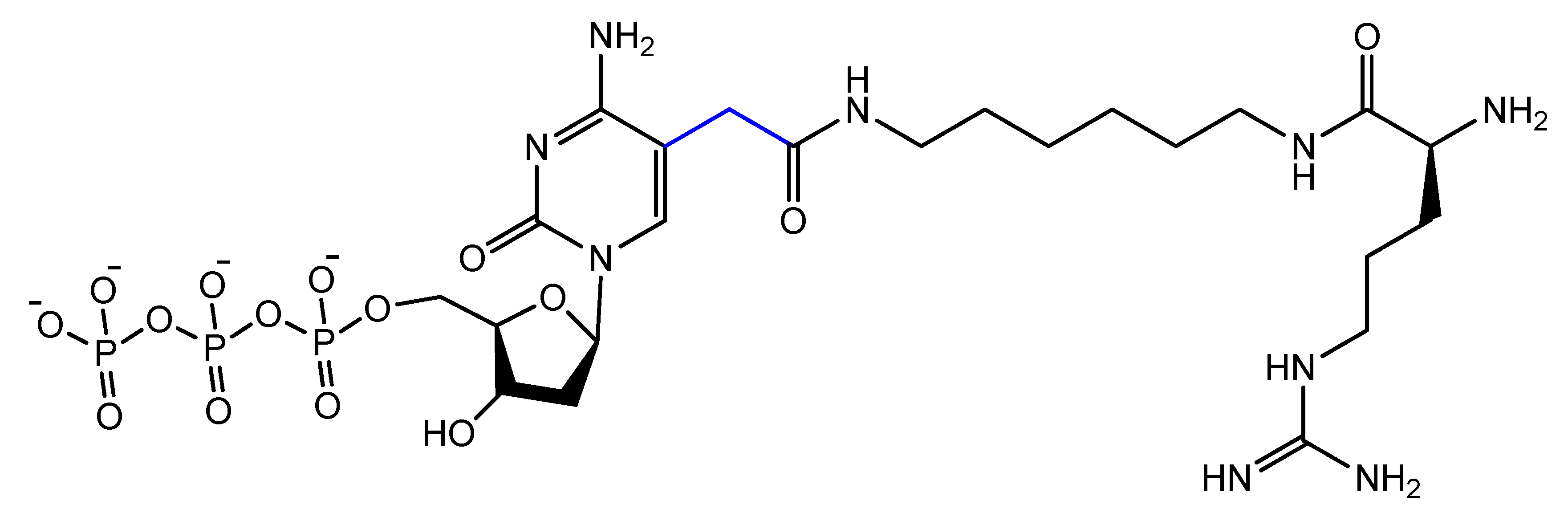
3.1.1. Terminal 3′–3′ and/or 5′–5′ Internucleotide, 3′ and 5′-Biotin Conjugates
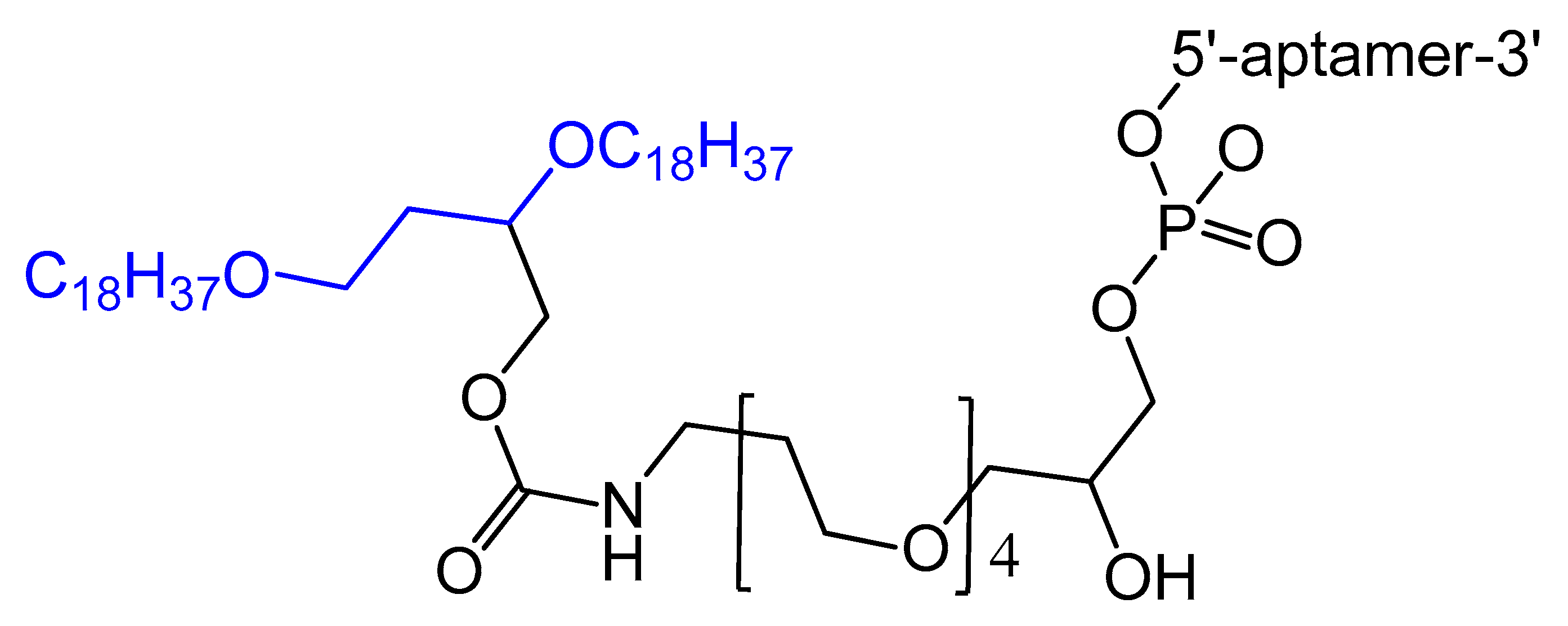
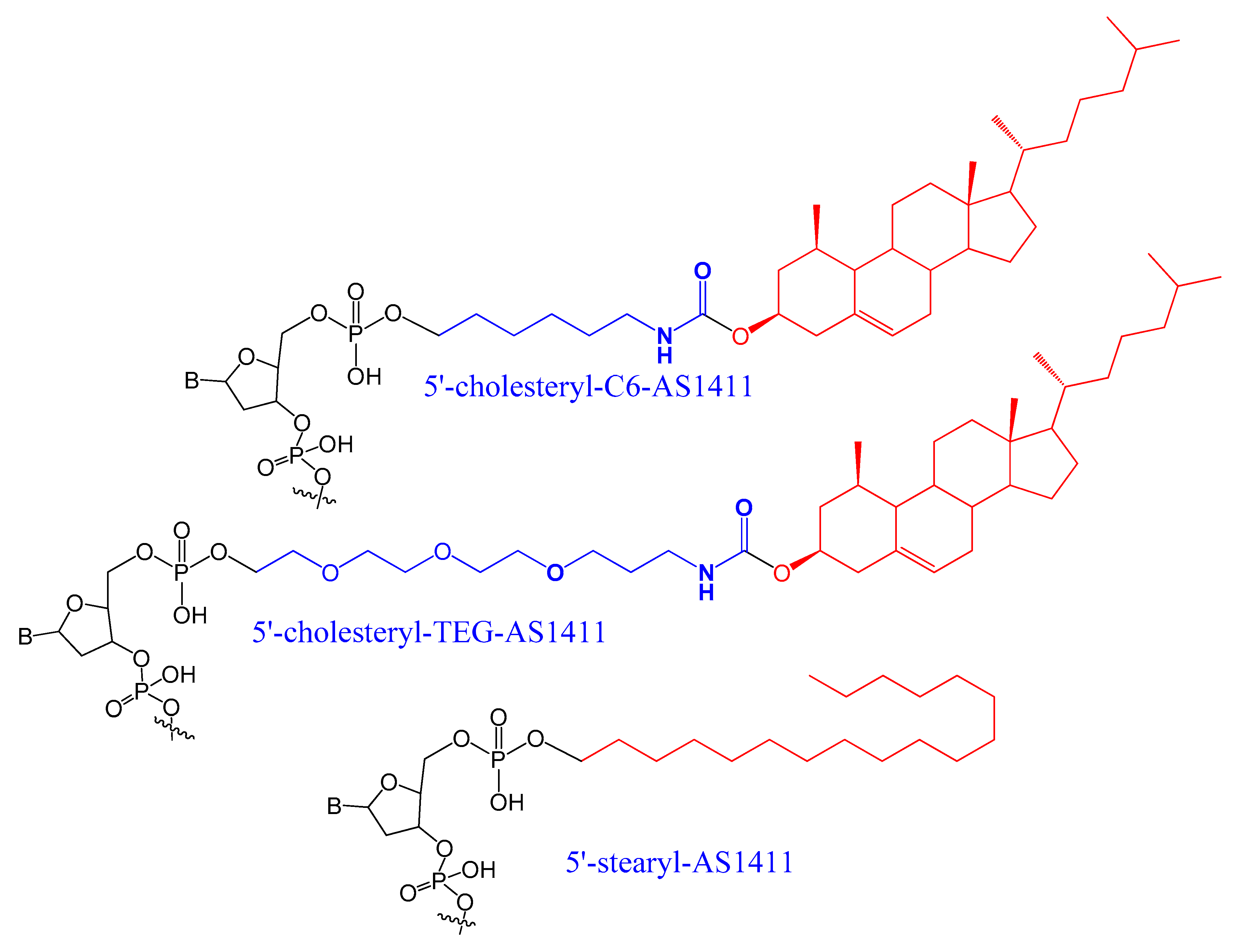
3.1.2. 5′-End with Cholesterol and Other Lipid Moieties
3.1.3. 5′-End PEGylation
3.2. Modifications on the Sugar Ring
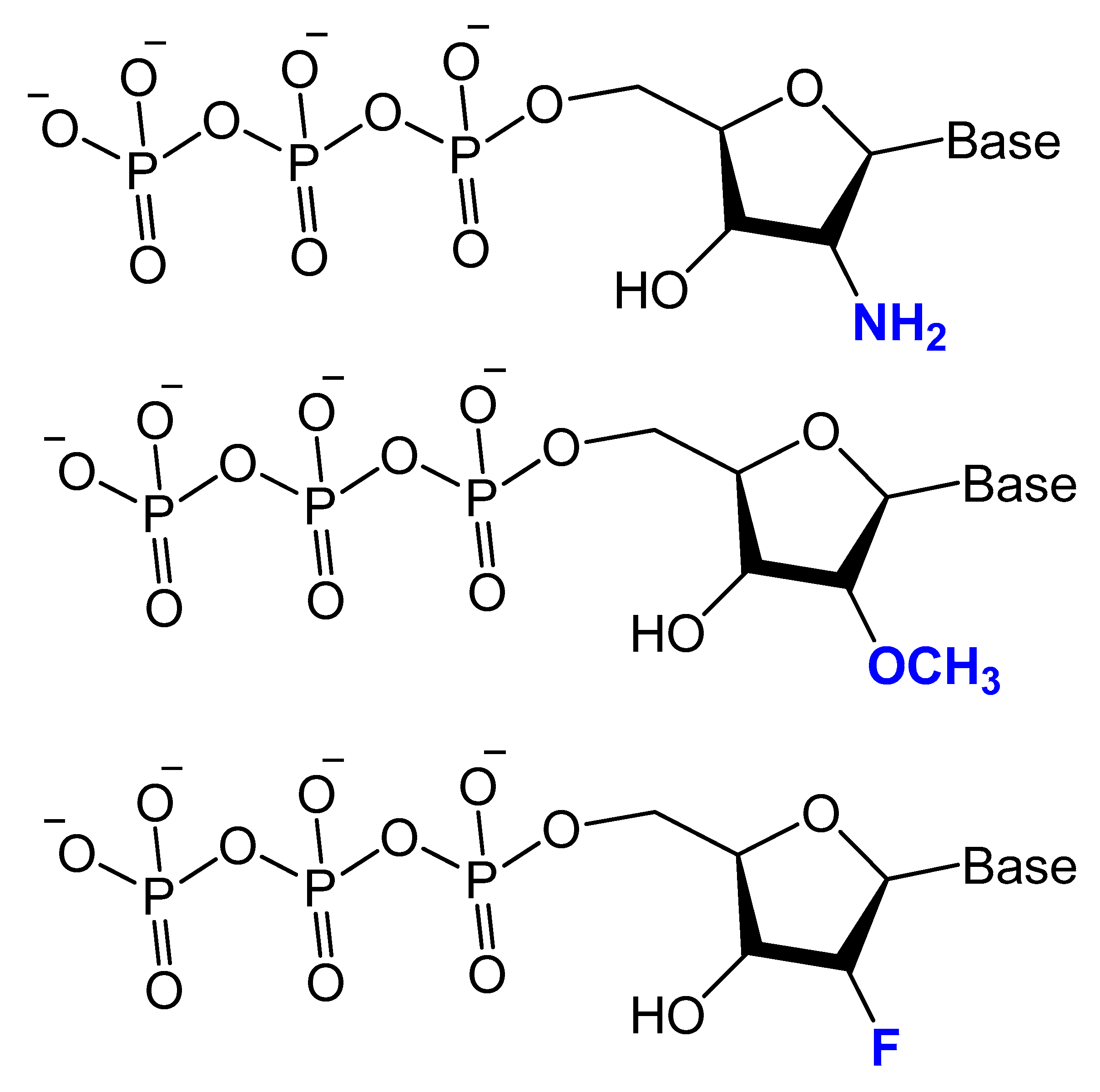
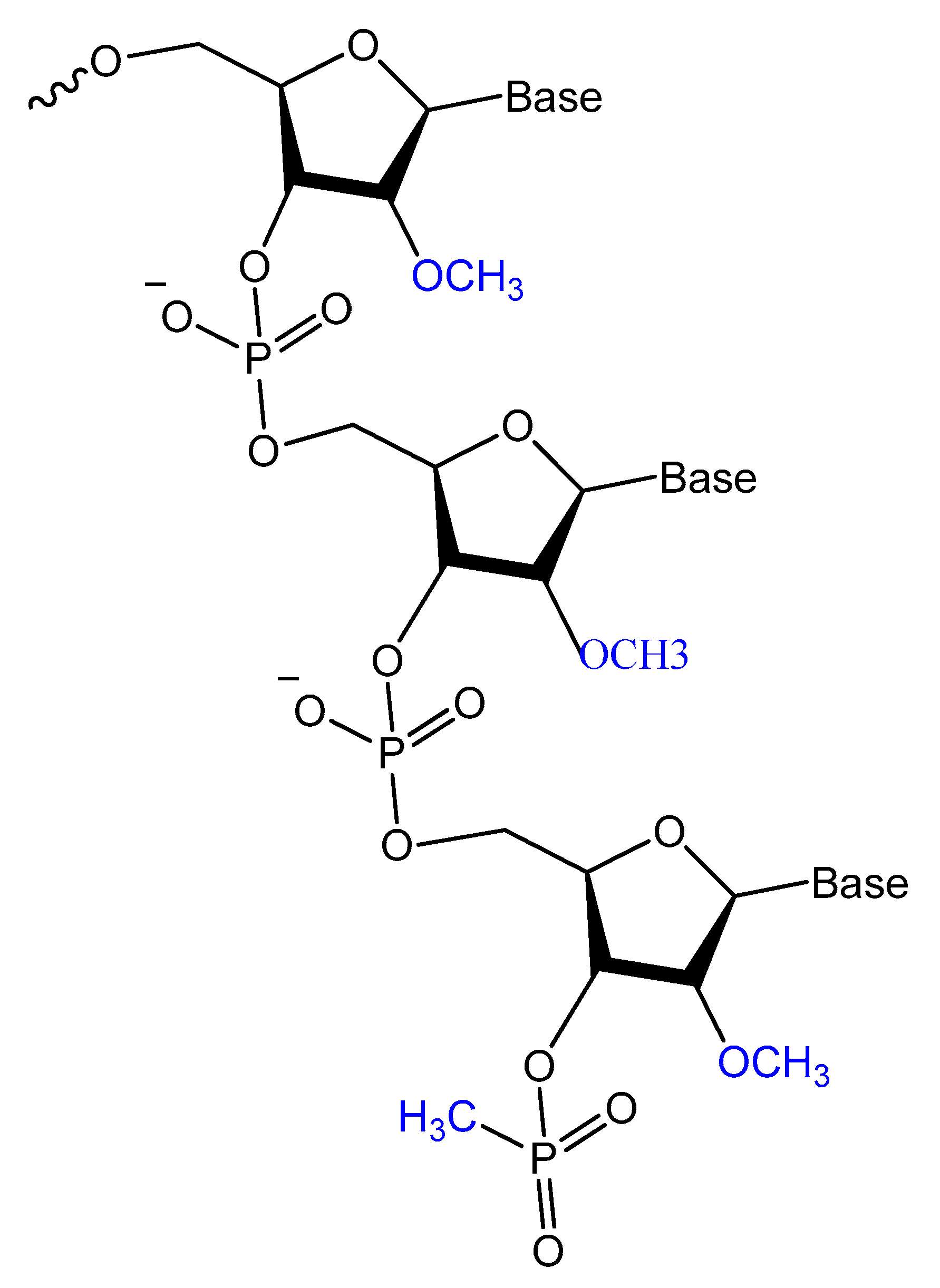
3.2.1. 2′-Substitutions
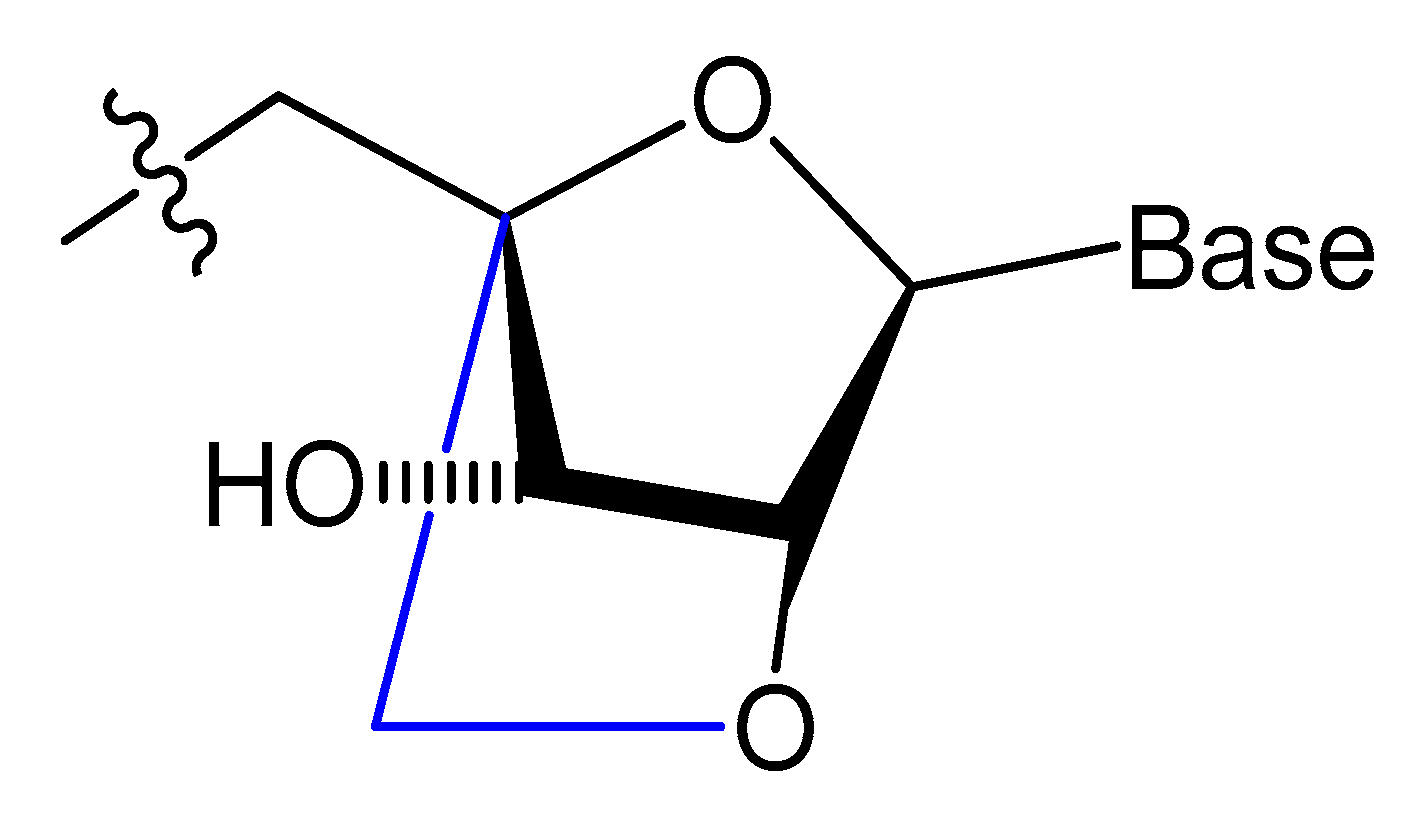
3.2.2. 4′-Oxygen Replacement
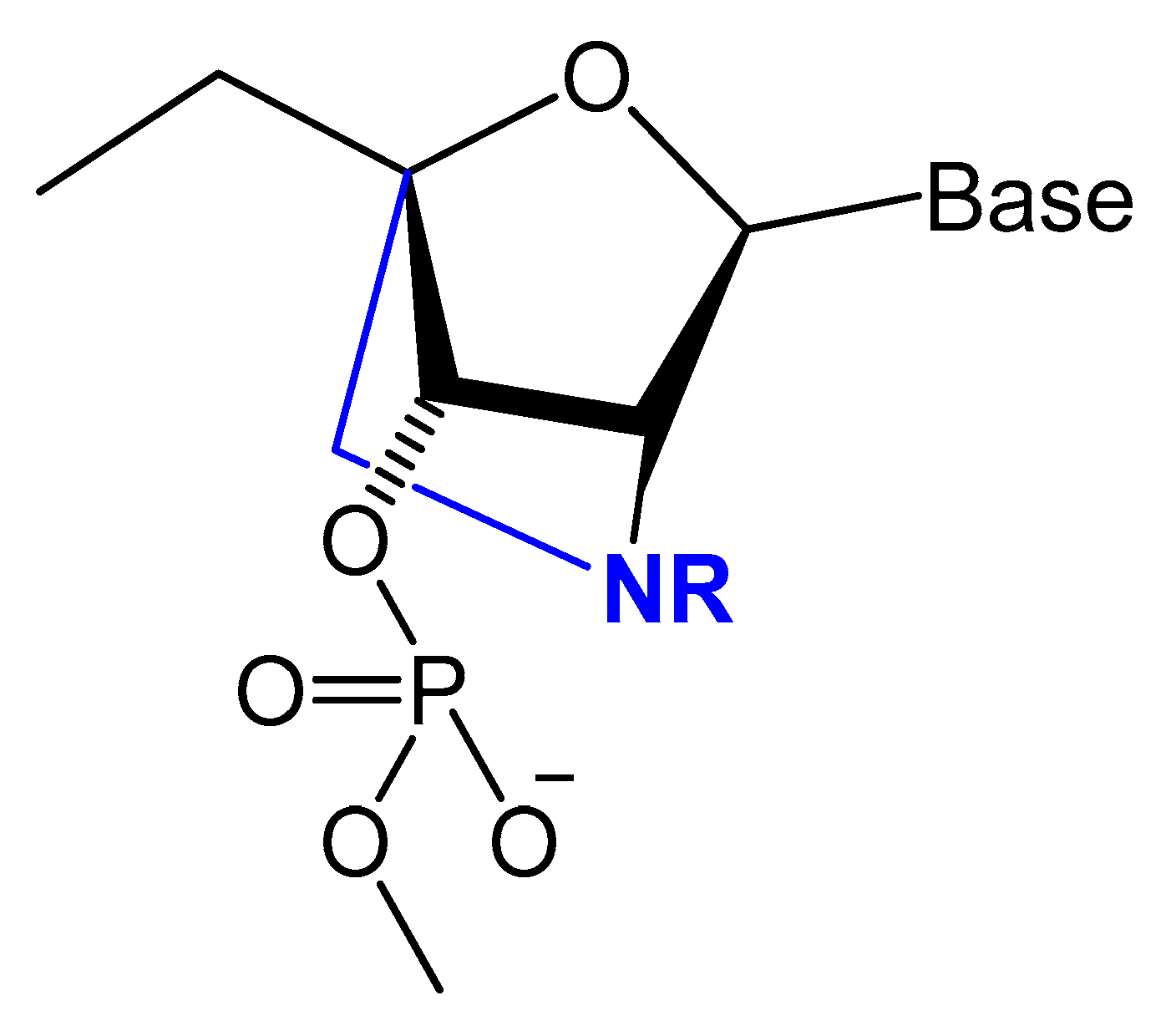
3.2.3. Locked and Unlocked Nucleic Acid
3.3. Modifications on the Phosphodiester Linkage
3.3.1. Methylphosphonate or Phosphorothioate
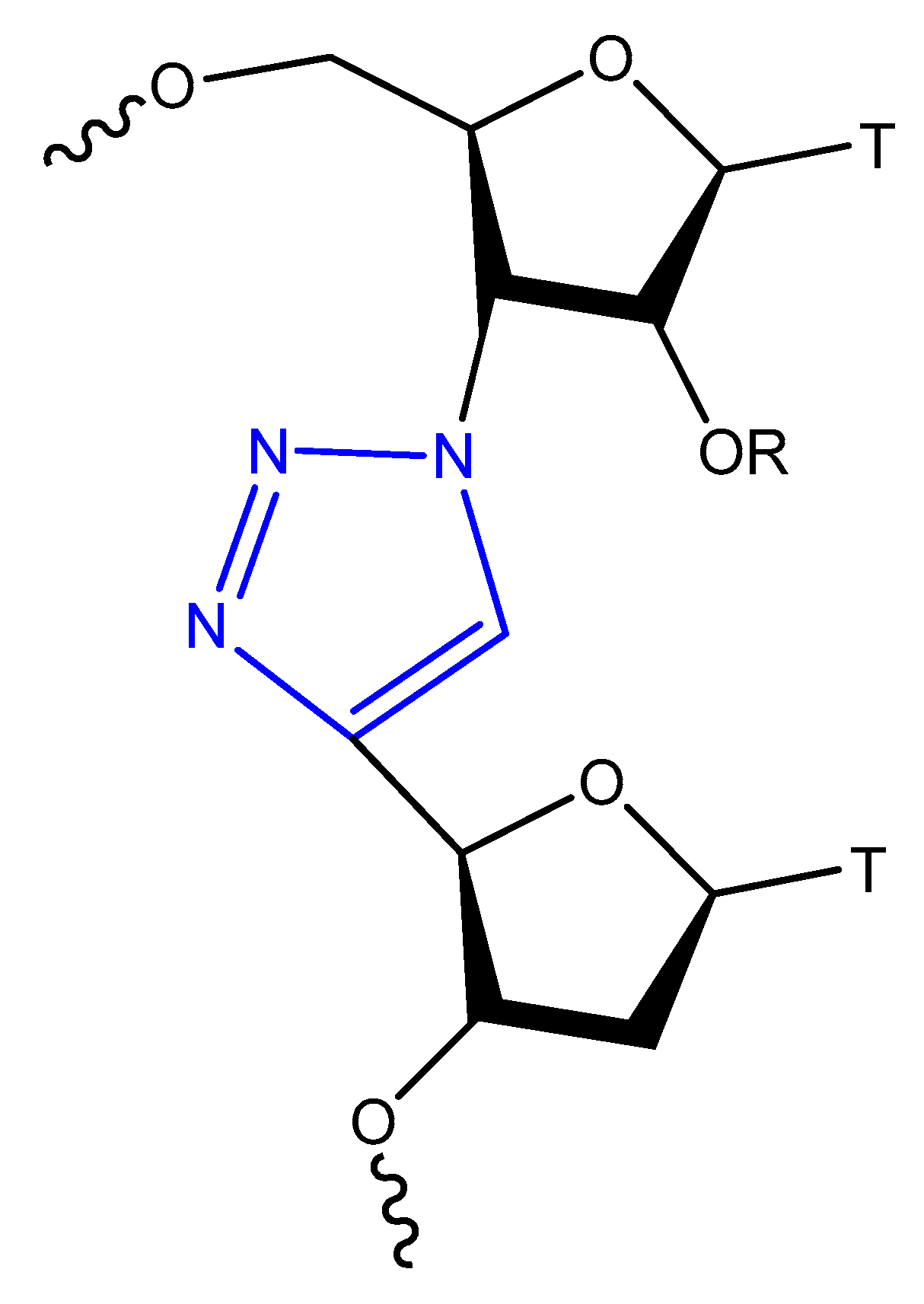
3.3.2. Triazole Modification
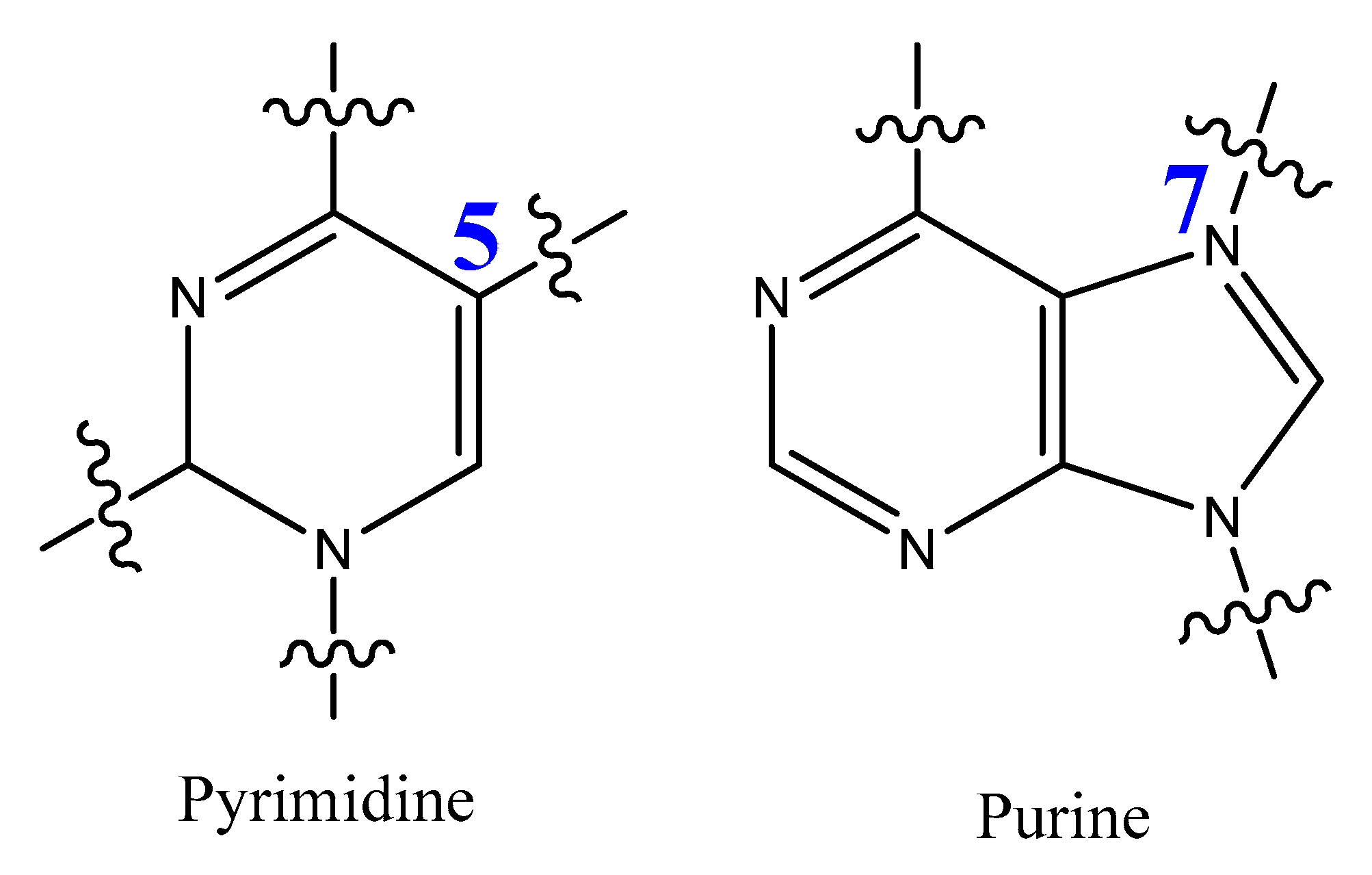
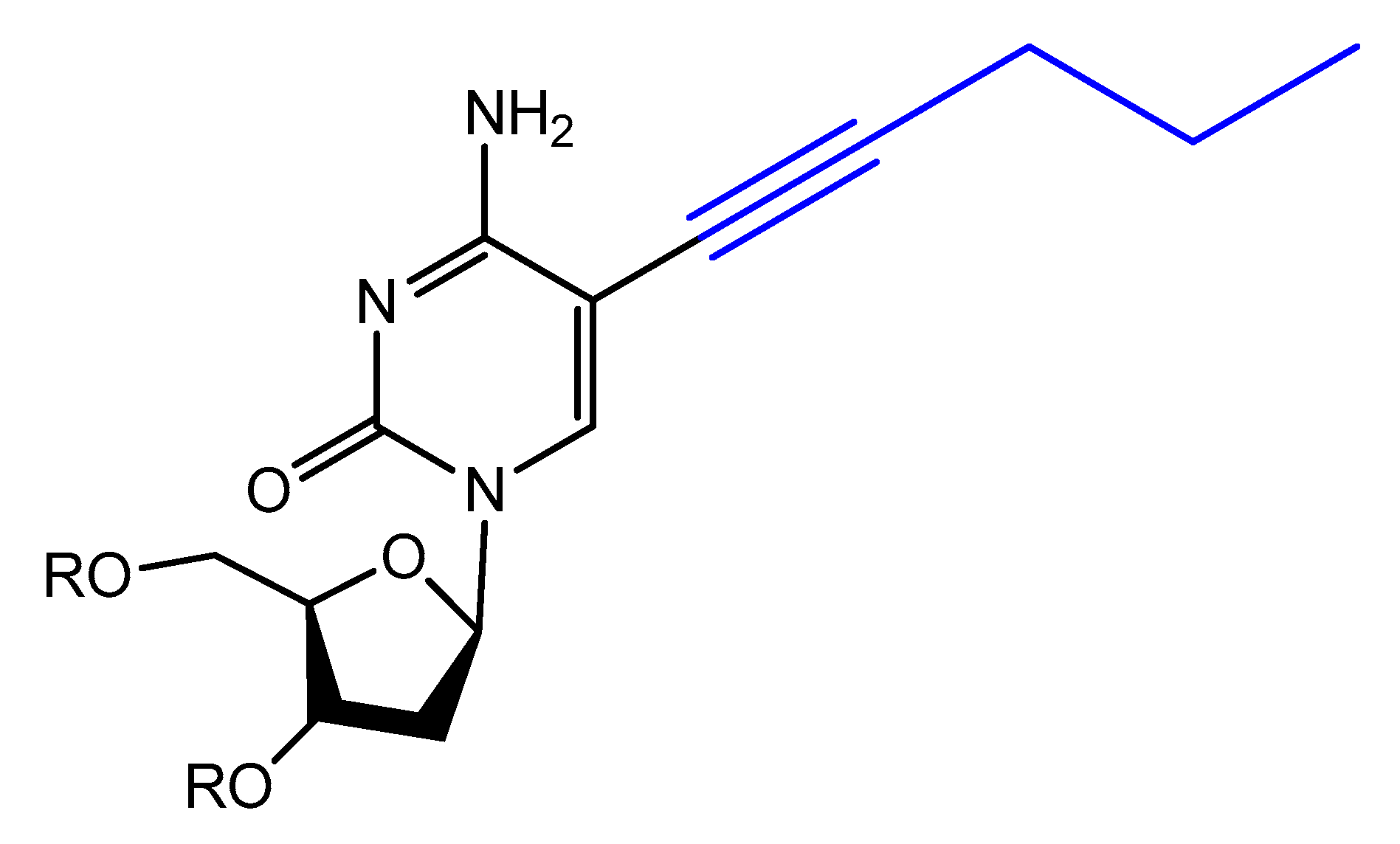
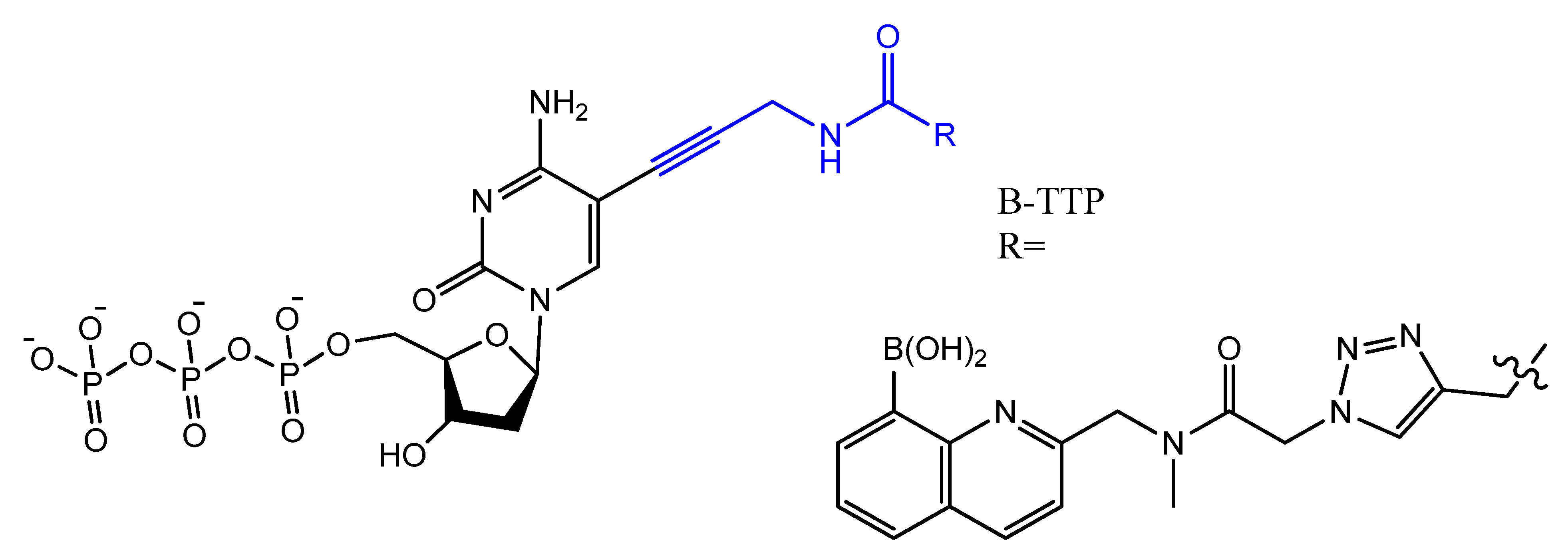
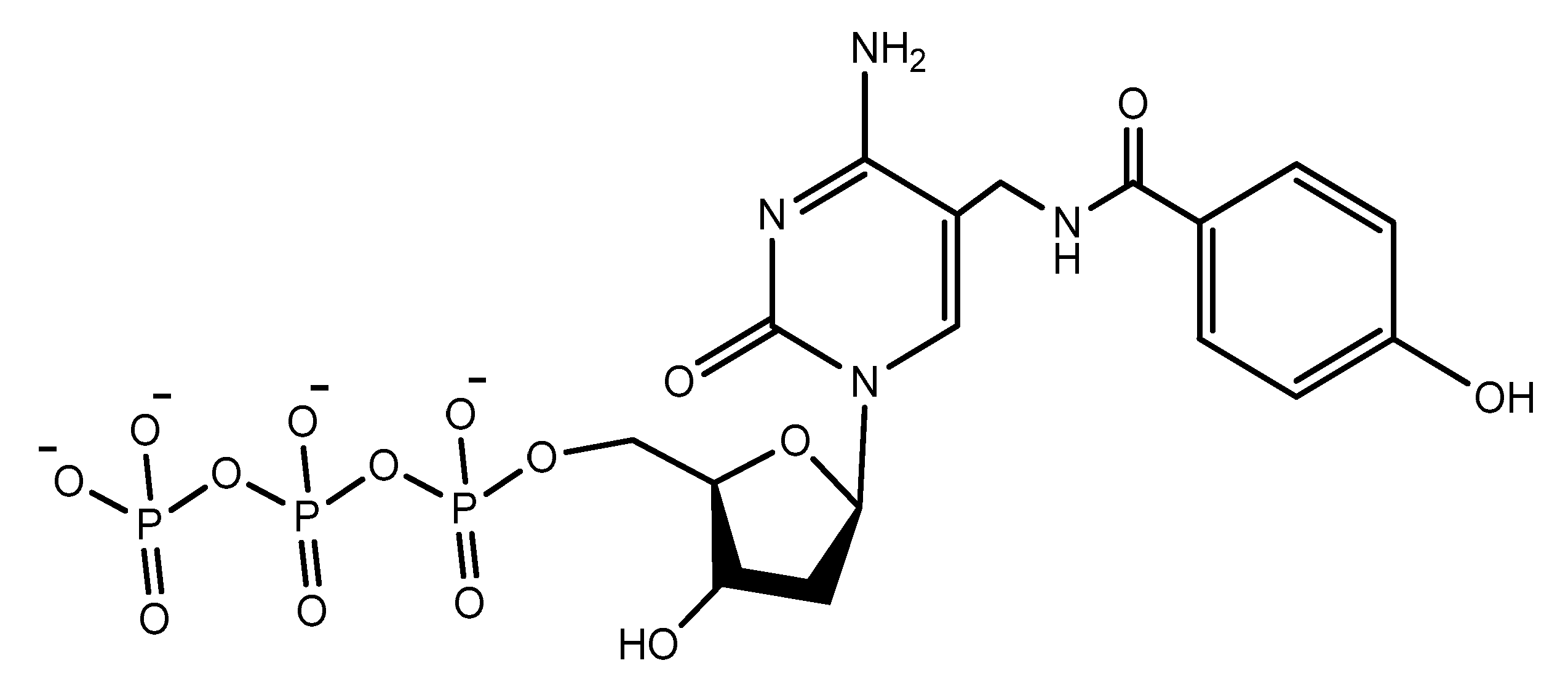
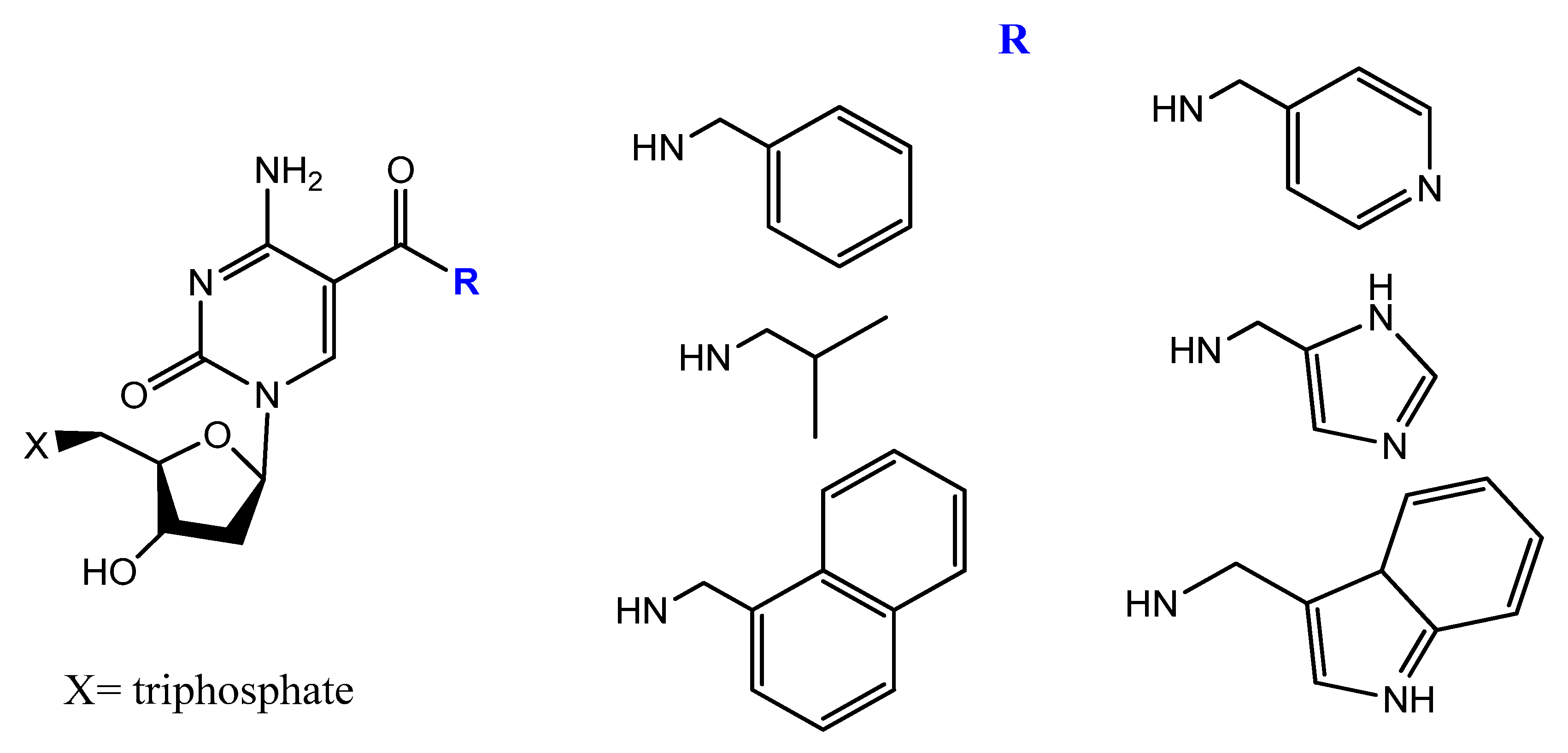
3.4. Modifications on the Bases and SOMAmers
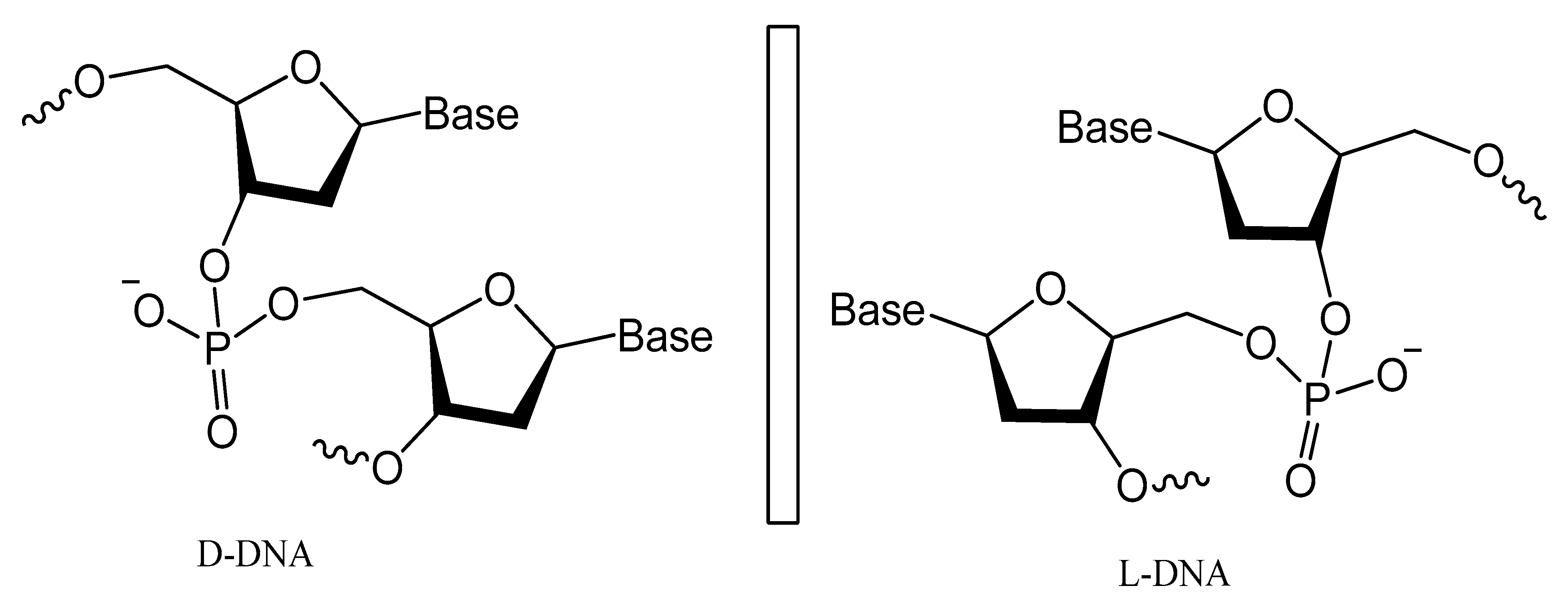
3.5. Spiegelmers
3.6. Circular Aptamers
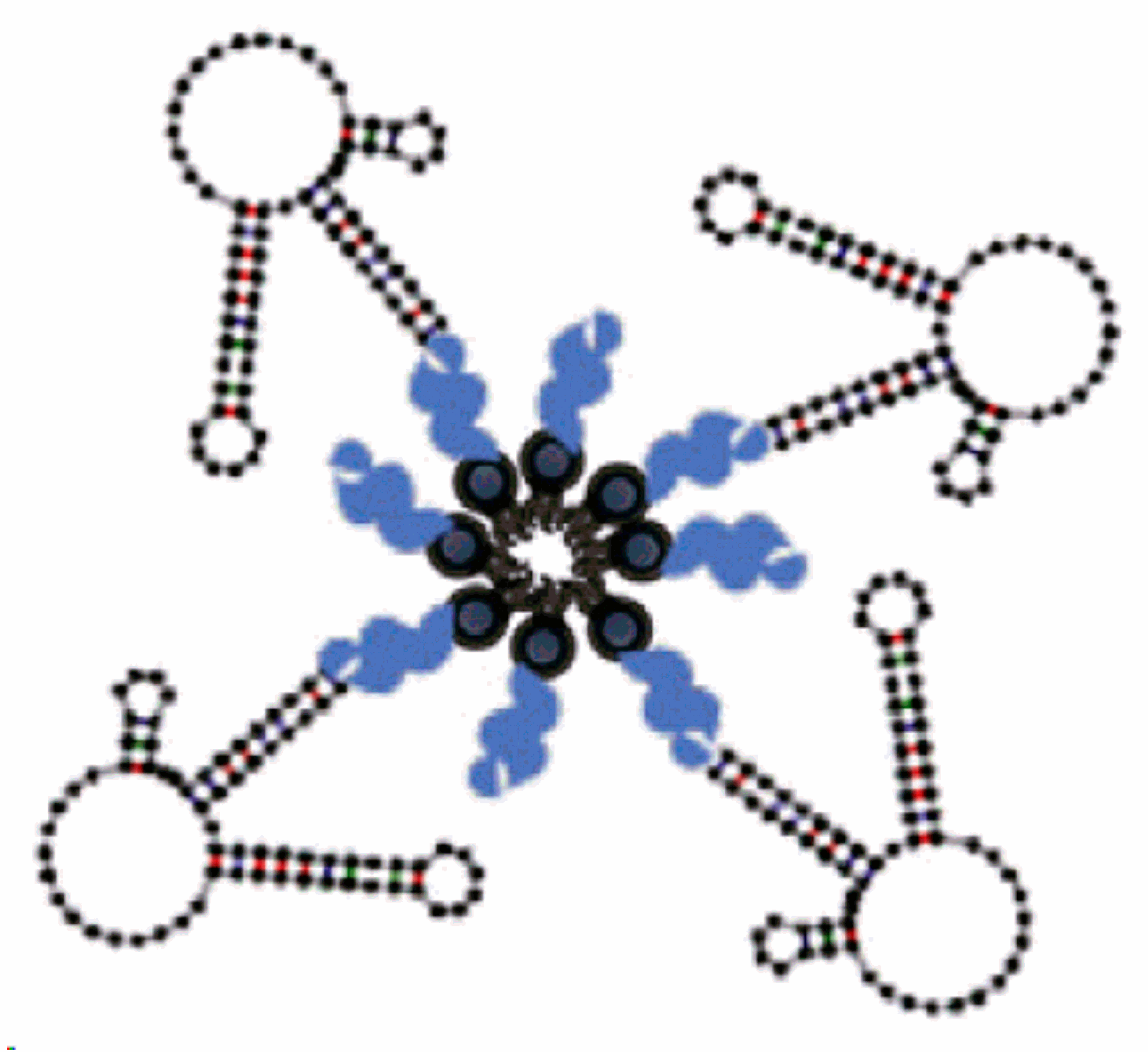
3.7. Multivalent and Dimerization of Aptamers
4. Chemical and Physical Conjugation Strategies of Aptamers to Nanoparticles
4.1. Direct and Post-Insertion
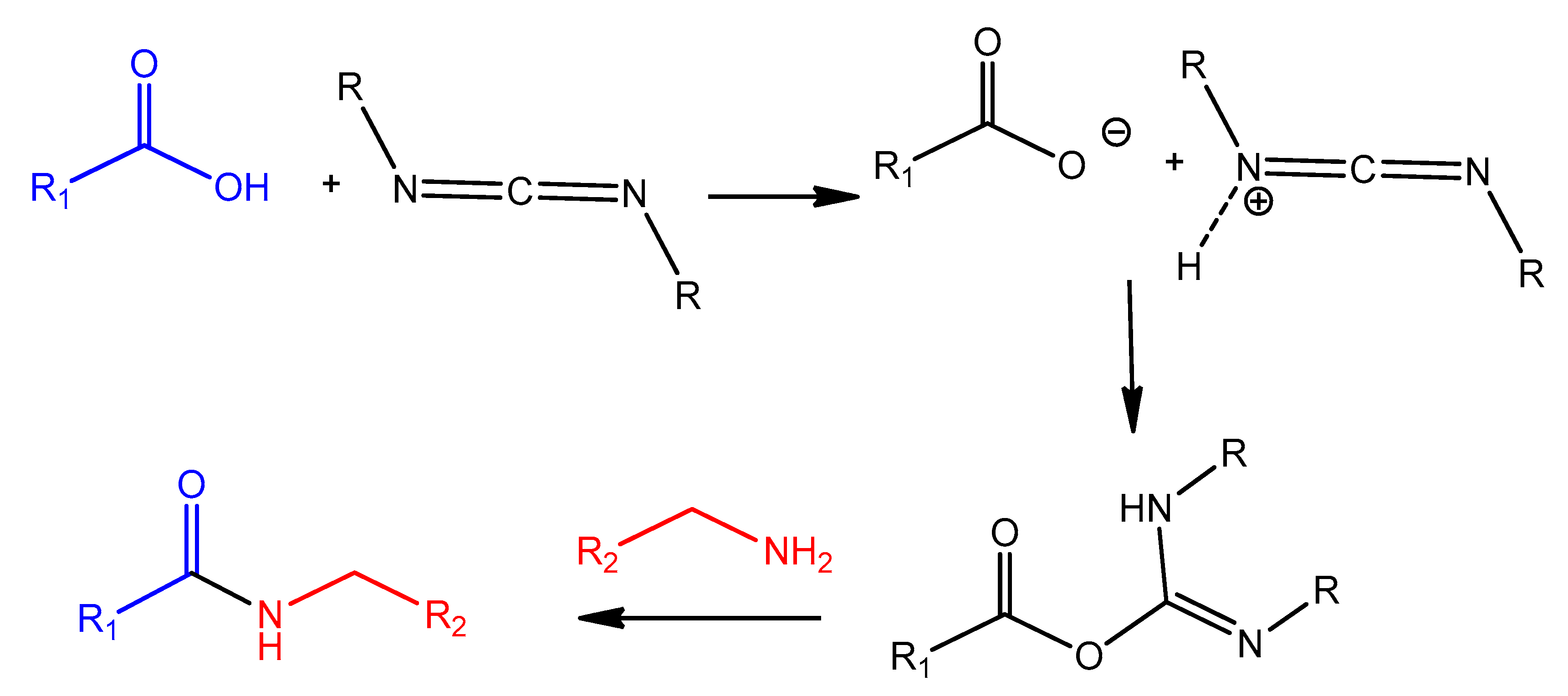
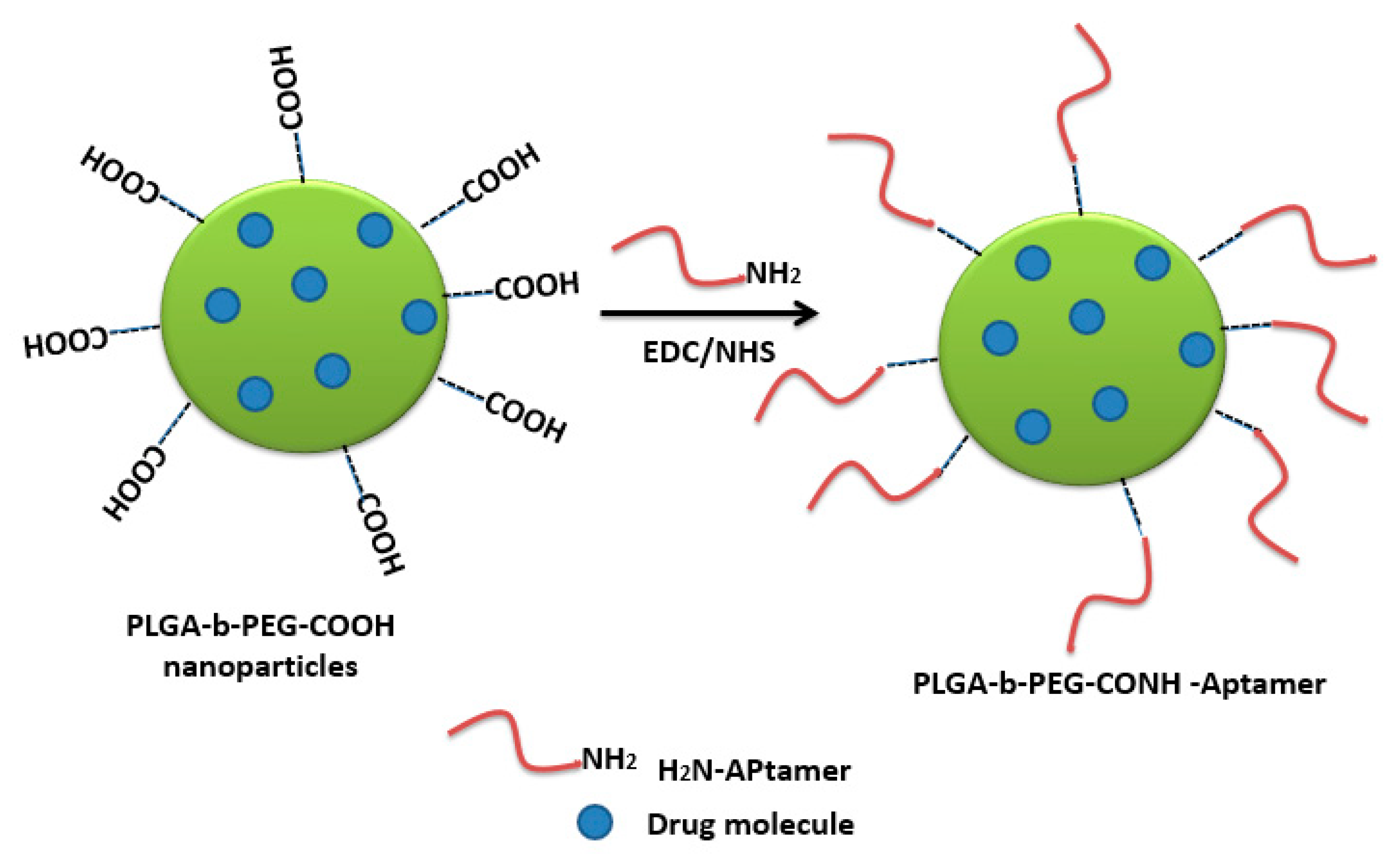
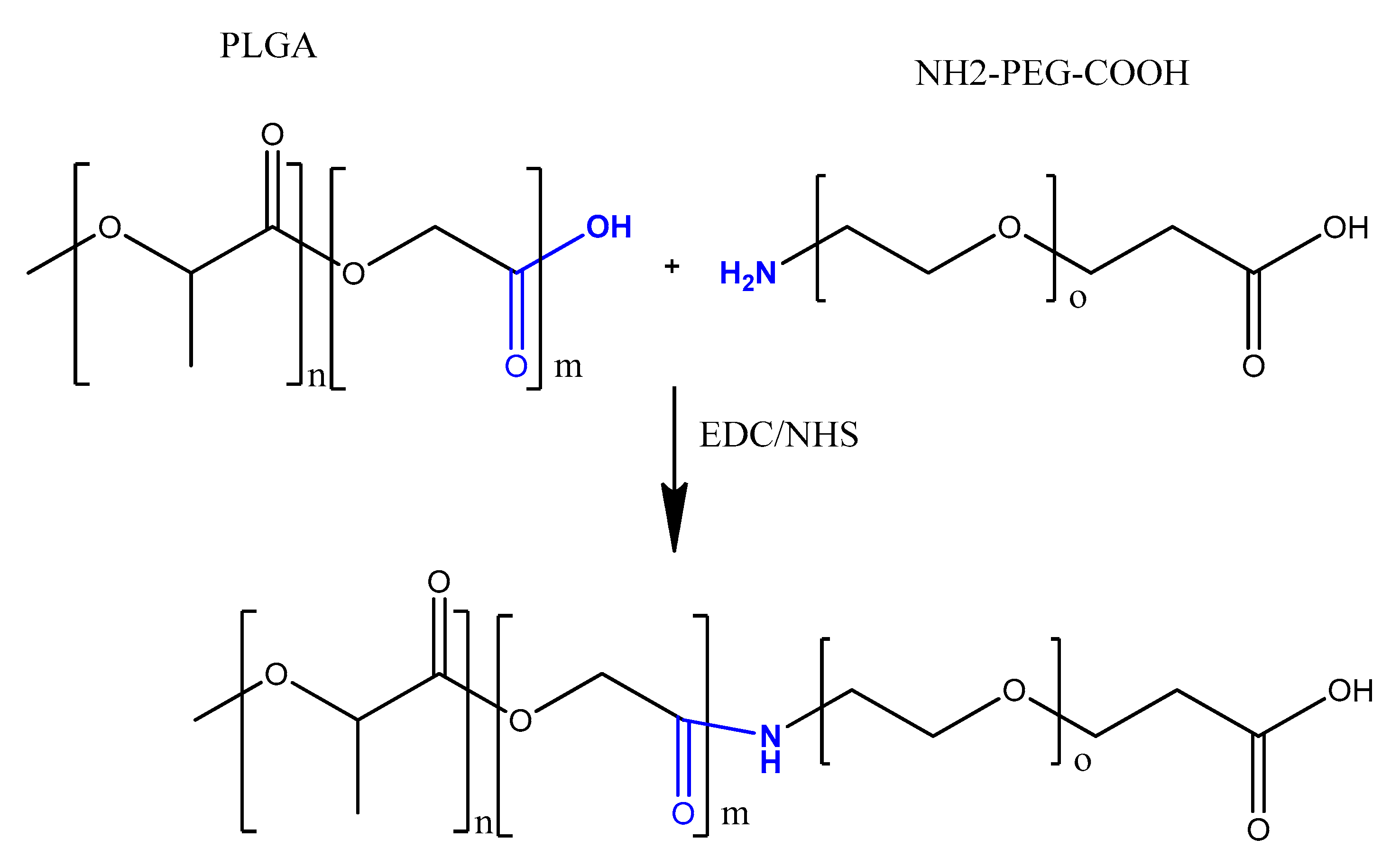
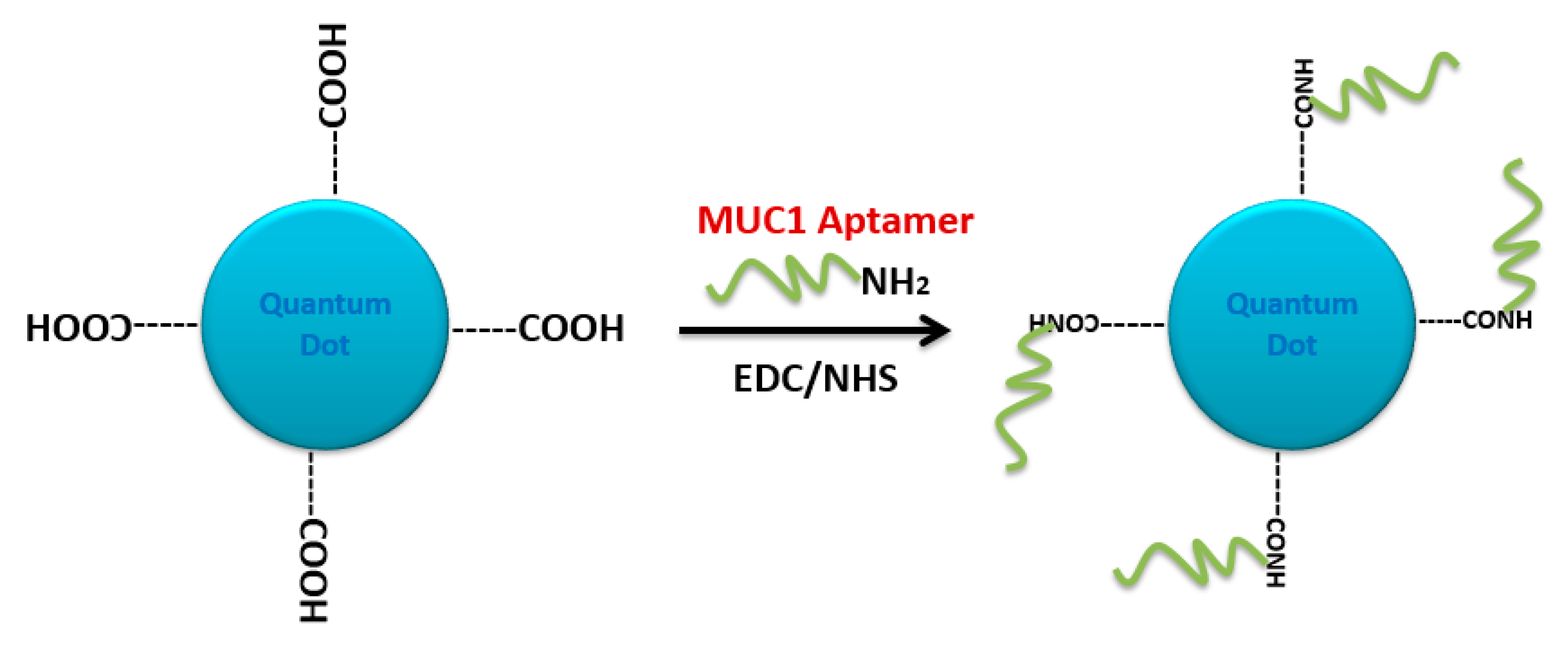
4.2. Carbodiimide Chemistry
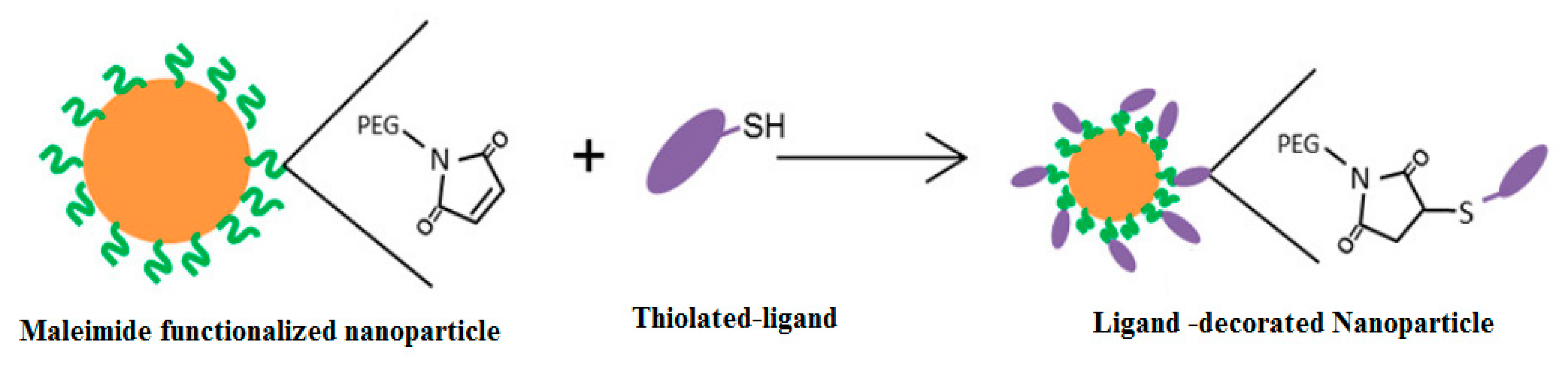
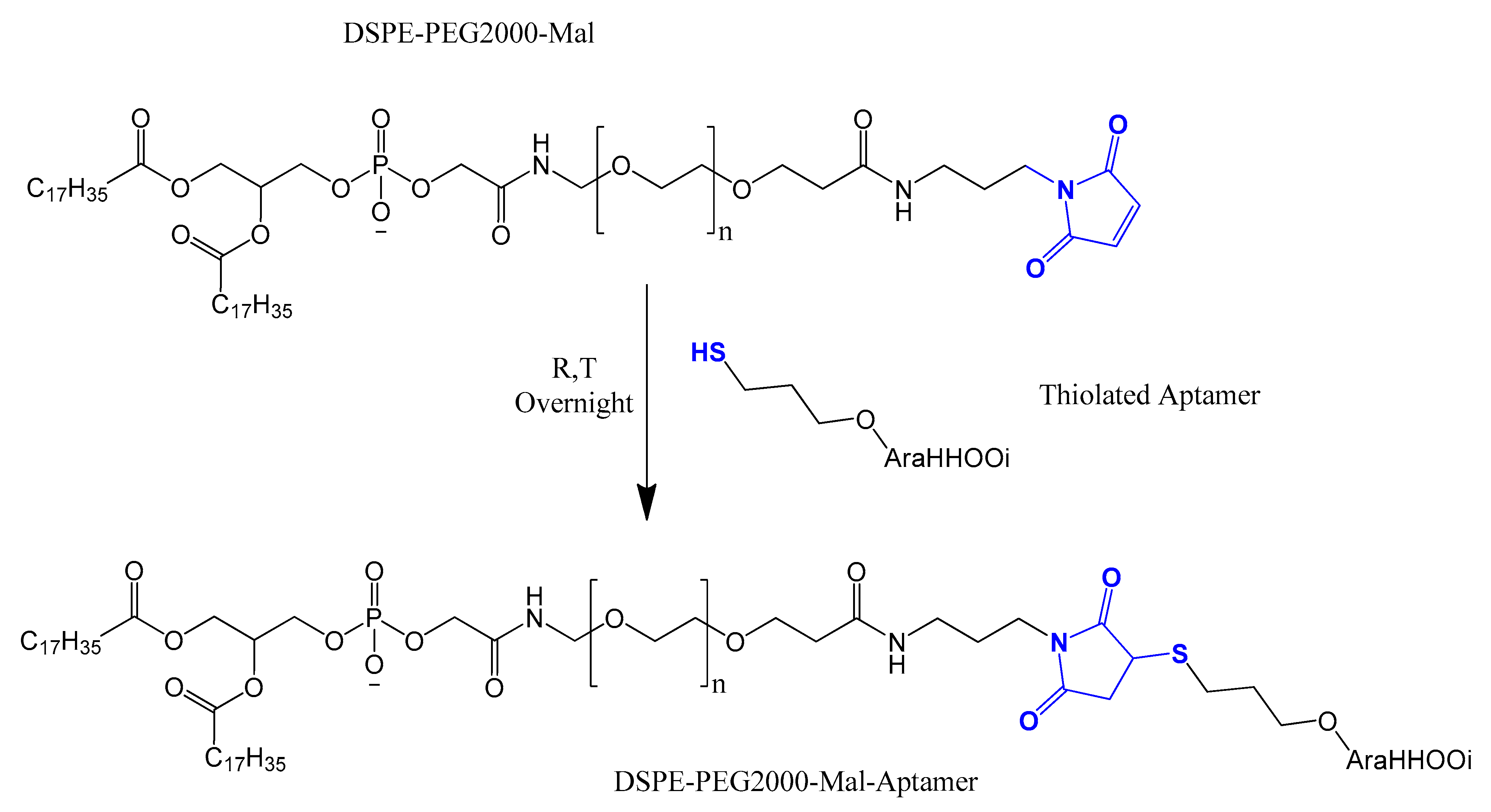
4.3. Thiol Maleiimide and Related Chemistry
4.4. Electrostatic and cDNA Strand Conjugation
4.5. Avidin–Biotin Coupling
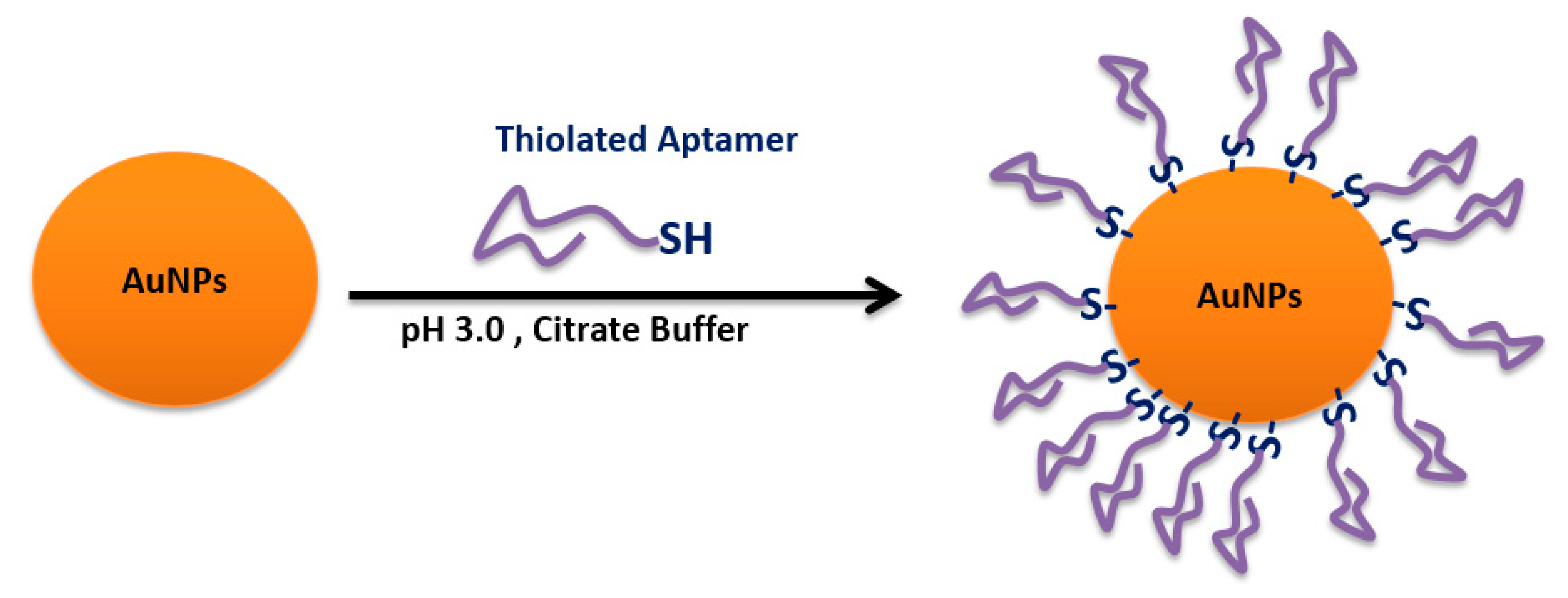
4.6. Sulfhydryl-Aptamer Gold Coordination
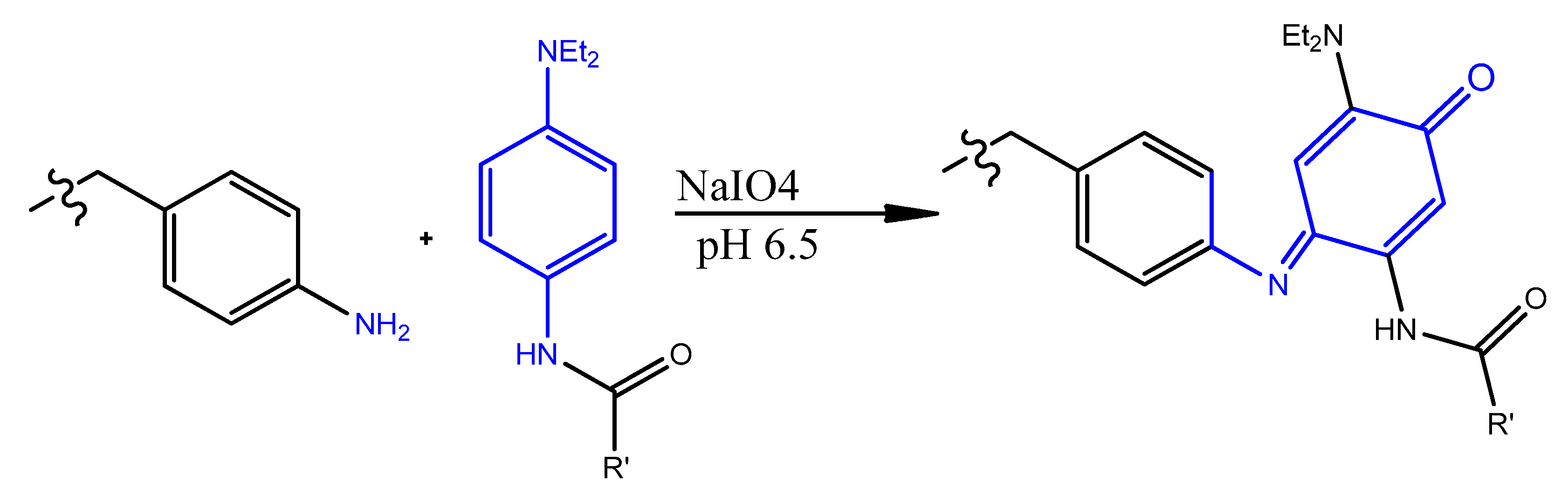
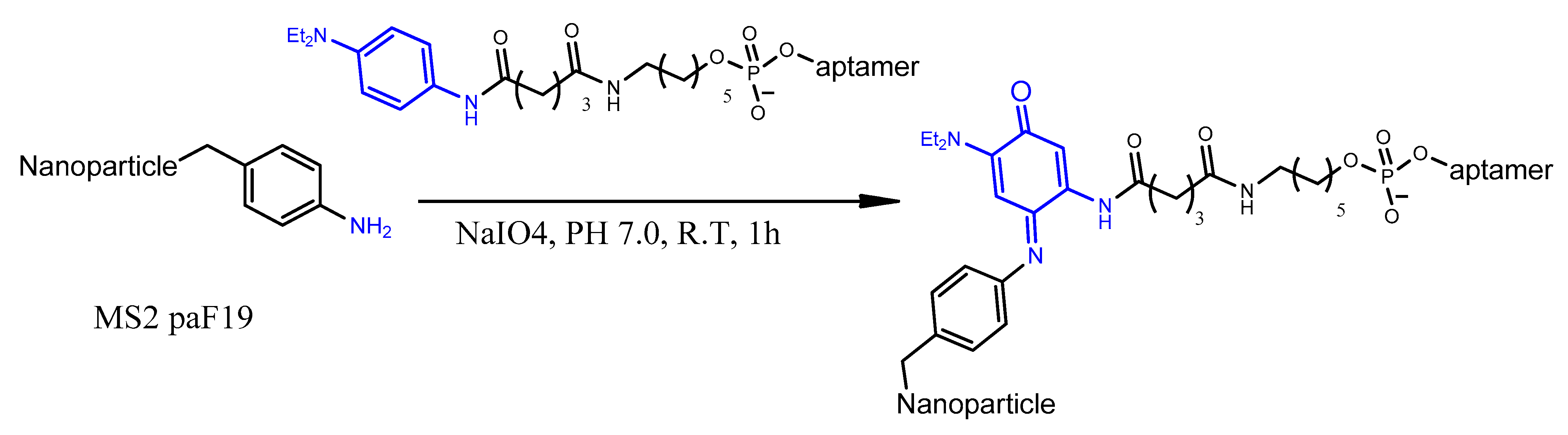
4.7. Oxidative Coupling
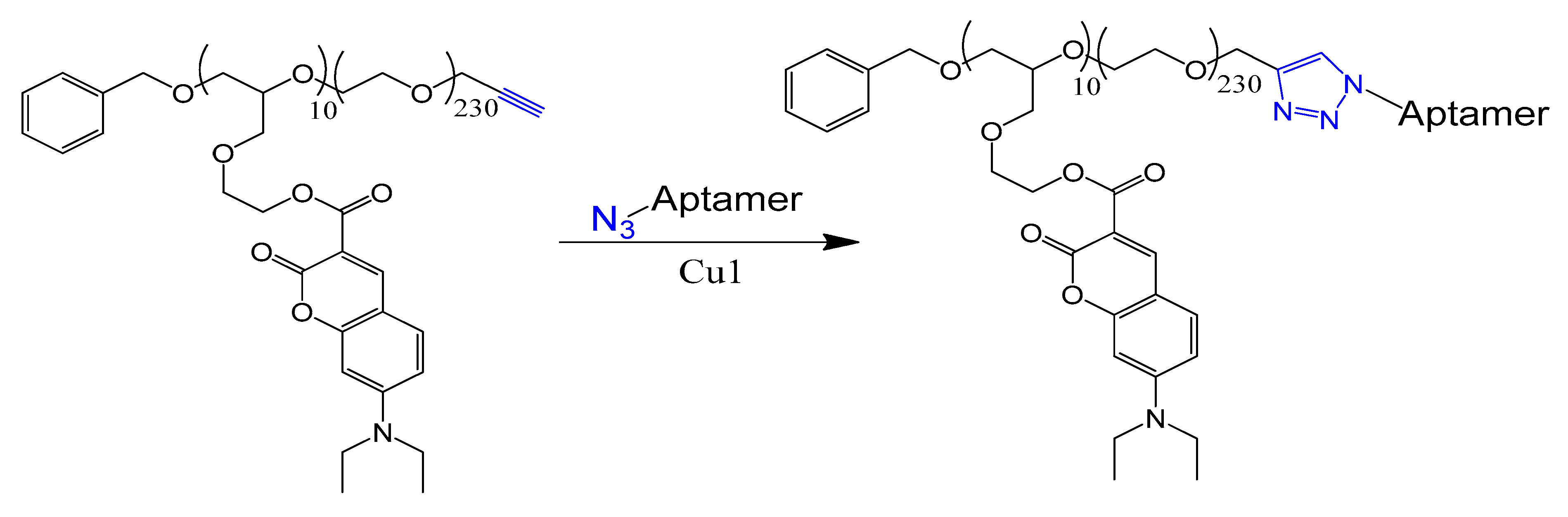
4.8. Click Chemistry
5. Aptamer Toxicity and Immunogenicity
6. Conclusions
Conflicts of Interest
References
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic acid aptamers: Clinical applications and promising new horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In Vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Wilbanks, B.; Smestad, J.; Heider, R.M.; Warrington, A.E.; Rodriguez, M.; Maher, L.J., III. Optimization of a 40-mer Antimyelin DNA Aptamer Identifies a 20-mer with Enhanced Properties for Potential Multiple Sclerosis Therapy. Nucleic Acid Ther. 2019, 29, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Amato, T.; Virgilio, A.; Pirone, L.; Vellecco, V.; Bucci, M.; Pedone, E.; Esposito, V.; Galeone, A. Investigating the properties of TBA variants with twin thrombin binding domains. Sci. Rep. 2019, 9, 9184. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Bahreyni, A.; Ramezani, M.; Alibolandi, M.; Hassanzadeh, P.; Abnous, K.; Taghdisi, S.M. High affinity of AS1411 toward copper; its application in a sensitive aptasensor for copper detection. Anal. Biochem. 2019, 575, 1–9. [Google Scholar] [CrossRef]
- Wu, Y.; Midinov, B.; White, R.J. Electrochemical Aptamer-Based Sensor for Real-Time Monitoring of Insulin. ACS Sens. 2019, 4, 498–503. [Google Scholar] [CrossRef]
- Zhao, H.; Ma, C.; Chen, M. A novel fluorometric method for inorganic pyrophosphatase detection based on G-quadruplex-thioflavin T. Mol. Cell. Probes 2019, 43, 29–33. [Google Scholar] [CrossRef]
- Soto Rodriguez, P.E.D.; Nash, V.I.C. Chapter 6—Aptamer-Based Strategies for Diagnostics. In Nucleic Acid Nanotheranostics; Filice, M., Ruiz-Cabello, J., Eds.; Elsevie: Amsterdam, The Netherlands, 2019; pp. 189–211. [Google Scholar]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt B, 1429–1447. [Google Scholar] [CrossRef]
- Nimjee, M.S.; Rusconi, C.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Campbell, M.A.; Wengel, J. Locked vs. unlocked nucleic acids (LNA vs. UNA): Contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 2011, 40, 5680–5689. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.G.; Damha, M.J. G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-D-arabinonucleic acids (2′F-ANA). Nucleic Acids Res. 2007, 35, 4977–4988. [Google Scholar] [CrossRef] [PubMed]
- Sacca, B.; Lacroix, L.; Mergny, J.L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005, 33, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Zaitseva, M.; Kaluzhny, D.; Shchyolkina, A.; Borisova, O.; Smirnov, I.; Pozmogova, G. Conformation and thermostability of oligonucleotide d(GGTTGGTGTGGTTGG) containing thiophosphoryl internucleotide bonds at different positions. Biophys. Chem. 2010, 146, 1–6. [Google Scholar] [CrossRef]
- Dougan, H.; Lyster, D.M.; Vo, C.V.; Stafford, A.; Weitz, J.I.; Hobbs, J.B. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nuclear Med. Biol. 2000, 27, 289–297. [Google Scholar] [CrossRef]
- de Smidt, P.C.; Le Doan, T.; de Falco, S.; van Berkel, T.J. Association of antisense oligonucleotides with lipoproteins prolongs the plasma half-life and modifies the tissue distribution. Nucleic Acids Res. 1991, 19, 4695–4700. [Google Scholar] [CrossRef]
- Obika, S.; Nanbu, D.; Hari, Y.; Morio, K.-I.; In, Y.; Ishida, T.; Imanishi, T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997, 38, 8735–8738. [Google Scholar] [CrossRef]
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630. [Google Scholar] [CrossRef]
- Dong, H.; Han, L.; Wang, J.; Xie, J.; Gao, Y.; Xie, F.; Jia, L. In Vivo inhibition of circulating tumor cells by two apoptosis-promoting circular aptamers with enhanced specificity. J. Control Release 2018, 280, 99–112. [Google Scholar] [CrossRef]
- Kang, Y.Y.; Song, J.; Jung, H.S.; Kwak, G.; Yu, G.; Ahn, J.-H.; Kim, S.H.; Mok, H. Implication of multivalent aptamers in DNA and DNA–RNA hybrid structures for efficient drug delivery in vitro and in vivo. J. Ind. Eng. Chem. 2018, 60, 250–258. [Google Scholar] [CrossRef]
- Bouhedda, F.; Fam, K.T.; Collot, M.; Autour, A.; Marzi, S.; Klymchenko, A.; Ryckelynck, M. A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells. Nat. Chem. Biol. 2019, 16, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Padilla, R.; Sousa, R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutantT7 RNA polymerase (RNAP). Nucleic Acids Res. 1999, 27, 1561–1563. [Google Scholar] [CrossRef] [PubMed]
- Maio, G.; Enweronye, O.; Zumrut, H.E.; Batool, S.; Van, N.; Mallikaratchy, P. Systematic optimization and modification of a DNA aptamer with 2′-O-methyl RNA analogues. ChemistrySelect 2017, 2, 2335–2340. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Lopes-Nunes, J.; Lopes, A.C.; Cabral Campello, M.P.; Paulo, A.; Queiroz, J.A.; Cruz, C. Aptamer-guided acridine derivatives for cervical cancer. Org. Biomol. Chem. 2019, 17, 2992–3002. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Miao, X.; Fan, G.-C.; Xu, T.; Jiang, L.-P.; Wu, P.; Cai, C.; Zhu, J.-J. Aptamer-Conjugated Au Nanocage/SiO2 Core–Shell Bifunctional Nanoprobes with High Stability and Biocompatibility for Cellular SERS Imaging and Near-Infrared Photothermal Therapy. ACS Sens. 2019, 4, 301–308. [Google Scholar] [CrossRef]
- Martínez-Jothar, L.; Doulkeridou, S.; Schiffelers, R.M.; Sastre Torano, J.; Oliveira, S.; van Nostrum, C.F.; Hennink, W.E. Insights into maleimide-thiol conjugation chemistry: Conditions for efficient surface functionalization of nanoparticles for receptor targeting. J. Control. Release 2018, 282, 101–109. [Google Scholar] [CrossRef]
- Savla, R.; Taratula, O.; Garbuzenko, O.; Minko, T. Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin conjugate for imaging and treatment of cancer. J. Control. Release 2011, 153, 16–22. [Google Scholar] [CrossRef]
- Stephanopoulos, N.; Tong, G.J.; Hsiao, S.C.; Francis, M.B. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS Nano 2010, 4, 6014–6020. [Google Scholar] [CrossRef]
- Danesh, N.M.; Lavaee, P.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Targeted and controlled release delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer-modified gold nanoparticles. Int. J. Pharm. 2015, 489, 311–317. [Google Scholar] [CrossRef]
- Ninomiya, K.; Yamashita, T.; Kawabata, S.; Shimizu, N. Targeted and ultrasound-triggered drug delivery using liposomes co-modified with cancer cell-targeting aptamers and a thermosensitive polymer. Ultrason Sonochem. 2014, 21, 1482–1488. [Google Scholar] [CrossRef]
- Presolski, I.S.; Hong, V.; Finn, M.G. Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol. 2011, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H. Recent developments in cell-SELEX technology for aptamer selection. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2323–2329. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, J.; Imanaka, H.; Yokota, Y.; Sugimoto, N. In Vitro selection of aptamers that act with Zn2+. J. Inorg. Biochem. 2000, 82, 197–206. [Google Scholar] [CrossRef]
- Hicke, B.J.; Marion, C.; Chang, Y.F.; Gould, T.; Lynott, C.K.; Parma, D.; Schmidt, P.G.; Warren, S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001, 276, 48644–48654. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, M.; Yang, G.; Zhang, D.; Ding, H.; Wang, H.; Fan, M.; Shen, B.; Shao, N. Single-stranded DNA aptamers that bind differentiated but not parental cells: Subtractive systematic evolution of ligands by exponential enrichment. J. Biotechnol. 2003, 102, 15–22. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33 Pt 2, 1141–1161. [Google Scholar] [CrossRef]
- Asai, R.; Nishimura, S.I.; Aita, T.; Takahashi, K. In Vitro Selection of DNA Aptamers on Chips Using a Method for Generating Point Mutations. Anal. Lett. 2004, 37, 645–656. [Google Scholar] [CrossRef]
