Revealing Drug-Target Interactions with Computational Models and Algorithms
Abstract
1. Introduction
2. Data Representation and Repositories
2.1. Benchmark Data Set
2.2. Flowchart
2.3. DTI Relevant Databases
- Step 1
- Extracting related data from different databases: (i) drugs, DTIs and drug-drug interactions from DrugBank; (ii) proteins, and protein-protein interactions from HPRD [12]; (iii) diseases, drug-disease and protein-disease associations from the Comparative Toxicogenomics Database [13]; (iv) side-effects and drug-side-effect associations from SIDER [14].
- Step 2
- Excluding isolated entities (nodes) which have no edges in the network.
- Step 3
- Integrating four types of nodes and six types of associations (edges) in Step 1 and constructing a heterogeneous network.
- Step 4
- Building multiple similarity networks to further increase the network heterogeneity.
- Step 5
- Removing homologous proteins or similar drugs from constructed heterogeneous networks to reduce the potential redundancy in the DTIs: (i) removing the DTIs involving homologous proteins with sequence identity scores larger than ; (2) removing the DTIs involving similar drugs with Tanimoto coefficients larger than ; (3) removing the DTIs involving the drugs with Jaccard similarity scores of side effects larger than ; (4) removing the DTIs involving the proteins or drugs associated with similar diseases (Jaccard similarity scores larger than ; (5) removing the DTIs involving either homologous proteins with sequence identity scores larger than or similar drugs with Tanimoto coefficients larger than .
2.3.1. DrugBank
2.3.2. SuperTarget
2.3.3. STITCH
2.3.4. ZINC
2.3.5. IUPHAR/BPS Guide to PHARMACOLOGY
2.3.6. SIDER
2.3.7. BindingDB
2.3.8. TTD
2.3.9. MATADOR
2.3.10. ChEMBL
2.3.11. DCDB
2.4. DTI Relevant Software Packages
2.4.1. RDKit
2.4.2. ChemDes
2.4.3. OpenBabel
2.4.4. Rchemcpp
2.4.5. PyDPI
2.4.6. Rcpi
2.4.7. KeBABS
2.4.8. PROFEAT
2.4.9. Pse-in-One
2.4.10. ProtrWeb
2.5. On-Line Tools/Web-Service for DTI Prediction
2.5.1. DrugE-Rank
2.5.2. DINIES
2.5.3. Drug2Gene
2.5.4. iGPCR-Drug
2.5.5. SynSysNet
2.5.6. SDTNBI
2.5.7. DTome
2.5.8. PharmMapper
2.5.9. SwissTargetPrediction
2.5.10. TargetNet
2.5.11. DT-Web
3. Network-Based Methods
3.1. DSSI
3.2. MTOI
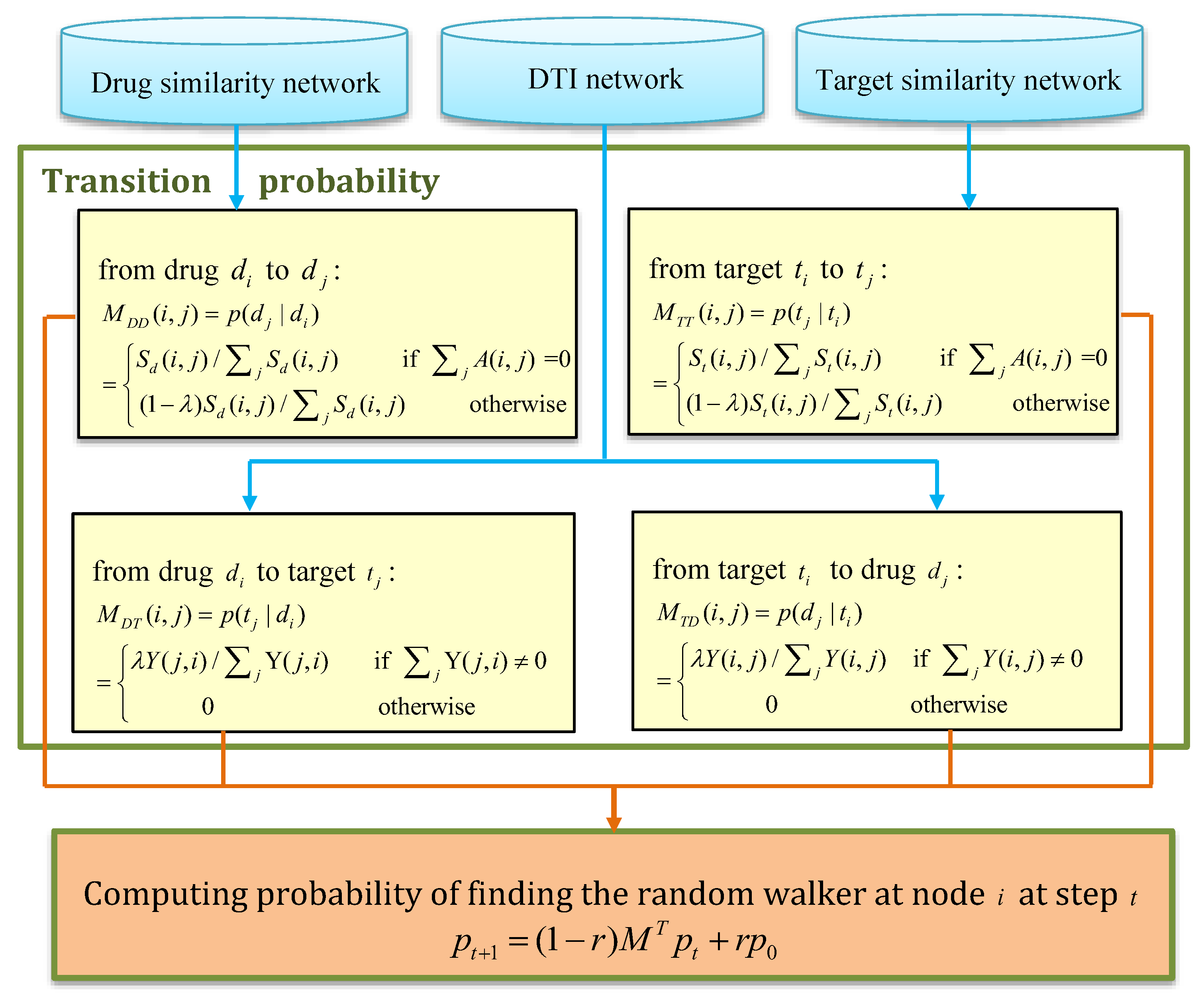
3.3. NRWRH
3.4. DBSI, TBSI, and NBI
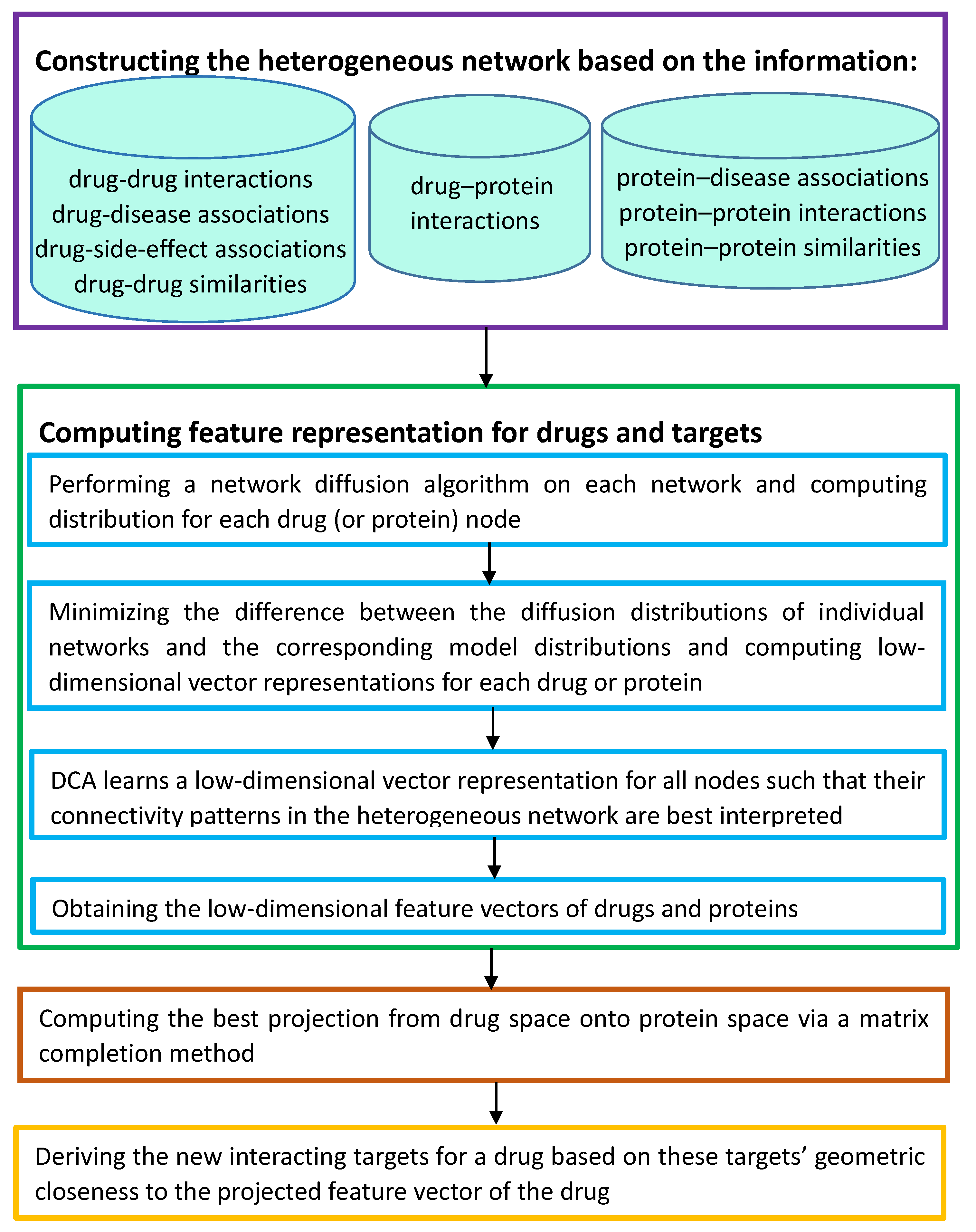
3.5. DTINet
4. Machine Learning-Based Methods
4.1. BLM
4.1.1. KRM
4.1.2. BLM
4.1.3. BLM-NII
4.2. Regularized Least Squares
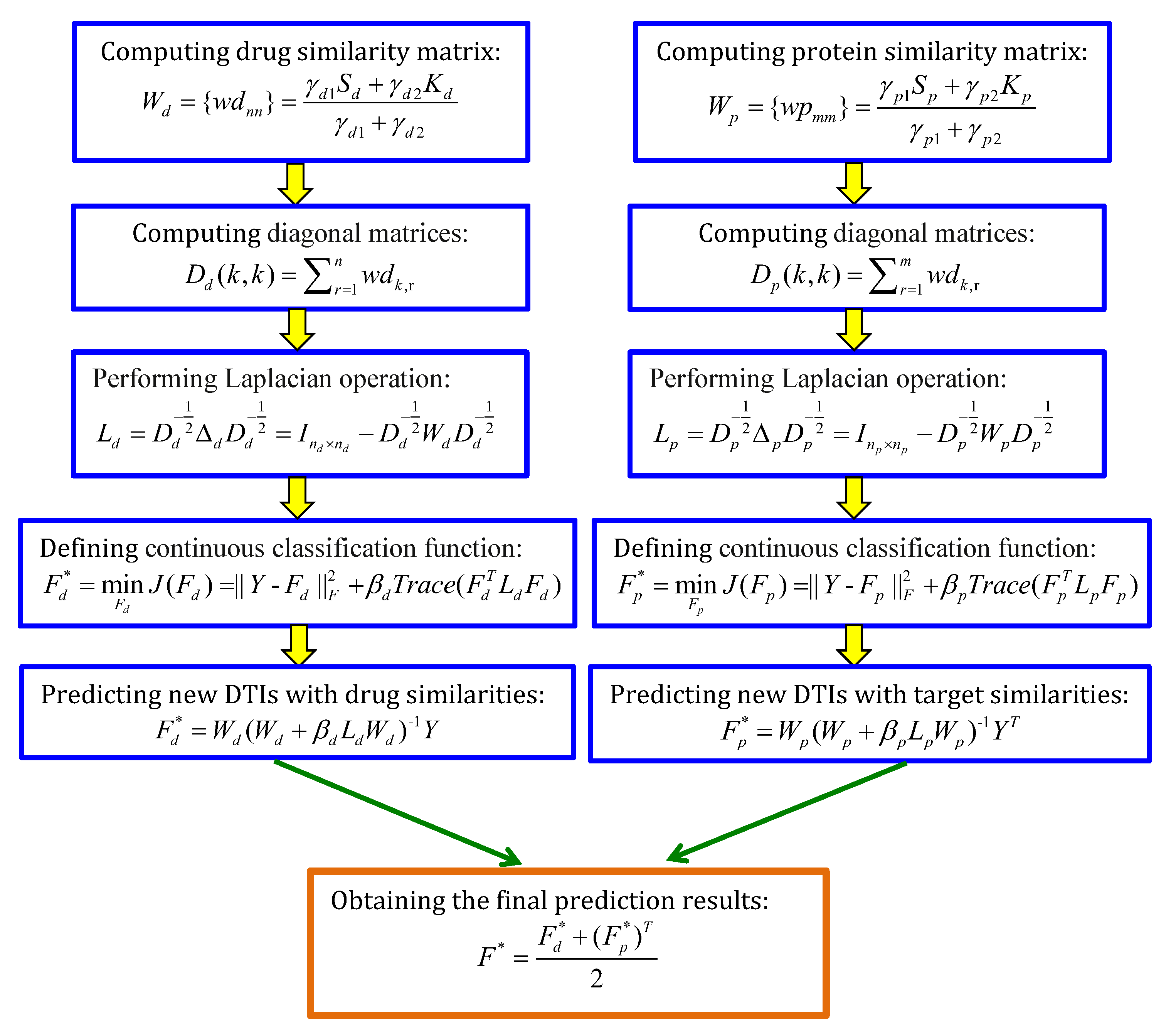
4.2.1. LapRLS, NetLapRLS
4.2.2. RLS
4.2.3. WNN, WNN-GIP
4.2.4. Kron-RLS
4.2.5. KMDR
4.3. Matrix Factorization
4.3.1. KBMF2K
4.3.2. PMF
4.3.3. MSCMF
4.3.4. NRLMF
4.3.5. DNILMF
4.4. Deep Learning
4.4.1. DeepDTIs
4.4.2. EENN
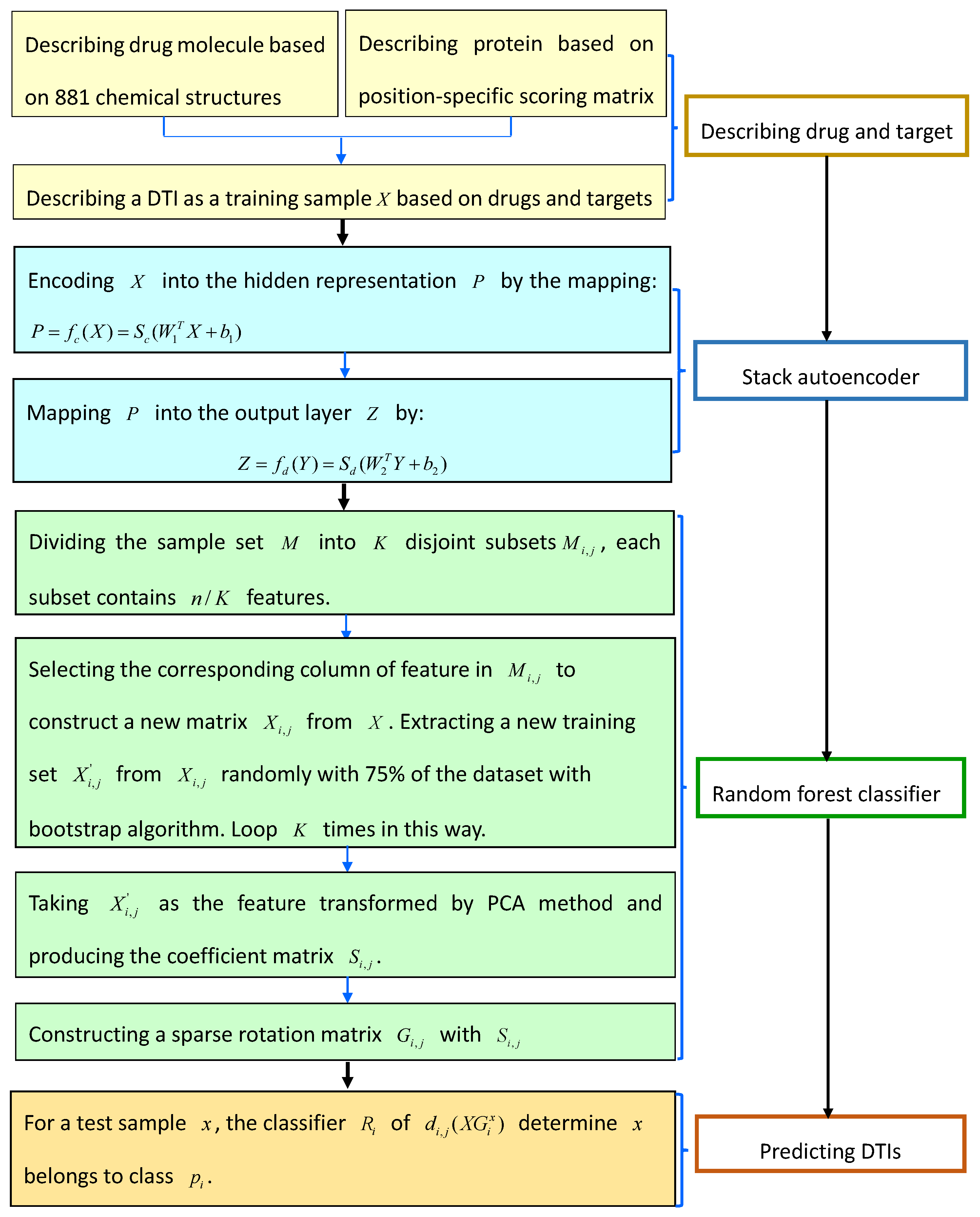
4.4.3. Stacked Autoencoder
4.5. Other Methods
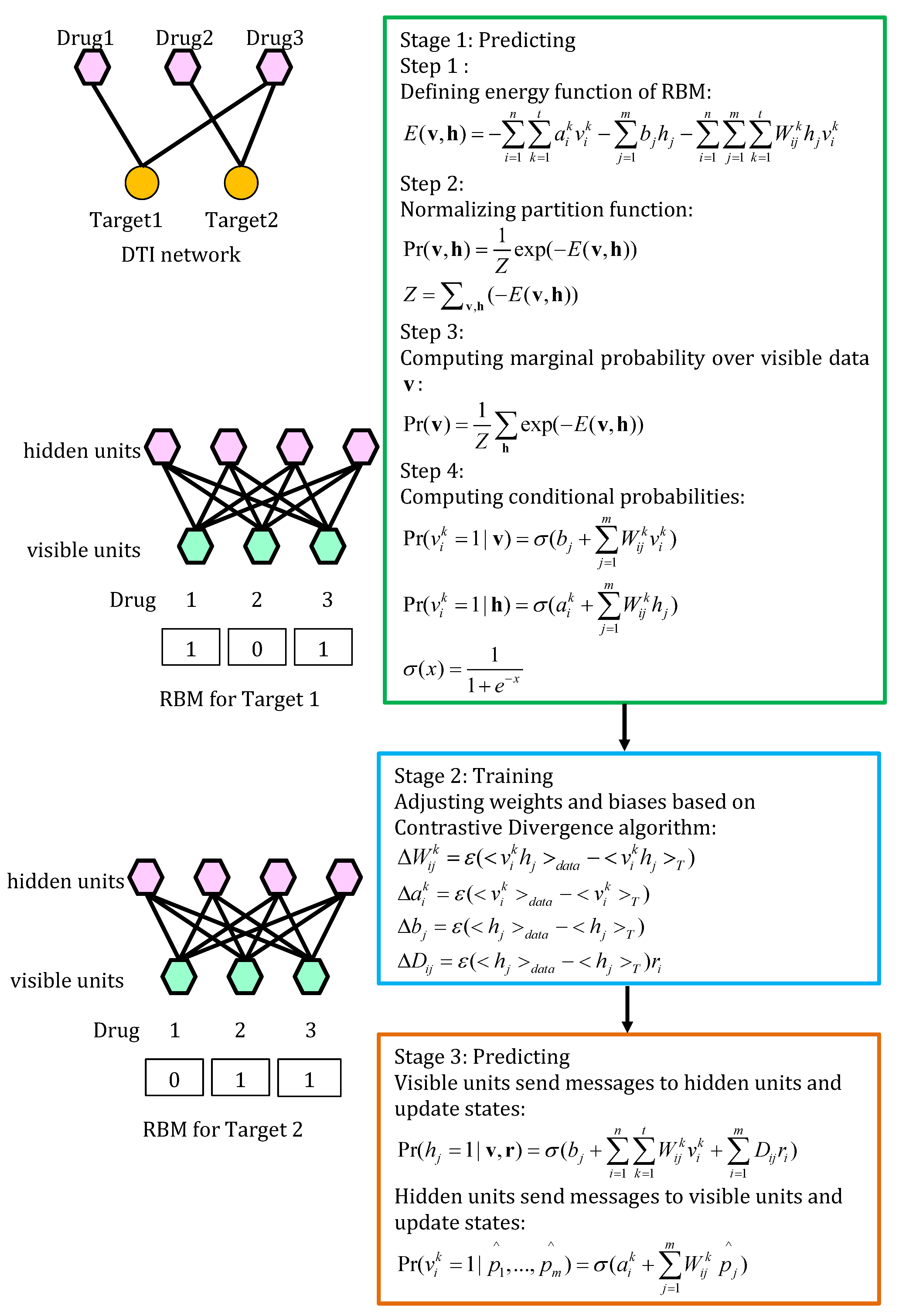
4.5.1. RBM
4.5.2. NetCBP
5. Discussion
6. Conclusions and Further Research
6.1. Heterogeneous Data Integration
6.2. Reliable Negative Sample Selection
6.3. Noncoding RNAs as Targets
6.4. Environmental Factors and Genetic Factors
6.5. Deep Learning
6.6. Sparse Representation
6.7. Types of DTI
6.8. Personalized Medicine
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Polamreddy, P.; Gattu, N. The drug repurposing landscape from 2012 to 2017: Evolution, challenges, and possible solutions. Drug Discov. Today 2018, 24, 789–795. [Google Scholar] [CrossRef]
- Yella, J.; Yaddanapudi, S.; Wang, Y.; Jegga, A. Changing trends in computational drug repositioning. Pharmaceuticals 2018, 11, 57. [Google Scholar] [CrossRef]
- Chen, X.; Yan, C.C.; Zhang, X.; Zhang, X.; Dai, F.; Yin, J.; Zhang, Y. Drug–target interaction prediction: Databases, web servers and computational models. Brief. Bioinform. 2015, 17, 696–712. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.C.; Ekins, S.; Williams, A.J.; Tropsha, A. A bibliometric review of drug repurposing. Drug Discov. Today 2018, 23, 661–672. [Google Scholar] [CrossRef]