- Park, J.-W.; Tatavarty, R.; Kim, D.W.; Jung, H.-T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. Chem. Commun. 2012, 48, 2071–2073. [Google Scholar] [CrossRef]
- Li, S.H.; Ding, H.; Huang, Y.; Cao, X.; Yang, G.; Li, J.; Xie, Z.; Meng, Y.; Li, X.; Zhao, Q.; et al. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J. Pathol. 2009, 218, 327–336. [Google Scholar] [CrossRef]
- Dong, L.; Tan, Q.; Ye, W.; Liu, D.; Chen, H.; Hu, H.; Wen, D.; Liu, Y.; Cao, Y.; Kang, J.; et al. Screening and Identifying a Novel ssDNA Aptamer against Alpha-fetoprotein Using CE-SELEX. Sci. Rep. 2015, 5, 15552. [Google Scholar] [CrossRef]
- Hamedani, N.S.; Muller, J. Capillary Electrophoresis for the Selection of DNA Aptamers Recognizing Activated Protein C. Methods Mol. Biol. 2016, 1380, 61–75. [Google Scholar] [PubMed]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef]
- Quang, N.N.; Perret, G.; Duconge, F. Applications of High-Throughput Sequencing for in vitro Selection and Characterization of Aptamers. Pharmaceuticals 2016, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Yang, H.; Qin, M.; Ding, X.; Liu, R.; Jiang, Y. In Vivo SELEX of an Inhibitory NSCLC-Specific RNA Aptamer from PEGylated RNA Library. Mol. Ther. Nucleic Acids 2018, 10, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Bel, N.; Das, A.T.; Berkhout, B. In Vivo SELEX of single-stranded domains in the HIV-1 leader RNA. J. Virol. 2014, 88, 1870–1880. [Google Scholar] [PubMed]
- Cheng, C.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In Vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Alshaer, W.; Hillaireau, H.; Fattal, E. Aptamer-guided nanomedicines for anticancer drug delivery. Adv. Drug Deliv. Rev. 2018, 134, 122–137. [Google Scholar] [CrossRef]
- Thiviyanathan, V.; Gorenstein, D.G. Aptamers and the next generation of diagnostic reagents. Proteom. Clin. Appl. 2012, 6, 563–573. [Google Scholar] [CrossRef]
- Ulrich, H.; Trujillo, C.A.; Nery, A.A.; Alves, J.M.; Majumder, P.; Resende, R.R.; Martins, A.H. DNA and RNA aptamers: From tools for basic research towards therapeutic applications. Comb. Chem. High. Throughput Screen 2006, 9, 619–632. [Google Scholar] [CrossRef]
- Diafa, S.; Hollenstein, M. Generation of Aptamers with an Expanded Chemical Repertoire. Molecules 2015, 20, 16643–16671. [Google Scholar] [CrossRef] [PubMed]
- Kratschmer, C.; Levy, M. Effect of Chemical Modifications on Aptamer Stability in Serum. Nucleic Acid Ther. 2017, 27, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Tolle, F.; Mayer, G. Dressed for success—applying chemistry to modulate aptamer functionality. Chem. Sci. 2013, 4, 60–67. [Google Scholar] [CrossRef]
- Jellinek, D.; Green, L.S.; Bell, C.; Lynott, C.K.; Gill, N.; Vargeese, C.; Kirschenheuter, G.; McGee, D.P.; Abesinghe, P.; Pieken, W.A.; et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 1995, 34, 11363–11372. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Sugimoto, N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules 2010, 15, 5423–5444. [Google Scholar] [CrossRef]
- Shum, K.T.; Tanner, J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem 2008, 9, 3037–3045. [Google Scholar] [CrossRef]
- Caruthers, M.H.; Barone, A.D.; Beaucage, S.L.; Dodds, D.R.; Fisher, E.F.; McBride, L.J.; Matteucci, M.; Stabinsky, Z.; Tang, J.Y. Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods Enzymol. 1987, 154, 287–313. [Google Scholar]
- Riccardi, C.; Russo Krauss, I.; Musumeci, D.; Morvan, F.; Meyer, A.; Vasseur, J.J.; Paduano, L.; Montesarchio, D. Fluorescent Thrombin Binding Aptamer-Tagged Nanoparticles for an Efficient and Reversible Control of Thrombin Activity. ACS Appl. Mater. Interfaces 2017, 9, 35574–35587. [Google Scholar] [CrossRef]
- Ortigao, J.F.R.; ÖSch, H.; Montenarh, M.; FrÖHlich, A.; Seliger, H. Oligonucleotide Analogs with Terminal 3′,3′- and 5′,5′-Internucleotidic Linkages as Antisense Inhibitors of Viral Replication. Antisense Res. Dev. 1991, 1, 380. [Google Scholar] [CrossRef]
- Healy, J.M.; Lewis, S.D.; Kurz, M.; Boomer, R.M.; Thompson, K.M.; Wilson, C.; McCauley, T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. [Google Scholar] [CrossRef]
- Prodeus, A.; Abdul-Wahid, A.; Fischer, N.W.; Huang, E.H.; Cydzik, M.; Gariepy, J. Targeting the PD-1/PD-L1 Immune Evasion Axis With DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers. Mol. Ther. Nucleic Acids 2015, 4, e237. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Lee, S.H.; Kim, J.H.; Noh, Y.H.; Noh, G.J.; Lee, S.W. Pharmacokinetics of a Cholesterol-conjugated Aptamer Against the Hepatitis C Virus (HCV) NS5B Protein. Mol. Ther. Nucleic Acids 2015, 4, e254. [Google Scholar] [CrossRef] [PubMed]
- Willis, M.C.; Collins, B.D.; Zhang, T.; Green, L.S.; Sebesta, D.P.; Bell, C.; Kellogg, E.; Gill, S.C.; Magallanez, A.; Knauer, S.; et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug. Chem. 1998, 9, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Musumeci, D.; Russo Krauss, I.; Piccolo, M.; Irace, C. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018, 118 Pt B, 1384–1399. [Google Scholar] [CrossRef]
- Veronese, F.M.; Pasut, G. PEGylation, successful approach to drug delivery. Drug Discov. Today 2005, 10, 1451–1458. [Google Scholar] [CrossRef]
- Hoffmann, S.; Hoos, J.; Klussmann, S.; Vonhoff, S. RNA Aptamers and Spiegelmers: Synthesis, Purification, and Post-Synthetic PEG Conjugation. Curr. Protoc. Nucleic Acid Chem. 2011, 46, 4–46. [Google Scholar] [CrossRef]
- Kazuhiko, H.; Natsuki, O.; Masakazu, N.; Tomoyoshi, K.; Asako, S.; Shinsuke, H.; Masayuki, T.; Kuniyoshi, H.; Hideaki, S.; Hiroaki, Y. A Novel PEGylation Method for Improving the Pharmacokinetic Properties of Anti-Interleukin-17A RNA Aptamers. Nucleic Acid Ther. 2017, 27, 36–44. [Google Scholar]
- Da Pieve, C.; Blackshaw, E.; Missailidis, S.; Perkins, A.C. PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice. Bioconjug. Chem. 2012, 23, 1377–1381. [Google Scholar] [CrossRef]
- Tan, L.; Neoh, K.G.; Kang, E.T.; Choe, W.S.; Su, X. PEGylated anti-MUC1 aptamer-doxorubicin complex for targeted drug delivery to MCF7 breast cancer cells. Macromol. Biosci. 2011, 11, 1331–1335. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef]
- Lin, Y.; Qiu, Q.; Gill, S.C.; Jayasena, S.D. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 1994, 22, 5229–5234. [Google Scholar] [CrossRef] [PubMed]
- Green, L.S.; Jellinek, D.; Bell, C.; Beebe, L.A.; Feistner, B.D.; Gill, S.C.; Jucker, F.M.; Janjic, N. Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem. Biol. 1995, 2, 683–695. [Google Scholar] [CrossRef]
- Pagratis, N.C.; Bell, C.; Chang, Y.F.; Jennings, S.; Fitzwater, T.; Jellinek, D.; Dang, C. Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997, 15, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Li, N.; Nguyen, H.H.; Byrom, M.; Ellington, A.D. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS ONE 2011, 6, e20299. [Google Scholar] [CrossRef]
- Esposito, C.L.; Passaro, D.; Longobardo, I.; Condorelli, G.; Marotta, P.; Affuso, A.; de Franciscis, V.; Cerchia, L. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS ONE 2011, 6, e24071. [Google Scholar] [CrossRef]
- Svobodova, M.; Bunka, D.H.; Nadal, P.; Stockley, P.G.; O’Sullivan, C.K. Selection of 2′F-modified RNA aptamers against prostate-specific antigen and their evaluation for diagnostic and therapeutic applications. Anal. Bioanal. Chem. 2013, 405, 9149–9157. [Google Scholar] [CrossRef]
- Lupold, S.E.; Hicke, B.J.; Lin, Y.; Coffey, D.S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002, 62, 4029–4033. [Google Scholar]
- Thirunavukarasu, D.; Chen, T.; Liu, Z.; Hongdilokkul, N.; Romesberg, F.E. Selection of 2′-Fluoro-Modified Aptamers with Optimized Properties. J. Am. Chem. Soc. 2017, 139, 2892–2895. [Google Scholar] [CrossRef]
- Lauridsen, H.L.; Rothnagel, J.A.; Veedu, R.N. Enzymatic Recognition of 2′-Modified Ribonucleoside 5′-Triphosphates: Towards the Evolution of Versatile Aptamers. ChemBioChem 2012, 13, 19–25. [Google Scholar] [CrossRef]
- Waters, E.K.; Genga, R.M.; Schwartz, M.C.; Nelson, J.A.; Schaub, R.G.; Olson, K.A.; Kurz, J.C.; McGinness, K.E. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood 2011, 117, 5514–5522. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Levy-Nissenbaum, E.; Alexis, F.; Luptak, A.; Teply, B.A.; Chan, J.M.; Shi, J.; Digga, E.; Cheng, J.; Langer, R.; et al. Engineering of targeted nanoparticles for cancer therapy using internalizing aptamers isolated by cell-uptake selection. ACS Nano 2012, 6, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Wilds, C.J.; Damha, M.J. 2′-Deoxy-2′-fluoro-beta-D-arabinonucleosides and oligonucleotides (2′F-ANA): Synthesis and physicochemical studies. Nucleic Acids Res. 2000, 28, 3625–3635. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.K.; Katolik, A.; Viladoms, J.; Damha, M.J. Studies on the hydrolytic stability of 2′-fluoroarabinonucleic acid (2′F-ANA). Org. Biomol. Chem. 2009, 7, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.K.; Katolik, A.; Viladoms, J.; Damha, M.J. Differential stability of 2′F-ANA*RNA and ANA*RNA hybrid duplexes: Roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010, 38, 2498–2511. [Google Scholar] [CrossRef] [PubMed]
- Minakawa, N.; Sanji, M.; Kato, Y.; Matsuda, A. Investigations toward the selection of fully-modified 4′-thioRNA aptamers: Optimization of in vitro transcription steps in the presence of 4′-thioNTPs. Bioorg. Med. Chem. 2008, 16, 9450–9456. [Google Scholar] [CrossRef]
- Kato, Y.; Minakawa, N.; Komatsu, Y.; Kamiya, H.; Ogawa, N.; Harashima, H.; Matsuda, A. New NTP analogs: The synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res. 2005, 33, 2942–2951. [Google Scholar] [CrossRef]