- Brazil, R. Repurposing viagra: The ‘little blue pill’ for all ills? Pathophysiology 2018, 14, 20. [Google Scholar]
- Jin, G.; Wong, S.T. Toward better drug repositioning: Prioritizing and integrating existing methods into efficient Pipelines. Drug Discov. Today 2014, 69, 637–644. [Google Scholar] [CrossRef]
- Shahreza, M.L.; Ghadiri, N.; Mousavi, S.R.; Varshosaz, J.; Green, J.R. A review of network-based approaches to drug repositioning. Brief. Bioinform. 2017, 19, 878–892. [Google Scholar] [CrossRef]
- Yamanishi, Y.; Araki, M.; Gutteridge, A.; Honda, W.; Kanehisa, M. Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2008, 24, i232–i240. [Google Scholar] [CrossRef]
- Liu, H.; Sun, J.; Guan, J.; Zheng, J.; Zhou, S. Improving compound–protein interaction prediction by building up highly credible negative samples. Bioinformatics 2015, 31, i221–i229. [Google Scholar] [CrossRef]
- Luo, Y.; Zhao, X.; Zhou, J.; Yang, J.; Zhang, Y.; Kuang, W.; Peng, J.; Chen, L.; Zeng, J. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 2017, 8, 573. [Google Scholar] [CrossRef]
- Prasad, T.K.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human protein reference database—2009 update. Nucleic Acids Res. 2008, 37, D767–D772. [Google Scholar] [CrossRef]
- Davis, A.P.; Murphy, C.G.; Johnson, R.; Lay, J.M.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Rosenstein, M.C.; Wiegers, T.C.; et al. The comparative toxicogenomics database: Update 2013. Nucleic Acids Res. 2012, 41, D1104–D1114. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Campillos, M.; Letunic, I.; Jensen, L.J.; Bork, P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010, 6, 343. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. Drugbank 5.0: A major update to the drugbank database for 2018. Nucleic Acids Res. 2017, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Günther, S.; Kuhn, M.; Dunkel, M.; Campillos, M.; Senger, C.; Petsalaki, E.; Ahmed, J.; Urdiales, E.G.; Gewiess, A.; Jensen, L.J.; et al. Supertarget and matador: Resources for exploring drug-target relationships. Nucleic Acids Res. 2007, 36, D919–D922. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Szklarczyk, D.; Franceschini, A.; Mering, C.V.; Jensen, L.J.; Bork, P. Stitch 3: Zooming in on protein–chemical interactions. Nucleic Acids Res. 2011, 40, D876–D880. [Google Scholar] [CrossRef]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. Zinc: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef]
- Harding, S.D.; Sharman, J.L.; Faccenda, E.; Southan, C.; Pawson, A.J.; Ireland, S.; Gray, A.J.; Bruce, L.; Alexander, S.P.; Anderton, S.; et al. The iuphar/bps guide to pharmacology in 2018: Updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Res. 2017, 46, D1091–D1106. [Google Scholar] [CrossRef]
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The sider database of drugs and side effects. Nucleic Acids Res. 2015, 44, D1075–D1079. [Google Scholar] [CrossRef]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. Bindingdb in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2015, 44, D1045–D1053. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Yu, C.Y.; Li, X.X.; Zhang, P.; Tang, J.; Yang, Q.; Fu, T.; Zhang, X.; Cui, X.; Tu, G.; et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2017, 46, D1121–D1127. [Google Scholar]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. Chembl: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011, 40, D1100–D1107. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, Q.; Yu, G.; Gai, W.; Li, Y.; Chen, X. Dcdb 2.0: A major update of the drug combination database. Database 2014, 2014, bau124. [Google Scholar] [CrossRef] [PubMed]
- Landrum, G. Rdkit: Open-Source Cheminformatics; YAeHMOP: Cumming, GA, USA, 2006. [Google Scholar]