- Darfeuille, F.; Hansen, J.B.; Orum, H.; Di Primo, C.; Toulme, J.J. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004, 32, 3101–3107. [Google Scholar] [CrossRef]
- Shi, H.; He, X.; Cui, W.; Wang, K.; Deng, K.; Li, D.; Xu, F. Locked nucleic acid/DNA chimeric aptamer probe for tumor diagnosis with improved serum stability and extended imaging window in vivo. Anal. Chim. Acta 2014, 812, 138–144. [Google Scholar] [CrossRef]
- Schmidt, K.S.; Borkowski, S.; Kurreck, J.; Stephens, A.W.; Bald, R.; Hecht, M.; Friebe, M.; Dinkelborg, L.; Erdmann, V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004, 32, 5757–5765. [Google Scholar] [CrossRef]
- Hernandez, F.J.; Kalra, N.; Wengel, J.; Vester, B. Aptamers as a model for functional evaluation of LNA and 2′-amino LNA. Bioorg. Med. Chem. Lett. 2009, 19, 6585–6587. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.; Hernandez, F.J.; Rasmussen, L.M.; Vester, B.; Wengel, J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011, 39, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Veedu, N.R.; Vester, B.; Wengel, J. Efficient enzymatic synthesis of LNA-modified DNA duplexes using KOD DNA polymerase. Org. Biomol. Chem. 2009, 7, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Kotkowiak, W.; Wengel, J.; Scotton, C.J.; Pasternak, A. Improved RE31 Analogues Containing Modified Nucleic Acid Monomers: Thermodynamic, Structural, and Biological Effects. J. Med. Chem. 2019, 62, 2499–2507. [Google Scholar] [CrossRef] [PubMed]
- Pozmogova, G.E.; Zaitseva, M.A.; Smirnov, I.P.; Shvachko, A.G.; Murina, M.A.; Sergeenko, V.I. Anticoagulant effects of thioanalogs of thrombin-binding DNA-aptamer and their stability in the plasma. Bull. Exp. Biol. Med. 2010, 150, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Abeydeera, N.D.; Egli, M.; Cox, N.; Mercier, K.; Conde, J.N.; Pallan, P.S.; Mizurini, D.M.; Sierant, M.; Hibti, F.E.; Hassell, T.; et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016, 44, 8052–8064. [Google Scholar] [CrossRef]
- Mann, A.P.; Somasunderam, A.; Nieves-Alicea, R.; Li, X.; Hu, A.; Sood, A.K.; Ferrari, M.; Gorenstein, D.G.; Tanaka, T. Identification of thioaptamer ligand against E-selectin: Potential application for inflamed vasculature targeting. PLoS ONE 2010, 5, e13050. [Google Scholar] [CrossRef]
- King, D.J.; Bassett, S.E.; Li, X.; Fennewald, S.A.; Herzog, N.K.; Luxon, B.A.; Shope, R.; Gorenstein, D.G. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-kappa B RelA(p65) and p50. Biochemistry 2002, 41, 9696–9706. [Google Scholar] [CrossRef]
- Somasunderam, A.; Ferguson, M.R.; Rojo, D.R.; Thiviyanathan, V.; Li, X.; O’Brien, W.A.; Gorenstein, D.G. Combinatorial selection, inhibition, and antiviral activity of DNA thioaptamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 2005, 44, 10388–10395. [Google Scholar] [CrossRef]
- Somasunderam, A.; Thiviyanathan, V.; Tanaka, T.; Li, X.; Neerathilingam, M.; Lokesh, G.L.; Mann, A.; Peng, Y.; Ferrari, M.; Klostergaard, J.; et al. Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of human CD44. Biochemistry 2010, 49, 9106–9112. [Google Scholar] [CrossRef]
- Gandham, S.H.; Volk, D.E.; Lokesh, G.L.; Neerathilingam, M.; Gorenstein, D.G. Thioaptamers targeting dengue virus type-2 envelope protein domain III. Biochem. Biophys. Res. Commun. 2014, 453, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Leonard, F.; Ha, N.P.; Sule, P.; Alexander, J.F.; Volk, D.E.; Lokesh, G.L.R.; Liu, X.; Cirillo, J.D.; Gorenstein, D.G.; Yuan, J.; et al. Thioaptamer targeted discoidal microparticles increase self immunity and reduce Mycobacterium tuberculosis burden in mice. J. Control. Release 2017, 266, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Xianbin, Y.; David, G.G. Progress in Thioaptamer Development. Curr. Drug Targets 2004, 5, 705–715. [Google Scholar]
- Lokesh, G.L.; Wang, H.; Lam, C.H.; Thiviyanathan, V.; Ward, N.; Gorenstein, D.G.; Volk, D.E. X-Aptamer Selection and Validation. Methods Mol. Biol. 2017, 1632, 151–174. [Google Scholar]
- He, W.; Elizondo-Riojas, M.A.; Li, X.; Lokesh, G.L.; Somasunderam, A.; Thiviyanathan, V.; Volk, D.E.; Durland, R.H.; Englehardt, J.; Cavasotto, C.N.; et al. X-aptamers: A bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry 2012, 51, 8321–8323. [Google Scholar] [CrossRef]
- Prater, C.E.; Miller, P.S. 3′-Methylphosphonate-Modified Oligo-2′-O-methylribonucleotides and Their Tat Peptide Conjugates: Uptake and Stability in Mouse Fibroblasts in Culture. Bioconjugate Chem. 2004, 15, 498–507. [Google Scholar] [CrossRef]
- Kibler-Herzog, L.; Zon, G.; Uznanski, B.; Whittier, G.; Wilson, W.D. Duplex stabilities of phosphorothioate, methylphosphonate, and RNA analogs of two DNA 14-mers. Nucleic Acids Res. 1991, 19, 2979–2986. [Google Scholar] [CrossRef]
- Mutisya, D.; Selvam, C.; Kennedy, S.D.; Rozners, E. Synthesis and properties of triazole-linked RNA. Bioorg. Med. Chem. Lett 2011, 21, 3420–3422. [Google Scholar] [CrossRef]
- Sau, S.; Hrdlicka, P.J. C2′-pyrene-functionalized triazole-linked DNA: Universal DNA/RNA hybridization probes. J. Org. Chem. 2012, 77, 5–16. [Google Scholar] [CrossRef]
- Varizhuk, A.M.; Kaluzhny, D.N.; Novikov, R.A.; Chizhov, A.O.; Smirnov, I.P.; Chuvilin, A.N.; Tatarinova, O.N.; Fisunov, G.Y.; Pozmogova, G.E.; Florentiev, V.L. Synthesis of triazole-linked oligonucleotides with high affinity to DNA complements and an analysis of their compatibility with biosystems. J. Org. Chem. 2013, 78, 5964–5969. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click chemistry with DNA. Chem. Soc. Rev. 2010, 39, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. Click nucleic acid ligation: Applications in biology and nanotechnology. Acc. Chem. Res. 2012, 45, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, S.; Srihari, P.; Nagesh, C.; Kiranmai, N.; Nagesh, N.; Idris, M.M. Synthesis of Readily Accessible Triazole-Linked Dimer Deoxynucleoside Phosphoramidite for Solid-Phase Oligonucleotide Synthesis. Synthesis 2010, 2010, 3710–3714. [Google Scholar] [CrossRef]
- Varizhuk, A.M.; Tsvetkov, V.B.; Tatarinova, O.N.; Kaluzhny, D.N.; Florentiev, V.L.; Timofeev, E.N.; Shchyolkina, A.K.; Borisova, O.F.; Smirnov, I.P.; Grokhovsky, S.L.; et al. Synthesis, characterization and in vitro activity of thrombin-binding DNA aptamers with triazole internucleotide linkages. Eur. J. Med. Chem. 2013, 67, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Hocek, M. Synthesis of base-modified 2′-deoxyribonucleoside triphosphates and their use in enzymatic synthesis of modified DNA for applications in bioanalysis and chemical biology. J. Org. Chem. 2014, 79, 9914–9921. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. Synthesis of Deoxynucleoside Triphosphates that Include Proline, Urea, or Sulfonamide Groups and Their Polymerase Incorporation into DNA. Chem. A Eur. J. 2012, 18, 13320–13330. [Google Scholar] [CrossRef]
- Latham, A.J.; Johnson, R.; Toole, J.J. The application of a modified nucleotide in aptamer selection: Novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res. 1994, 22, 2817–2822. [Google Scholar] [CrossRef]
- Gupta, S.; Drolet, D.W.; Wolk, S.K.; Waugh, S.M.; Rohloff, J.C.; Carter, J.D.; Mayfield, W.S.; Otis, M.R.; Fowler, C.R.; Suzuki, T.; et al. Pharmacokinetic Properties of DNA Aptamers with Base Modifications. Nucleic Acid Ther. 2017, 27, 345–353. [Google Scholar] [CrossRef]
- Jensen, K.B.; Atkinson, B.L.; Willis, M.C.; Koch, T.H.; Gold, L. Using in vitro selection to direct the covalent attachment of human immunodeficiency virus type 1 Rev protein to high-affinity RNA ligands. Proc. Natl. Acad. Sci. USA 1995, 92, 12220–12224. [Google Scholar] [CrossRef]
- Li, M.; Lin, N.; Huang, Z.; Du, L.; Altier, C.; Fang, H.; Wang, B. Selecting aptamers for a glycoprotein through the incorporation of the boronic acid moiety. J. Am. Chem. Soc. 2008, 130, 12636–12638. [Google Scholar] [CrossRef]
- Imaizumi, Y.; Kasahara, Y.; Fujita, H.; Kitadume, S.; Ozaki, H.; Endoh, T.; Kuwahara, M.; Sugimoto, N. Efficacy of base-modification on target binding of small molecule DNA aptamers. J. Am. Chem. Soc. 2013, 135, 9412–9419. [Google Scholar] [CrossRef] [PubMed]
- Shoji, A.; Kuwahara, M.; Ozaki, H.; Sawai, H. Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J. Am. Chem. Soc. 2007, 129, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, K.; Kasamatsu, T.; Nagashima, J.; Hanawa, K.; Kuwahara, M.; Ozaki, H.; Sawai, H. Arginine-modified DNA aptamers that show enantioselective recognition of the dicarboxylic acid moiety of glutamic acid. Anal. Sci. 2008, 24, 167–172. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, I.S.; Temme, J.S.; Habeshian, S.; Felczak, K.; Pankiewicz, K.; Hedstrom, L.; Krauss, I.J. Multivalent glycocluster design through directed evolution. Angew. Chem. Int. Ed. Engl. 2011, 50, 11238–11242. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; Drzyzga, M.G.; MacPherson, I.S.; Krauss, I.J. Directed evolution of 2G12-targeted nonamannose glycoclusters by SELMA. Chemistry 2013, 19, 17291–17295. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; MacPherson, I.S.; DeCourcey, J.F.; Krauss, I.J. High temperature SELMA: Evolution of DNA-supported oligomannose clusters which are tightly recognized by HIV bnAb 2G12. J. Am. Chem. Soc. 2014, 136, 1726–1729. [Google Scholar] [CrossRef]
- Lee, K.Y.; Kang, H.; Ryu, S.H.; Lee, D.S.; Lee, J.H.; Kim, S. Bioimaging of nucleolin aptamer-containing 5-(N-benzylcarboxyamide)-2′-deoxyuridine more capable of specific binding to targets in cancer cells. J. Biomed. Biotechnol. 2010, 2010, 168306. [Google Scholar] [CrossRef]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jimenez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D.; et al. In Vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef]