- Dong, J.; Cao, D.-S.; Miao, H.-Y.; Liu, S.; Deng, B.-C.; Yun, Y.-H.; Wang, N.-N.; Lu, A.-P.; Zeng, W.-B.; Chen, A.F. Chemdes: An integrated web-based platform for molecular descriptor and fingerprint computation. J. Cheminform. 2015, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Klambauer, G.; Wischenbart, M.; Mahr, M.; Unterthiner, T.; Mayr, A.; Hochreiter, S. Rchemcpp: A web service for structural analoging in chembl, drugbank and the connectivity map. Bioinformatics 2015, 31, 3392–3394. [Google Scholar] [CrossRef]
- Cao, D.-S.; Xu, Q.-S.; Liang, Y.-Z. propy: A tool to generate various modes of chou’s pseaac. Bioinformatics 2013, 9, 960–962. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.-S.; Xiao, N.; Xu, Q.-S.; Chen, A.F. Rcpi: R/bioconductor package to generate various descriptors of proteins, compounds and their interactions. Bioinformatics 2014, 31, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Palme, J.; Hochreiter, S.; Bodenhofer, U. Kebabs: An r package for kernel-based analysis of biological sequences. Bioinformatics 2015, 31, 2574–2576. [Google Scholar] [CrossRef]
- Li, Z.-R.; Lin, H.H.; Han, L.; Jiang, L.; Chen, X.; Chen, Y.Z. Profeat: A web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2006, 34, W32–W37. [Google Scholar] [CrossRef]
- Liu, B.; Liu, F.; Wang, X.; Chen, J.; Fang, L.; Chou, K.-C. Pse-in-one: A web server for generating various modes of pseudo components of dna, rna, and protein sequences. Nucleic Acids Res. 2015, 43, W65–W71. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Cao, D.-S.; Zhu, M.-F.; Xu, Q.-S. protr/protrweb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics 2015, 31, 1857–1859. [Google Scholar] [CrossRef]
- Cheng, T.; Hao, M.; Takeda, T.; Bryant, S.H.; Wang, Y. Large-scale prediction of drug-target interaction: A data-centric review. AAPS J. 2017, 19, 1264–1275. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Gao, J.; Wu, D.; Zhang, S.; Mamitsuka, H.; Zhu, S. Druge-rank: Improving drug–target interaction prediction of new candidate drugs or targets by ensemble learning to rank. Bioinformatics 2016, 32, i18–i27. [Google Scholar] [CrossRef]