- Betz, K.; Kimoto, M.; Diederichs, K.; Hirao, I.; Marx, A. Structural Basis for Expansion of the Genetic Alphabet with an Artificial Nucleobase Pair. Angew. Chem. Int. Ed. 2017, 56, 12000–12003. [Google Scholar] [CrossRef]
- Renders, M.; Miller, E.; Lam, C.H.; Perrin, D.M. Whole cell-SELEX of aptamers with a tyrosine-like side chain against live bacteria. Org. Biomol. Chem. 2017, 15, 1980–1989. [Google Scholar] [CrossRef] [PubMed]
- Balintová, J.; Simonova, A.; Białek-Pietras, M.; Olejniczak, A.; Lesnikowski, Z.J.; Hocek, M. Carborane-linked 2′-deoxyuridine 5′-O-triphosphate as building block for polymerase synthesis of carborane-modified DNA. Bioorg. Med. Chem. Lett. 2017, 27, 4786–4788. [Google Scholar] [CrossRef] [PubMed]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the Chemistry of DNA for in vitro Selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Tolle, F.; Rosenthal, M.; Brandle, G.M.; Ewers, J.; Mayer, G. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 2018, 13, 1153–1180. [Google Scholar] [CrossRef]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D.; et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef]
- Hopfield, J.J. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA 1974, 71, 4135–4139. [Google Scholar] [CrossRef]
- Hathout, Y.; Brody, E.; Clemens, P.R.; Cripe, L.; DeLisle, R.K.; Furlong, P.; Gordish-Dressman, H.; Hache, L.; Henricson, E.; Hoffman, E.P.; et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 2015, 112, 7153–7158. [Google Scholar] [CrossRef]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef]
- Duo, J.; Chiriac, C.; Huang, R.Y.C.; Mehl, J.; Chen, G.; Tymiak, A.; Sabbatini, P.; Pillutla, R.; Zhang, Y. Slow Off-Rate Modified Aptamer (SOMAmer) as a Novel Reagent in Immunoassay Development for Accurate Soluble Glypican-3 Quantification in Clinical Samples. Anal. Chem. 2018, 90, 5162–5170. [Google Scholar] [CrossRef]
- Gawande, B.N.; Rohloff, J.C.; Carter, J.D.; von Carlowitz, I.; Zhang, C.; Schneider, D.J.; Janjic, N. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. USA 2017, 114, 2898–2903. [Google Scholar] [CrossRef]
- Wang, Y.; Ng, N.; Liu, E.; Lam, C.H.; Perrin, D.M. Systematic study of constraints imposed by modified nucleoside triphosphates with protein-like side chains for use in in vitro selection. Org. Biomol. Chem. 2017, 15, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Sczepanski, J.T.; Joyce, G.F. Binding of a structured D-RNA molecule by an L-RNA aptamer. J. Am. Chem. Soc. 2013, 135, 13290–13293. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Sell, S.; Kaczmarek, P.; Maasch, C.; Buchner, K.; Pruszynska-Oszmalek, E.; Kolodziejski, P.; Purschke, W.G.; Nowak, K.W.; Strowski, M.Z.; et al. A mixed mirror-image DNA/RNA aptamer inhibits glucagon and acutely improves glucose tolerance in models of type 1 and type 2 diabetes. J. Biol. Chem. 2013, 288, 21136–21147. [Google Scholar] [CrossRef]
- Wlotzka, B.; Leva, S.; Eschgfaller, B.; Burmeister, J.; Kleinjung, F.; Kaduk, C.; Muhn, P.; Hess-Stumpp, H.; Klussmann, S. In Vivo properties of an anti-GnRH Spiegelmer: An example of an oligonucleotide-based therapeutic substance class. Proc. Natl. Acad. Sci. USA 2002, 99, 8898–8902. [Google Scholar] [CrossRef]
- Leva, S.; Lichte, A.; Burmeister, J.; Muhn, P.; Jahnke, B.; Fesser, D.; Erfurth, J.; Burgstaller, P.; Klussmann, S. GnRH Binding RNA and DNA Spiegelmers: A Novel Approach toward GnRH Antagonism. Chem. Biol. 2002, 9, 351–359. [Google Scholar] [CrossRef]
- Purschke, W.G.; Radtke, F.; Kleinjung, F.; Klussmann, S. A DNA Spiegelmer to staphylococcal enterotoxin B. Nucleic Acids Res. 2003, 31, 3027–3032. [Google Scholar] [CrossRef]
- Kabza, A.M.; Sczepanski, J.T. An l-RNA Aptamer with Expanded Chemical Functionality that Inhibits MicroRNA Biogenesis. ChemBioChem 2017, 18, 1824–1827. [Google Scholar] [CrossRef]
- Taylor, A.I.; Holliger, P. Selecting Fully-Modified XNA Aptamers Using Synthetic Genetics. Curr. Protoc. Chem. Biol. 2018, 10, e44. [Google Scholar] [CrossRef]
- Biondi, E.; Benner, S.A. Artificially Expanded Genetic Information Systems for New Aptamer Technologies. Biomedicines 2018, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.R.; Shao, Y.; Hoshika, S.; Yang, Z.; Shelke, S.A.; Herrou, J.; Kim, H.-J.; Kim, M.-J.; Piccirilli, J.A.; Benner, S.A. A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair. Angew. Chem. 2015, 127, 9991–9994. [Google Scholar] [CrossRef]
- Biondi, E.; Lane, J.D.; Das, D.; Dasgupta, S.; Piccirilli, J.A.; Hoshika, S.; Bradley, K.M.; Krantz, B.A.; Benner, S.A. Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen. Nucleic Acids Res. 2016, 44, 9565–9577. [Google Scholar] [CrossRef] [PubMed]
- Di Giusto, D.A.; King, G.C. Construction, stability, and activity of multivalent circular anticoagulant aptamers. J. Biol. Chem. 2004, 279, 46483–46489. [Google Scholar] [CrossRef]
- Kuai, H.; Zhao, Z.; Mo, L.; Liu, H.; Hu, X.; Fu, T.; Zhang, X.; Tan, W. Circular Bivalent Aptamers Enable in vivo Stability and Recognition. J. Am. Chem. Soc. 2017, 139, 9128–9131. [Google Scholar] [CrossRef]
- Riccardi, C.; Meyer, A.; Vasseur, J.-J.; Russo Krauss, I.; Paduano, L.; Morvan, F.; Montesarchio, D. Fine-tuning the properties of the thrombin binding aptamer through cyclization: Effect of the 5′-3′ connecting linker on the aptamer stability and anticoagulant activity. Bioorg. Chem. 2019, 103379. [Google Scholar] [CrossRef]
- Riccardi, C.; Meyer, A.; Vasseur, J.-J.; Russo Krauss, I.; Paduano, L.; Oliva, R.; Petraccone, L.; Morvan, F.; Montesarchio, D. Stability Is Not Everything: The Case of the Cyclisation of a Thrombin-Binding Aptamer. ChemBioChem 2019, 20, 1789–1794. [Google Scholar] [CrossRef]
- Shi, S.H.; Hayashi, Y.; Petralia, R.S.; Zaman, S.H.; Wenthold, R.J.; Svoboda, K.; Malinow, R. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 1999, 284, 1811–1816. [Google Scholar] [CrossRef]
- Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules 2016, 21, 1613. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef]
- Hasegawa, H.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers against VEGF165 using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008, 30, 829–834. [Google Scholar] [CrossRef]
- Batool, S.; Argyropoulos, K.V.; Azad, R.; Okeoma, P.; Zumrut, H.; Bhandari, S.; Dekhang, R.; Mallikaratchy, P.R. Dimerization of an aptamer generated from Ligand-guided selection (LIGS) yields a high affinity scaffold against B-cells. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 232–240. [Google Scholar] [CrossRef]
- Wu, Y.; Sefah, K.; Liu, H.; Wang, R.; Tan, W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Tang, L.; Yang, X.; Hwang, K.; Wang, W.; Yin, Q.; Wong, N.Y.; Dobrucki, L.W.; Yasui, N.; Katzenellenbogen, J.A.; et al. Selective Delivery of an Anticancer Drug with Aptamer-Functionalized Liposomes to Breast Cancer Cells in vitro and in Vivo. J. Mater. Chem. B 2013, 1, 5288–5297. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Tong, R.; Mishra, A.; Xu, W.; Wong, G.C.; Cheng, J.; Lu, Y. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew. Chem. Int. Ed. Engl. 2009, 48, 6494–6498. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Fàbrega, C.; Grijalvo, S.; Vitiello, G.; D’Errico, G.; Eritja, R.; Montesarchio, D. AS1411-decorated niosomes as effective nanocarriers for Ru(iii)-based drugs in anticancer strategies. J. Mater. Chem. B 2018, 6, 5368–5384. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.; Yin, M.; Yang, X.; Yuan, Q.; Ren, J.; Qu, X. Aptamer-capped multifunctional mesoporous strontium hydroxyapatite nanovehicle for cancer-cell-responsive drug delivery and imaging. Biomacromolecules 2012, 13, 4257–4263. [Google Scholar] [CrossRef]
- Cammarata, C.R.; Hughes, M.E.; Ofner, C.M., 3rd. Carbodiimide induced cross-linking, ligand addition, and degradation in gelatin. Mol. Pharm. 2015, 12, 783–793. [Google Scholar] [CrossRef]
- Mann, A.P.; Bhavane, R.C.; Somasunderam, A.; Liz Montalvo-Ortiz, B.; Ghaghada, K.B.; Volk, D.; Nieves-Alicea, R.; Suh, K.S.; Ferrari, M.; Annapragada, A.; et al. Thioaptamer conjugated liposomes for tumor vasculature targeting. Oncotarget 2011, 2, 298–304. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, M.; Tong, X.; Sun, N.; Zhou, L.; Cao, Y.; Wang, J.; Zhang, H.; Pei, R. Aptamer-Modified Temperature-Sensitive Liposomal Contrast Agent for Magnetic Resonance Imaging. Biomacromolecules 2015, 16, 2618–2623. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, R.; Fang, X.; Chen, F.; Wang, Y.; Chen, M. Nucleolin targeting AS1411 aptamer modified pH-sensitive micelles for enhanced delivery and antitumor efficacy of paclitaxel. Nano Res. 2015, 8, 201–218. [Google Scholar] [CrossRef]
- Li, X.; Yu, Y.; Ji, Q.; Qiu, L. Targeted delivery of anticancer drugs by aptamer AS1411 mediated Pluronic F127/cyclodextrin-linked polymer composite micelles. Nanomedicine 2015, 11, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Siddiqui, I.A.; Nihal, M.; Pilla, S.; Rosenthal, K.; Mukhtar, H.; Gong, S. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials 2013, 34, 5244–5253. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, H.R.; Patravale, V.B.; Joshi, M.D. Polymeric nanoparticles for targeted treatment in oncology: Current insights. Int. J. Nanomed. 2015, 10, 1001–1018. [Google Scholar]
- Cheng, J.; Teply, B.A.; Sherifi, I.; Sung, J.; Luther, G.; Gu, F.X.; Levy-Nissenbaum, E.; Radovic-Moreno, A.F.; Langer, R.; Farokhzad, O.C. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials 2007, 28, 869–876. [Google Scholar] [CrossRef]
- Tong, R.; Yala, L.; Fan, T.M.; Cheng, J. The formulation of aptamer-coated paclitaxel-polylactide nanoconjugates and their targeting to cancer cells. Biomaterials 2010, 31, 3043–3053. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef]
- Guo, J.; Gao, X.; Su, L.; Xia, H.; Gu, G.; Pang, Z.; Jiang, X.; Yao, L.; Chen, J.; Chen, H. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials 2011, 32, 8010–8020. [Google Scholar] [CrossRef]
- Yu, C.; Hu, Y.; Duan, J.; Yuan, W.; Wang, C.; Xu, H.; Yang, X.D. Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitro. PLoS ONE 2011, 6, e24077. [Google Scholar] [CrossRef]
- Das, M.; Duan, W.; Sahoo, S.K. Multifunctional nanoparticle-EpCAM aptamer bioconjugates: A paradigm for targeted drug delivery and imaging in cancer therapy. Nanomedicine 2015, 11, 379–389. [Google Scholar] [CrossRef]
- Alibolandi, M.; Ramezani, M.; Sadeghi, F.; Abnous, K.; Hadizadeh, F. Epithelial cell adhesion molecule aptamer conjugated PEG-PLGA nanopolymersomes for targeted delivery of doxorubicin to human breast adenocarcinoma cell line in vitro. Int. J. Pharm. 2015, 479, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Xiong, M.; Zhang, X.; Cai, G.; Chen, H.; Zeng, Q.; Yu, Z. Poly(lactic-co-glycolic acid) nanoparticles conjugated with CD133 aptamers for targeted salinomycin delivery to CD133+ osteosarcoma cancer stem cells. Int. J. Nanomed. 2015, 10, 2537–2554. [Google Scholar]
- Aravind, A.; Jeyamohan, P.; Nair, R.; Veeranarayanan, S.; Nagaoka, Y.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. AS1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnol. Bioeng. 2012, 109, 2920–2931. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, Z.; Dinarvand, R.; Mottaghitalab, F.; Esfandyari-Manesh, M.; Sayari, E.; Atyabi, F. Aptamer decorated hyaluronan/chitosan nanoparticles for targeted delivery of 5-fluorouracil to MUC1 overexpressing adenocarcinomas. Carbohydr. Polym. 2015, 121, 190–198. [Google Scholar] [CrossRef]
- Sayari, E.; Dinarvand, M.; Amini, M.; Azhdarzadeh, M.; Mollarazi, E.; Ghasemi, Z.; Atyabi, F. MUC1 aptamer conjugated to chitosan nanoparticles, an efficient targeted carrier designed for anticancer SN38 delivery. Int. J. Pharm. 2014, 473, 304–315. [Google Scholar] [CrossRef]
- Li, J.; You, J.; Dai, Y.; Shi, M.; Han, C.; Xu, K. Gadolinium oxide nanoparticles and aptamer-functionalized silver nanoclusters-based multimodal molecular imaging nanoprobe for optical/magnetic resonance cancer cell imaging. Anal. Chem. 2014, 86, 11306–11311. [Google Scholar] [CrossRef]
- Bagalkot, V.; Zhang, L.; Levy-Nissenbaum, E.; Jon, S.; Kantoff, P.W.; Langer, R.; Farokhzad, O.C. Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007, 7, 3065–3070. [Google Scholar] [CrossRef]
- Cai, L.; Chen, Z.Z.; Chen, M.Y.; Tang, H.W.; Pang, D.W. MUC-1 aptamer-conjugated dye-doped silica nanoparticles for MCF-7 cells detection. Biomaterials 2013, 34, 371–381. [Google Scholar] [CrossRef]
- Zhang, H. Thermally cross-linked superparamagnetic iron oxide nanoparticle-A10 RNA aptamer-doxorubicin conjugate. In Molecular Imaging and Contrast Agent Database (MICAD); National Center for Biotechnology Information (US): Bethesda, MD, USA, 2004. [Google Scholar]
- Deng, K.; Hou, Z.; Li, X.; Li, C.; Zhang, Y.; Deng, X. Aptamer-Mediated Up-conversion Core/MOF Shell Nanocomposites for Targeted Drug Delivery and Cell Imaging. Sci. Rep. 2015, 5, 7851. [Google Scholar] [CrossRef]
- Yang, X.; Liu, X.; Liu, Z.; Pu, F.; Ren, J.; Qu, X. Near-Infrared Light-Triggered, Targeted Drug Delivery to Cancer Cells by Aptamer Gated Nanovehicles. Adv. Mater. 2012, 24, 2890–2895. [Google Scholar] [CrossRef]
- Chen, T.; Shukoor, M.I.; Wang, R.; Zhao, Z.; Yuan, Q.; Bamrungsap, S.; Xiong, X.; Tan, W. Smart Multifunctional Nanostructure for Targeted Cancer Chemotherapy and Magnetic Resonance Imaging. ACS Nano 2011, 5, 7866–7873. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; O’Donoghue, M.B.; Liu, H.; Tan, W. A liposome-based nanostructure for aptamer directed delivery. Chem. Commun. 2010, 46, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Ara, M.N.; Matsuda, T.; Hyodo, M.; Sakurai, Y.; Hatakeyama, H.; Ohga, N.; Hida, K.; Harashima, H. An aptamer ligand based liposomal nanocarrier system that targets tumor endothelial cells. Biomaterials 2014, 35, 7110–7120. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hou, J.; Liu, X.; Guo, Y.; Wu, Y.; Zhang, L.; Yang, Z. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials 2014, 35, 3840–3850. [Google Scholar] [CrossRef]
- Ababneh, N.; Alshaer, W.; Allozi, O.; Mahafzah, A.; El-Khateeb, M.; Hillaireau, H.; Noiray, M.; Fattal, E.; Ismail, S. In Vitro selection of modified RNA aptamers against CD44 cancer stem cell marker. Nucleic Acid Ther. 2013, 23, 401–407. [Google Scholar] [CrossRef]
- Alshaer, W.; Hillaireau, H.; Vergnaud, J.; Ismail, S.; Fattal, E. Functionalizing Liposomes with anti-CD44 Aptamer for Selective Targeting of Cancer Cells. Bioconjug. Chem. 2015, 26, 1307–1313. [Google Scholar] [CrossRef]
- Alshaer, W.; Hillaireau, H.; Vergnaud, J.; Mura, S.; Delomenie, C.; Sauvage, F.; Ismail, S.; Fattal, E. Aptamer-guided siRNA-loaded nanomedicines for systemic gene silencing in CD-44 expressing murine triple-negative breast cancer model. J. Control. Release 2018, 271, 98–106. [Google Scholar] [CrossRef]
- Liao, Z.X.; Chuang, E.Y.; Lin, C.C.; Ho, Y.C.; Lin, K.J.; Cheng, P.Y.; Chen, K.J.; Wei, H.J.; Sung, H.W. An AS1411 aptamer-conjugated liposomal system containing a bubble-generating agent for tumor-specific chemotherapy that overcomes multidrug resistance. J. Control. Release 2015, 208, 42–51. [Google Scholar] [CrossRef]
- Yu, S.; Dong, R.; Chen, J.; Chen, F.; Jiang, W.; Zhou, Y.; Zhu, X.; Yan, D. Synthesis and self-assembly of amphiphilic aptamer-functionalized hyperbranched multiarm copolymers for targeted cancer imaging. Biomacromolecules 2014, 15, 1828–1836. [Google Scholar] [CrossRef]
- Wu, X.; Ding, B.; Gao, J.; Wang, H.; Fan, W.; Wang, X.; Zhang, W.; Wang, X.; Ye, L.; Zhang, M.; et al. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int. J. Nanomed. 2011, 6, 1747–1756. [Google Scholar]
- Pilapong, C.; Sitthichai, S.; Thongtem, S.; Thongtem, T. Smart magnetic nanoparticle-aptamer probe for targeted imaging and treatment of hepatocellular carcinoma. Int. J. Pharm. 2014, 473, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Kishore, V.; Paderi, J.E.; Akkus, A.; Smith, K.M.; Balachandran, D.; Beaudoin, S.; Panitch, A.; Akkus, O. Incorporation of a decorin biomimetic enhances the mechanical properties of electrochemically aligned collagen threads. Acta Biomater. 2011, 7, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Pala, K.; Serwotka, A.; Jelen, F.; Jakimowicz, P.; Otlewski, J. Tumor-specific hyperthermia with aptamer-tagged superparamagnetic nanoparticles. Int. J. Nanomed. 2014, 9, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yao, H.; Meng, Y.; Wang, Y.; Yan, X.; Huang, R. Specific aptamer-conjugated mesoporous silica-carbon nanoparticles for HER2-targeted chemo-photothermal combined therapy. Acta Biomater. 2015, 16, 196–205. [Google Scholar] [CrossRef]
- Baek, S.E.; Lee, K.H.; Park, Y.S.; Oh, D.K.; Oh, S.; Kim, K.S.; Kim, D.E. RNA aptamer-conjugated liposome as an efficient anticancer drug delivery vehicle targeting cancer cells in vivo. J. Control. Release 2014, 196, 234–242. [Google Scholar] [CrossRef]
- Charoenphol, P.; Bermudez, H. Aptamer-Targeted DNA Nanostructures for Therapeutic Delivery. Mol. Pharm. 2014, 11, 1721–1725. [Google Scholar] [CrossRef]
- Chang, M.; Yang, C.S.; Huang, D.M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano 2011, 5, 6156–6163. [Google Scholar] [CrossRef]
- Kurosaki, T.; Higuchi, N.; Kawakami, S.; Higuchi, Y.; Nakamura, T.; Kitahara, T.; Hashida, M.; Sasaki, H. Self-assemble gene delivery system for molecular targeting using nucleic acid aptamer. Gene 2012, 491, 205–209. [Google Scholar] [CrossRef]
- Zhao, N.; Bagaria, H.G.; Wong, M.S.; Zu, Y. A nanocomplex that is both tumor cell-selective and cancer gene-specific for anaplastic large cell lymphoma. J. Nanobiotechnol. 2011, 9, 2. [Google Scholar] [CrossRef]
- Subramanian, N.; Kanwar, J.R.; Athalya, P.K.; Janakiraman, N.; Khetan, V.; Kanwar, R.K.; Eluchuri, S.; Krishnakumar, S. EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex. J. Biomed. Sci. 2015, 22, 4. [Google Scholar] [CrossRef]
- Lee, I.H.; An, S.; Yu, M.K.; Kwon, H.K.; Im, S.H.; Jon, S. Targeted chemoimmunotherapy using drug-loaded aptamer-dendrimer bioconjugates. J. Control. Release 2011, 155, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, Y.; Xie, Y.; An, Y.; Huang, Y.; Zhu, Z.; Yang, C.J. A controllable aptamer-based self-assembled DNA dendrimer for high affinity targeting, bioimaging and drug delivery. Sci. Rep. 2015, 5, 10099. [Google Scholar] [CrossRef] [PubMed]
- Green, N. Avidin. 1. The USE of [14C] Biotin for Kinetic Studies and for Assay. Biochem. J. 1963, 89, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chen, T.; Wu, C.; Zhu, G.; Qiu, L.; Cui, C.; Hou, W.; Tan, W. Aptamer CaCO3 nanostructures: A facile, pH-responsive, specific platform for targeted anticancer theranostics. Chem. Asian J. 2015, 10, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Duan, S.; He, J.; Liang, W.; Su, J.; Zhu, J.; Hu, N.; Zhao, Y.; Lu, X. Highly sensitive detection of leukemia cells based on aptamer and quantum dots. Oncol. Rep. 2016, 36, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Li, X.; Li, H.; Zhang, W. Quantifying thiol–gold interactions towards the efficient strength control. Nat. Commun. 2014, 5, 4348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Servos, M.R.; Liu, J. Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a pH-Assisted and Surfactant-Free Route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef]