- Yamanishi, Y.; Kotera, M.; Moriya, Y.; Sawada, R.; Kanehisa, M.; Goto, S. Dinies: Drug–target interaction network inference engine based on supervised analysis. Nucleic Acids Res. 2014, 42, W39–W45. [Google Scholar] [CrossRef] [PubMed]
- Roider, H.G.; Pavlova, N.; Kirov, I.; Slavov, S.; Slavov, T.; Uzunov, Z.; Weiss, B. Drug2gene: An exhaustive resource to explore effectively the drug-target relation network. BMC Bioinform. 2014, 15, 68. [Google Scholar] [CrossRef]
- Xiao, X.; Min, J.-L.; Wang, P.; Chou, K.-C. igpcr-drug: A web server for predicting interaction between gpcrs and drugs in cellular networking. PLoS ONE 2013, 8, e72234. [Google Scholar] [CrossRef] [PubMed]
- von Eichborn, J.; Dunkel, M.; Gohlke, B.O.; Preissner, S.C.; Hoffmann, M.F.; Bauer, J.M.; Armstrong, J.D.; Schaefer, M.H.; Andrade-Navarro, M.A.; Novere, N.L.; et al. Synsysnet: Integration of experimental data on synaptic protein–protein interactions with drug-target relations. Nucleic Acids Res. 2012, 41, D834–D840. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, F.; Li, J.; Li, W.; Liu, G.; Tang, Y. Sdtnbi: An integrated network and chemoinformatics tool for systematic prediction of drug–target interactions and drug repositioning. Brief. Bioinform. 2016, 18, 333–347. [Google Scholar] [CrossRef]
- Sun, J.; Wu, Y.; Xu, H.; Zhao, Z. Dtome: A web-based tool for drug-target interactome construction. In BMC Bioinformatics; BioMed Central: London, UK, 2012. [Google Scholar]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. Pharmmapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. Swisstargetprediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Yao, Z.-J.; Dong, J.; Che, Y.-J.; Zhu, M.-F.; Wen, M.; Wang, N.-N.; Wang, S.; Lu, A.-P.; Cao, D.-S. Targetnet: A web service for predicting potential drug–target interaction profiling via multi-target sar models. J. Comput.