- Dam, H.D.; Culver, K.S.; Odom, T.W. Grafting aptamers onto gold nanostars increases in vitro efficacy in a wide range of cancer cell types. Mol. Pharm. 2014, 11, 580–587. [Google Scholar] [CrossRef]
- Shiao, Y.S.; Chiu, H.H.; Wu, P.H.; Huang, Y.F. Aptamer-functionalized gold nanoparticles as photoresponsive nanoplatform for co-drug delivery. ACS Appl. Mater. Interfaces 2014, 6, 21832–21841. [Google Scholar] [CrossRef]
- Luo, L.Y.; Shiao, Y.S.; Huang, Y.F. Release of photoactivatable drugs from plasmonic nanoparticles for targeted cancer therapy. ACS Nano 2011, 5, 7796–7804. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, G.; You, M.; Song, E.; Shukoor, M.I.; Zhang, K.; Altman, M.B.; Chen, Y.; Zhu, Z.; Huang, C.Z.; et al. Assembly of aptamer switch probes and photosensitizer on gold nanorods for targeted photothermal and photodynamic cancer therapy. ACS Nano 2012, 6, 5070–5077. [Google Scholar] [CrossRef]
- Huang, Y.F.; Sefah, K.; Bamrungsap, S.; Chang, H.T.; Tan, W. Selective photothermal therapy for mixed cancer cells using aptamer-conjugated nanorods. Langmuir 2008, 24, 11860–11865. [Google Scholar] [CrossRef]
- Yang, L.; Tseng, Y.T.; Suo, G.; Chen, L.; Yu, J.; Chiu, W.J.; Huang, C.C.; Lin, C.H. Photothermal therapeutic response of cancer cells to aptamer-gold nanoparticle-hybridized graphene oxide under NIR illumination. ACS Appl. Mater. Interfaces 2015, 7, 5097–5106. [Google Scholar] [CrossRef]
- Zhao, F.; Zhou, J.; Su, X.; Wang, Y.; Yan, X.; Jia, S.; Du, B. A Smart Responsive Dual Aptamers-Targeted Bubble-Generating Nanosystem for Cancer Triplex Therapy and Ultrasound Imaging. Small 2017, 13, 1603990. [Google Scholar] [CrossRef]
- Hooker, M.J.; Esser-Kahn, A.; Francis, M.B. Modification of aniline containing proteins using an oxidative coupling strategy. J. Am. Chem. Soc. 2006, 128, 15558–15559. [Google Scholar] [CrossRef]
- Mastico, A.R.; Talbot, S.J.; Stockley, P.G. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol. 1993, 74, 541–548. [Google Scholar] [CrossRef]
- Carrico, Z.M.; Romanini, D.W.; Mehl, R.A.; Francis, M.B. Oxidative coupling of peptides to a virus capsid containing unnatural amino acids. Chem. Commun. 2008, 10, 1205–1207. [Google Scholar] [CrossRef]
- Oh, S.S.; Lee, B.F.; Leibfarth, F.A.; Eisenstein, M.; Robb, M.J.; Lynd, N.A.; Hawker, C.J.; Soh, H.T. Synthetic Aptamer-Polymer Hybrid Constructs for Programmed Drug Delivery into Specific Target Cells. J. Am. Chem. Soc. 2014, 136, 15010–15015. [Google Scholar] [CrossRef]
- Aravind, A.; Varghese, S.H.; Veeranarayanan, S.; Mathew, A.; Nagaoka, Y.; Iwai, S.; Fukuda, T.; Hasumura, T.; Yoshida, Y.; Maekawa, T.; et al. Aptamer-labeled PLGA nanoparticles for targeting cancer cells. Cancer Nanotechnol. 2012, 3, 1–12. [Google Scholar] [CrossRef]
- Li, L.L.; Yin, Q.; Cheng, J.; Lu, Y. Polyvalent mesoporous silica nanoparticle-aptamer bioconjugates target breast cancer cells. Adv. Healthc. Mater. 2012, 1, 567–572. [Google Scholar] [CrossRef]
- Alibolandi, M.; Taghdisi, S.M.; Ramezani, P.; Hosseini Shamili, F.; Farzad, S.A.; Abnous, K.; Ramezani, M. Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int. J. Pharm. 2017, 519, 352–364. [Google Scholar] [CrossRef]
- Tao, W.; Zeng, X.; Wu, J.; Zhu, X.; Yu, X.; Zhang, X.; Zhang, J.; Liu, G.; Mei, L. Polydopamine-Based Surface Modification of Novel Nanoparticle-Aptamer Bioconjugates for in vivo Breast Cancer Targeting and Enhanced Therapeutic Effects. Theranostics 2016, 6, 470–484. [Google Scholar] [CrossRef]
- Mosafer, J.; Teymouri, M.; Abnous, K.; Tafaghodi, M.; Ramezani, M. Study and evaluation of nucleolin-targeted delivery of magnetic PLGA-PEG nanospheres loaded with doxorubicin to C6 glioma cells compared with low nucleolin-expressing L929 cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 72, 123–133. [Google Scholar] [CrossRef]
- Li, X.; Zhu, X.; Qiu, L. Constructing aptamer anchored nanovesicles for enhanced tumor penetration and cellular uptake of water soluble chemotherapeutics. Acta Biomater. 2016, 35, 269–279. [Google Scholar] [CrossRef]
- Barzegar Behrooz, A.; Nabavizadeh, F.; Adiban, J.; Shafiee Ardestani, M.; Vahabpour, R.; Aghasadeghi, M.R.; Sohanaki, H. Smart bomb AS1411 aptamer-functionalized/PAMAM dendrimer nanocarriers for targeted drug delivery in the treatment of gastric cancer. Clin. Exp. Pharmacol. Physiol. 2017, 44, 41–51. [Google Scholar] [CrossRef]
- Ayatollahi, S.; Salmasi, Z.; Hashemi, M.; Askarian, S.; Oskuee, R.K.; Abnous, K.; Ramezani, M. Aptamer-targeted delivery of Bcl-xL shRNA using alkyl modified PAMAM dendrimers into lung cancer cells. Int. J. Biochem. Cell Biol. 2017, 92, 210–217. [Google Scholar] [CrossRef]
- Bandekar, A.; Zhu, C.; Jindal, R.; Bruchertseifer, F.; Morgenstern, A.; Sofou, S. Anti-prostate-specific membrane antigen liposomes loaded with 225Ac for potential targeted antivascular alpha-particle therapy of cancer. J. Nucl. Med. 2014, 55, 107–114. [Google Scholar] [CrossRef]
- Dhar, S.; Gu, F.X.; Langer, R.; Farokhzad, O.C.; Lippard, S.J. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc. Natl. Acad. Sci. USA 2008, 105, 17356–17361. [Google Scholar] [CrossRef]
- Yu, M.K.; Kim, D.; Lee, I.H.; So, J.S.; Jeong, Y.Y.; Jon, S. Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles. Small 2011, 7, 2241–2249. [Google Scholar] [CrossRef]
- Leach, J.C.; Wang, A.; Ye, K.; Jin, S. A RNA-DNA Hybrid Aptamer for Nanoparticle-Based Prostate Tumor Targeted Drug Delivery. Int. J. Mol. Sci. 2016, 17, 380. [Google Scholar] [CrossRef]
- Hao, Z.; Fan, W.; Hao, J.; Wu, X.; Zeng, G.Q.; Zhang, L.J.; Nie, S.F.; Wang, X.D. Efficient delivery of micro RNA to bone-metastatic prostate tumors by using aptamer-conjugated atelocollagen in vitro and in vivo. Drug Deliv. 2016, 23, 874–881. [Google Scholar] [CrossRef]
- Lin, Z.; Ma, Q.; Fei, X.; Zhang, H.; Su, X. A novel aptamer functionalized CuInS2 quantum dots probe for daunorubicin sensing and near infrared imaging of prostate cancer cells. Anal. Chim. Acta 2014, 818, 54–60. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, X.; Zhang, Y.; Zhou, G.; Ke, X.; Wang, H.; Tinnefeld, P.; He, Z. One-pot synthesized aptamer-functionalized CdTe:Zn2+ quantum dots for tumor-targeted fluorescence imaging in vitro and in vivo. Anal. Chem. 2013, 85, 5843–5849. [Google Scholar] [CrossRef]
- Guo, F.; Hu, Y.; Yu, L.; Deng, X.; Meng, J.; Wang, C.; Yang, X.D. Enhancement of Thermal Damage to Adenocarcinoma Cells by Iron Nanoparticles Modified with MUC1 Aptamer. J. Nanosci. Nanotechnol. 2016, 16, 2246–2253. [Google Scholar] [CrossRef]
- Esfandyari-Manesh, M.; Mohammadi, A.; Atyabi, F.; Nabavi, S.M.; Ebrahimi, S.M.; Shahmoradi, E.; Varnamkhasti, B.S.; Ghahremani, M.H.; Dinarvand, R. Specific targeting delivery to MUC1 overexpressing tumors by albumin-chitosan nanoparticles conjugated to DNA aptamer. Int. J. Pharm. 2016, 515, 607–615. [Google Scholar] [CrossRef]
- Perepelyuk, M.; Maher, C.; Lakshmikuttyamma, A.; Shoyele, S.A. Aptamer-hybrid nanoparticle bioconjugate efficiently delivers miRNA-29b to non-small-cell lung cancer cells and inhibits growth by downregulating essential oncoproteins. Int. J. Nanomed. 2016, 11, 3533–3544. [Google Scholar]
- Azhdarzadeh, M.; Atyabi, F.; Saei, A.A.; Varnamkhasti, B.S.; Omidi, Y.; Fateh, M.; Ghavami, M.; Shanehsazzadeh, S.; Dinarvand, R. Theranostic MUC-1 aptamer targeted gold coated superparamagnetic iron oxide nanoparticles for magnetic resonance imaging and photothermal therapy of colon cancer. Colloids Surf. B Biointerfaces 2016, 143, 224–232. [Google Scholar] [CrossRef]
- Charbgoo, F.; Alibolandi, M.; Taghdisi, S.M.; Abnous, K.; Soltani, F.; Ramezani, M. MUC1 aptamer-targeted DNA micelles for dual tumor therapy using doxorubicin and KLA peptide. Nanomedicine 2018, 14, 685–697. [Google Scholar] [CrossRef]
- Taghavi, S.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. Chitosan-modified PLGA nanoparticles tagged with 5TR1 aptamer for in vivo tumor-targeted drug delivery. Cancer Lett. 2017, 400, 1–8. [Google Scholar] [CrossRef]
- Xu, C.; Han, X.; Jiang, Y.; Yuan, S.; Wu, Z.; Wu, Z.; Qi, X. Microenvironmental Control of MUC1 Aptamer-Guided Acid-Labile Nanoconjugate within Injectable Microporous Hydrogels. Bioconjug. Chem. 2017, 28, 2530–2537. [Google Scholar] [CrossRef]
- Zhang, Z.; Ali, M.M.; Eckert, M.A.; Kang, D.K.; Chen, Y.Y.; Sender, L.S.; Fruman, D.A.; Zhao, W. A polyvalent aptamer system for targeted drug delivery. Biomaterials 2013, 34, 9728–9735. [Google Scholar] [CrossRef]
- Huang, F.Y.; Chang, H.-T.; Tan, W. Cancer Cell Targeting Using Multiple Aptamers Conjugated on Nanorods. Anal. Chem. 2008, 80, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Lavaee, P.; Ramezani, M.; Abnous, K. Reversible targeting and controlled release delivery of daunorubicin to cancer cells by aptamer-wrapped carbon nanotubes. Eur. J. Pharm. Biopharm. 2011, 77, 200–206. [Google Scholar] [CrossRef]
- Li, J.; Wu, S.; Wu, C.; Qiu, L.; Zhu, G.; Cui, C.; Liu, Y.; Hou, W.; Wang, Y.; Zhang, L.; et al. Versatile surface engineering of porous nanomaterials with bioinspired polyphenol coatings for targeted and controlled drug delivery. Nanoscale 2016, 8, 8600–8606. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Lavaee, P.; Emrani, A.S.; Hassanabad, K.Y.; Ramezani, M.; Abnous, K. Double targeting, controlled release and reversible delivery of daunorubicin to cancer cells by polyvalent aptamers-modified gold nanoparticles. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 61, 753–761. [Google Scholar] [CrossRef]