-Aided Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Alaimo, S.; Bonnici, V.; Cancemi, D.; Ferro, A.; Giugno, R.; Pulvirenti, A. Dt-web: A web-based application for drug-target interaction and drug combination prediction through domain-tuned network-based inference. BMC Syst. Biol. 2015, 9, S4. [Google Scholar] [CrossRef]
- Campillos, M.; Kuhn, M.; Gavin, A.-C.; Jensen, L.J.; Bork, P. Drug target identification using side-effect similarity. Science 2008, 321, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Bai, H.; Ouyang, Q.; Lai, L.; Tang, C. Finding multiple target optimal intervention in disease-related molecular network. Mol. Syst. Biol. 2008, 4, 228. [Google Scholar] [CrossRef]
- Chen, X.; Liu, M.-X.; Yan, G.-Y. Drug–target interaction prediction by random walk on the heterogeneous network. Mol. BioSyst. 2012, 8, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liu, C.; Jiang, J.; Lu, W.; Li, W.; Liu, G.; Zhou, W.; Huang, J.; Tang, Y. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol. 2012, 8, e1002503. [Google Scholar] [CrossRef]
- Bleakley, K.; Yamanishi, Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics 2009, 25, 2397–2403. [Google Scholar] [CrossRef]
- Mei, J.-P.; Kwoh, C.-K.; Yang, P.; Li, X.-L.; Zheng, J. Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics 2012, 29, 238–245. [Google Scholar] [CrossRef]
- Xia, Z.; Wu, L.-Y.; Zhou, X.; Wong, T.S. Semi-supervised drug-protein interaction prediction from heterogeneous biological spaces. In BMC Systems Biology; BioMed Central: London, UK, 2010; p. S6. [Google Scholar]
- Tvan, L.; Nabuurs, S.B.; Marchiori, E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 2011, 27, 3036–3043. [Google Scholar]
- Tvan, L.; Marchiori, E. Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS ONE 2013, 8, e66952. [Google Scholar]