- Kang, S.; Luo, Y.L.; Huang, Y.F.; Yeh, C.K. DNA-conjugated gold nanoparticles for ultrasound targeted drug delivery. In Proceedings of the 2012 IEEE International Ultrasonics Symposium, Dresden, Germany, 7–10 October 2012. [Google Scholar]
- Wang, C.H.; Kang, S.T.; Lee, Y.H.; Luo, Y.L.; Huang, Y.F.; Yeh, C.K. Aptamer-conjugated and drug-loaded acoustic droplets for ultrasound theranosis. Biomaterials 2012, 33, 1939–1947. [Google Scholar] [CrossRef]
- Zhou, J.; Soontornworajit, B.; Wang, Y. A temperature-responsive antibody-like nanostructure. Biomacromolecules 2010, 11, 2087–2093. [Google Scholar] [CrossRef]
- Fan, Z.; Shelton, M.; Singh, A.K.; Senapati, D.; Khan, S.A.; Ray, P.C. Multifunctional plasmonic shell-magnetic core nanoparticles for targeted diagnostics, isolation, and photothermal destruction of tumor cells. ACS Nano 2012, 6, 1065–1073. [Google Scholar] [CrossRef]
- Moosavian, S.A.; Abnous, K.; Badiee, A.; Jaafari, M.R. Improvement in the drug delivery and anti-tumor efficacy of PEGylated liposomal doxorubicin by targeting RNA aptamers in mice bearing breast tumor model. Colloids Surf. B Biointerfaces 2016, 139, 228–236. [Google Scholar] [CrossRef]
- Powell, D.; Chandra, S.; Dodson, K.; Shaheen, F.; Wiltz, K.; Ireland, S.; Syed, M.; Dash, S.; Wiese, T.; Mandal, T.; et al. Aptamer-functionalized hybrid nanoparticle for the treatment of breast cancer. Eur. J. Pharm. Biopharm. 2017, 114, 108–118. [Google Scholar] [CrossRef]
- Fan, W.; Wang, X.; Ding, B.; Cai, H.; Wang, X.; Fan, Y.; Li, Y.; Liu, S.; Nie, S.; Lu, Q. Thioaptamer-conjugated CD44-targeted delivery system for the treatment of breast cancer in vitro and in vivo. J. Drug Target. 2016, 24, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xiang, D.; Shigdar, S.; Yang, W.; Li, Q.; Lin, J.; Liu, K.; Duan, W. Epithelial cell adhesion molecule aptamer functionalized PLGA-lecithin-curcumin-PEG nanoparticles for targeted drug delivery to human colorectal adenocarcinoma cells. Int. J. Nanomed. 2014, 9, 1083–1096. [Google Scholar]
- Xie, X.; Li, F.; Zhang, H.; Lu, Y.; Lian, S.; Lin, H.; Gao, Y.; Jia, L. EpCAM aptamer-functionalized mesoporous silica nanoparticles for efficient colon cancer cell-targeted drug delivery. Eur. J. Pharm. Sci. 2016, 83, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, F.; Qi, X.; Dong, Y.; Zhang, Y.; Ge, Z.; Cai, G.; Zhang, X. Epidermal growth factor receptor aptamer-conjugated polymer-lipid hybrid nanoparticles enhance salinomycin delivery to osteosarcoma and cancer stem cells. Exp. Ther. Med. 2018, 15, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.J.; Li, K.F.; Zhang, L.X.; Wang, H.; Liu, L.S.; Zheng, Z.Z.; Han, N.Y.; Yang, Z.J.; Fan, T.Y. In Vitro study of novel gadolinium-loaded liposomes guided by GBI-10 aptamer for promising tumor targeting and tumor diagnosis by magnetic resonance imaging. Int. J. Nanomed. 2015, 10, 5187–5204. [Google Scholar]
- Chen, X.C.; Deng, Y.L.; Lin, Y.; Pang, D.W.; Qing, H.; Qu, F.; Xie, H.Y. Quantum dot-labeled aptamer nanoprobes specifically targeting glioma cells. Nanotechnology 2008, 19, 235105. [Google Scholar] [CrossRef]
- Monaco, I.; Camorani, S.; Colecchia, D.; Locatelli, E.; Calandro, P.; Oudin, A.; Niclou, S.; Arra, C.; Chiariello, M.; Cerchia, L.; et al. Aptamer Functionalization of Nanosystems for Glioblastoma Targeting through the Blood-Brain Barrier. J. Med. Chem. 2017, 60, 4510–4516. [Google Scholar] [CrossRef]
- Song, X.; Ren, Y.; Zhang, J.; Wang, G.; Han, X.; Zheng, W.; Zhen, L. Targeted delivery of doxorubicin to breast cancer cells by aptamer functionalized DOTAP/DOPE liposomes. Oncol. Rep. 2015, 34, 1953–1960. [Google Scholar] [CrossRef]
- Belyanina, I.V.; Zamay, T.N.; Zamay, G.S.; Zamay, S.S.; Kolovskaya, O.S.; Ivanchenko, T.I.; Denisenko, V.V.; Kirichenko, A.K.; Glazyrin, Y.E.; Garanzha, I.V.; et al. In Vivo Cancer Cells Elimination Guided by Aptamer-Functionalized Gold-Coated Magnetic Nanoparticles and Controlled with Low Frequency Alternating Magnetic Field. Theranostics 2017, 7, 3326–3337. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Lee, A.S.; Yap, L.W.; Jans, D.A.; Wagstaff, K.M.; Cheng, W. Tumor cell-specific photothermal killing by SELEX-derived DNA aptamer-targeted gold nanorods. Nanoscale 2016, 8, 187–196. [Google Scholar] [CrossRef]
- Jurek, P.M.; Zablocki, K.; Wasko, U.; Mazurek, M.P.; Otlewski, J.; Jelen, F. Anti-FGFR1 aptamer-tagged superparamagnetic conjugates for anticancer hyperthermia therapy. Int. J. Nanomed. 2017, 12, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Lavaee, P.; Jalalian, S.H.; Robati, R.Y.; Abnous, K. Double targeting and aptamer-assisted controlled release delivery of epirubicin to cancer cells by aptamers-based dendrimer in vitro and in vivo. Eur. J. Pharm. Biopharm. 2016, 102, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Pi, F.; Zhang, H.; Li, H. RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomedicine 2017, 13, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Yu, Z.C.; He, Y.N.; Zhang, T.; Du, L.B.; Dong, Y.M.; Chen, H.W.; Zhang, Y.Y.; Wang, W.Q. Salinomycin-loaded lipid-polymer nanoparticles with anti-CD20 aptamers selectively suppress human CD20+ melanoma stem cells. Acta Pharmacol. Sin. 2018, 39, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Eyetech Study Group. Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: Phase II study results. Ophthalmology 2003, 110, 979–986. [Google Scholar] [CrossRef]
- Retina, E.S.G.J. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina 2002, 22, 143–152. [Google Scholar]
- Drolet, D.W.; Nelson, J.; Tucker, C.E.; Zack, P.M.; Nixon, K.; Bolin, R.; Judkins, M.B.; Farmer, J.A.; Wolf, J.L.; Gill, S.C.; et al. Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharm. Res. 2000, 17, 1503–1510. [Google Scholar] [CrossRef]
- Ismail, S.I.; Alshaer, W. Therapeutic aptamers in discovery, preclinical and clinical stages. Adv. Drug Deliv. Rev. 2018, 134, 51–64. [Google Scholar] [CrossRef]
- Hassel, S.K.; Mayer, G. Aptamers as Therapeutic Agents: Has the Initial Euphoria Subsided? Mol. Diagn. Ther. 2019, 23, 301–309. [Google Scholar] [CrossRef]
- D’Amico, D.J. Pegaptanib Sodium for Neovascular Age-Related Macular Degeneration: Two-Year Safety Results of the Two Prospective, Multicenter, Controlled Clinical Trials. Ophthalmology 2006, 113, 992–1001.e6. [Google Scholar] [CrossRef]
- Goebl, N.; Berridge, B.; Wroblewski, V.J.; Brown-Augsburger, P.L. Development of a Sensitive and Specific in Situ Hybridization Technique for the Cellular Localization of Antisense Oligodeoxynucleotide Drugs in Tissue Sections. Toxicol. Pathol. 2007, 35, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Farman, C.A.; Kornbrust, D.J. Oligodeoxynucleotide Studies in Primates: Antisense and Immune Stimulatory Indications. Toxicol. Pathol. 2003, 31 (Suppl. S1), 119–122. [Google Scholar] [PubMed]
- Henry, S.P.; Giclas, P.C.; Leeds, J.; Pangburn, M.; Auletta, C.; Levin, A.A.; Kornbrust, D.J. Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: Potential mechanism of action. J. Pharmacol. Exp. Ther. 1997, 281, 810–816. [Google Scholar] [PubMed]
- Avci-Adali, M.; Steinle, H.; Michel, T.; Schlensak, C.; Wendel, H.P. Potential Capacity of Aptamers to Trigger Immune Activation in Human Blood. PLoS ONE 2013, 8, e68810. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G. Potential Inherent Stimulation of the Innate Immune System by Nucleic Acid Aptamers and Possible Corrective Approaches. Pharmaceuticals 2018, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Swayze, E.E.; Siwkowski, A.M.; Wancewicz, E.V.; Migawa, M.T.; Wyrzykiewicz, T.K.; Hung, G.; Monia, B.P.; Bennett, A.C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2006, 35, 687–700. [Google Scholar] [CrossRef]
- Burdick, A.D.; Sciabola, S.; Mantena, S.R.; Hollingshead, B.D.; Stanton, R.; Warneke, J.A.; Zeng, M.; Martsen, E.; Medvedev, A.; Makarov, S.S.; et al. Sequence motifs associated with hepatotoxicity of locked nucleic acid—modified antisense oligonucleotides. Nucleic Acids Res. 2014, 42, 4882–4891. [Google Scholar] [CrossRef]
- Waring, M.J. Lipophilicity in drug discovery. Expert Opin. Drug Discov. 2010, 5, 235–248. [Google Scholar] [CrossRef]
- Lee, Y.; Urban, J.H.; Xu, L.; Sullenger, B.A.; Lee, J. 2′Fluoro Modification Differentially Modulates the Ability of RNAs to Activate Pattern Recognition Receptors. Nucleic Acid Ther. 2016, 26, 173–182. [Google Scholar] [CrossRef]
- Ganson, N.J.; Povsic, T.J.; Sullenger, B.A.; Alexander, J.H.; Zelenkofske, S.L.; Sailstad, J.M.; Rusconi, C.P.; Hershfield, M.S. Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J. Allergy Clin. Immunol. 2016, 137, 1610–1613. [Google Scholar] [CrossRef]









| SELEX Type | Description | Ref. |
|---|---|---|
| Metal-Dependant Aptamers | Enrichment of oligonucleotide library with and without ion salts to generate aptamers that only function in the presence of metal ion salts. | [34] |
| Crossover-SELEX | First, oligonucleotides are enriched using cell-SELEX. The product of cell-SELEX is then enriched against its purified protein to yield a higher binding affinity. Crossover-SELEX is useful for targets that are rare in their original environment. | [35] |
| Subtractive SELEX | Selection of aptamers that have the ability to differentiate between two closely related targets (e.g., distinguishing between a normal cell structure and another disease-related one). This is obtained by adding rounds of negative selection against normal cells. | [36] |
| Conditional SELEX | Selection of aptamers that are affected by the presence of regulatory molecules; aptamer selection is performed in two stages here: The first stage in the presence of regulatory molecules and the second in the absence of regulatory molecules. Only the sequences that successfully bind to the target in either one of the stages but not the other is selected, depending on whether the aptamer is to be used in the presence or absence of regulatory molecules. | [37] |
| On-chip selection | This is similar to the microarray method. Single and double base variations are introduced using in silico methods to a pre-selected sequence with the highest affinity to its target and then embedded on a surface plasmon resonance (SPR) chip. On-chip selection is useful for aptamer selection against a large number of targets. | [38] |
| Immobilization-free SELEX or GO-SELEX | First, the library is incubated with the target, and graphene oxide (GO) is then added to the mix in order to bind the unbound sequences via p–p stacking. | [39] |
| Tissue slide-based SELEX | Selection of aptamers against clinical samples. Cancerous tissue is used in the first stage as a target. Then, the tissue is scraped from the slide with the bound sequences. These sequences are then eluted, and counter selection against normal tissue is performed to eliminate shared aptamers. | [40] |
| Capillary Electrophoresis SELEX (CE-SELEX) | CE-SELEX separates the target bounded from unbounded sequences by the difference in electrophoretic mobility, which is a highly efficient separation method. This method enables the selection of aptamer candidates with high affinity while reducing the selection rounds to 1 to 4 from nearly 20 in conventional SELEX | [41,42] |
| Microfluidic SELEX (M-SELEX) | Combining traditional SELEX with a microfluidic system. This system contains reagent-loaded micro-lines, a pressurized reagent reservoir manifold, a PCR thermocycler, and actuatable valves for selection and sample routing. | [43] |
| High-Throughput Sequencing SELEX (HTS-SELEX) | Aptamers are identified through an iterative process of evolutionary selection starting from a random pool containing billions of sequences. The most predominant characteristic of HTS-SELEX is that it firstly allows for sequencing of the library across all the selection rounds. Thus, enriched sequences are visible at a much earlier round, which is more time efficient. Fewer selection rounds also avoid the potential PCR bias caused by over selection. | [44] |
| Aptamers | Antibodies | |
|---|---|---|
| Synthesis | Chemically synthesized and easy to produce | High cost and complexity of production |
| Size | Small compared to antibodies | Large |
| Stability | Prone to nuclease degradation | Short biological half-life |
| Targets | Wide range of targets, starting from ions to whole living cells | Produced only against immunogenic molecules, which limits the range of targets |
| Toxicity and Immunogenicity | Low toxicity and non-immunogenic | Immunogenic |
| Binding Specificity | High binding specificity | High binding specificity |
| Binding Affinity | High binding affinity | High binding affinity |
| Clearance Rate | Rapid circulation clearance | Low clearance rate |
| Chemical Conjugation | Easy to conjugate to nanoparticles and drugs | More difficult to conjugate |
| Chemical modification | Tolerant to chemical modifications to enhance structural and functional propertie | Modifications often lead to reduced activity |
| Target | Aptamer | Nanoparticle | Drug/Imaging Molecule | Tumors | Conjugation Methodology | Ref. |
|---|---|---|---|---|---|---|
| Nucleolin | AS1411 | PLGA-b-PEG | Paclitaxel | Glioma | Carbodiimide chemistry | [231] |
| Polyvalent mesoporous nanoparticles | Doxorubicin | Breast | Thiol-maleimide chemistry | [232] | ||
| pegylated PAMAM dendrimer | Camptothecin | Colorectal | Thiol-maleimide chemistry | [233] | ||
| polydopamine were surface modify a PLGA-b-TPGS polymer | Docetaxel | Breast | Thiol-maleimide related chemistry/Michael addition on dihydroxyindole unit | [234] | ||
| PLGA-b-PEG | Doxorubicin and superparamagnetic iron oxide | Glioma | Carbodiimide chemistry | [235] | ||
| polymersome | Doxorubicin | Breast | 3′-Cholesterol AS1411/direct conjugation | [236] | ||
| PAMAM-PEG | 5-fluorouracil | Gastric cancer | Thiol-maleimide chemistry | [237] | ||
| Alkyl-modified PAMAM dendrimers | Bcl-xLshRNA | Lung Cancer | Carbodiimide chemistry | [238] | ||
| PSMA | A10 (F-RNA) | PEGylated liposomes | 225Ac | Prostate | Carbodiimide chemistry | [239] |
| PLGA-b-PEG | Cis-Pt(IV) | Prostate | Carbodiimide chemistry | [240] | ||
| TCL-SPION | Doxorubicin | Prostate | Carbodiimide chemistry of oligonucleotide linker followed by aptamer complementary base pair binding | [241] | ||
| A10-3-J1 | Superparamagnetic iron oxide | Doxorubicin | Prostate | Avidin-biotin DNA linker followed by aptamer complementary base pair binding | [242] | |
| A10-3.2 | Atelocollagen | miR-15a and miR-16- | Prostate | Thiol-maleimide chemistry | [243] | |
| MUC1 | DNA aptamer | CuInS2 quantum dot | Daunorubicin | Prostate | Carbodiimide chemistry of oligonucleotide linker followed by aptamer complementary base pair binding | [244] |
| MUC1 | DNA aptamer | Zn-doped CdTe QDs | Zn2+ doped CdTe QDs | Lung | Complementary DNA | [245] |
| iron oxide nanoparticles | Hyperthermia | Breast | Avidin-biotin coupling | [246] | ||
| Chitosan-coated human serum albumin | Paclitaxel | Breast | Carbodiimide chemistry | [247] | ||
| Poloxamer | miRNA-29b | Lung | Carbodiimide chemistry | [248] | ||
| Au@SPIONs | Photothermal therapy | Colon | SH-Aptamer gold coordination | [249] | ||
| Micelle | Doxorubicin and proapoptotic peptide (KLA) | Breast, Colon | Carbodiimide chemistry | [250] | ||
| 5TR1 DNA aptamer | PLGA modified with chitosan | Epirubicin | Breast | Electrostatic interaction | [251] | |
| DNA aptamer MA3 | Thermosensitive hydrogel | Doxorubicin | Breast | Thiol-maleimide chemistry | [252] | |
| PTK7 | Sgc8 (DNA) | Polyvalent aptamer system | Doxorubicin | T-cell acute lymphoblastic leukaemia | Complementary DNA | [253] |
| Au-Ag nanorods | Doxorubicin | T-cell acute lymphoblastic leukemia | SH-Aptamer gold coordination | [254] | ||
| Single-walled carbon nanotubes | Daunorubicin | T-cell acute lymphoblastic leukemia | direct conjugation | [255] | ||
| PTK7 | Sgc8 (DNA) | Mesoporus nanoparticles | Doxorubicin | T-cell acute lymphoblastic leukaemia | Carbodiimide chemistry | [256] |
| Gold nanoparticles | Daunorubicin | T-cell acute lymphoblastic leukemia | SH-Aptamer gold coordination | [257] | ||
| Au | Doxorubicin | T-cell acute lymphoblastic leukemia | SH-Aptamer gold coordination | [258] | ||
| Acoustic droplets | Daunorubicin | T-cell acute lymphoblastic leukemia | Thiol-maleimide chemistry | [259] | ||
| IgM | TDO5 (DNA) | PAMAM Dendrimer | Uptake study | Burkitt’s lymphoma | Carbodiimide chemistry | [260] |
| HER2 | S6 aptamer | Plasmonic gold coating on magnetic nanoparticles | Fe3O4 | Breast | SH-Aptamer gold coordination | [261] |
| TSA14 | PEGylated Liposomes | Doxorubicin | Breast | Thiol-maleimide chemistry | [262] | |
| A6 | hybrid nanoparticles (cationic lipids and PLGA-b-PEG) | siRNA | Breast | Thiol-maleimide chemistry | [263] | |
| CD44 | DNA thiolated aptamer | PEG-PAMAM | miRNA | Breast | Carbodiimide chemistry for PAMAM followed by Aptamer Thiol-maleimide chemistry | [264] |
| EpCAM | EpApt | PLGA-b-PEG | Lecithisn curcumin | Colorectal | Carbodiimide chemistry | [265] |
| DNA-EpCAM | mesoporous silica | Doxorubicin | colon | Carbodiimide chemistry | [266] | |
| EGFR | RNA | Lipid-polymer nanoparticle | Salinomycin | Osteosarcoma CSCs | Thiol-maleimide chemistry | [267] |
| Tenascin-C | GBI-10 | PEGylated Liposomes | Gadolinium Compounds | Glioma | Carbodiimide chemistry | [268] |
| GBI-10 | QD–Apt nanoprobes | CdSe/ZnS | Glioma | Carbodiimide chemistry | [269] | |
| PDGFR | Gint4.T | PLGA-b-PEG | PI3K-mTOR inhibitor | glioblastoma | Carbodiimide chemistry | [270] |
| Cell-SELEX | SRZ1 | Cationic-liposomes | Doxorubicin | Breast cancer | Avidin-biotin coupling | [271] |
| fibronectin protein | DNA aptamer AS-14 | gold-coated magnetic nanoparticles | Magnetodynamic nanotherapy | Ehrlich carcinoma | Thiol-maleimide chemistry | [272] |
| Cell-SELEX | KW16-13 | gold nanorods | Photothermal therapy | Breast | Thiol-maleimide chemistry | [273] |
| FGFR1 | DNA aptamer | Iron oxide nanoparticles | Hyperthermia | Osteosarcoma | Avidin-biotin coupling | [274] |
| Nucliolin MUC1 ATP | AS1411 MUC1 ATP | DNA dendrimers, pH sensitive release | Epirubicin | Breast, Colon | Electrostatic interaction | [275] |
| Annexin A2 | Annexin A2 aptamer | DNA/RNA hybrid Nanoparticles | Doxorubicin | Ovarian cancer | Complementary base pairing | [276] |
| CD20 | DNA aptamer | Lipid-polymer Nanoparticles | Salinomycin | Melanoma | Thiol-maleimide chemistry | [277] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2020, 25, 3. https://doi.org/10.3390/molecules25010003
Odeh F, Nsairat H, Alshaer W, Ismail MA, Esawi E, Qaqish B, Bawab AA, Ismail SI. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules. 2020; 25(1):3. https://doi.org/10.3390/molecules25010003
Chicago/Turabian StyleOdeh, Fadwa, Hamdi Nsairat, Walhan Alshaer, Mohammad A. Ismail, Ezaldeen Esawi, Baraa Qaqish, Abeer Al Bawab, and Said I. Ismail. 2020. "Aptamers Chemistry: Chemical Modifications and Conjugation Strategies" Molecules 25, no. 1: 3. https://doi.org/10.3390/molecules25010003
APA StyleOdeh, F., Nsairat, H., Alshaer, W., Ismail, M. A., Esawi, E., Qaqish, B., Bawab, A. A., & Ismail, S. I. (2020). Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules, 25(1), 3. https://doi.org/10.3390/molecules25010003