- Pahikkala, T.; Airola, A.; Pietilä, S.; Shakyawar, S.; Szwajda, A.; Tang, J.; Aittokallio, T. Toward more realistic drug–target interaction predictions. Brief. Bioinform. 2014, 16, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Q.; Li, Y.; Wu, Y.; Li, R.; Dong, Y.; Li, Y.; Xiong, Q.; Huang, Z.; Li, M. A kernel matrix dimension reduction method for predicting drug-target interaction. Chemom. Intell. Lab. Syst. 2017, 162, 104–110. [Google Scholar] [CrossRef]
- Gönen, M. Predicting drug–target interactions from chemical and genomic kernels using bayesian matrix factorization. Bioinformatics 2012, 28, 2304–2310. [Google Scholar] [CrossRef]
- Cobanoglu, M.C.; Liu, C.; Hu, F.; Oltvai, Z.N.; Bahar, I. Predicting drug–target interactions using probabilistic matrix factorization. J. Chem. Inf. Model. 2013, 53, 3399–3409. [Google Scholar] [CrossRef]
- Zheng, X.; Ding, H.; Mamitsuka, H.; Zhu, S. Collaborative matrix factorization with multiple similarities for predicting drug-target interactions. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 1025–1033. [Google Scholar]
- Liu, Y.; Wu, M.; Miao, C.; Zhao, P.; Li, X.-L. Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLoS Comput. Biol. 2016, 12, e1004760. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Bryant, S.H.; Wang, Y. Predicting drug-target interactions by dual-network integrated logistic matrix factorization. Sci. Rep. 2017, 7, 40376. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zhang, Z.; Niu, S.; Sha, H.; Yang, R.; Yun, Y.; Lu, H. Deep-learning-based drug–target interaction prediction. J. Proteome Res. 2017, 16, 1401–1409. [Google Scholar] [CrossRef]
- Gao, K.Y.; Fokoue, A.; Luo, H.; Iyengar, A.; Dey, S.; Zhang, P. Interpretable drug target prediction using deep neural representation. In Proceedings of the IJCAI, Stockholm, Sweden, 13–19 July 2018; pp. 3371–3377. [Google Scholar]
- Wang, L.; You, Z.-H.; Chen, X.; Xia, S.-X.; Liu, F.; Yan, X.; Zhou, Y.; Song, K.-J. A computational-based method for predicting drug–target interactions by using stacked autoencoder deep neural network. J. Comput. Biol. 2018, 25, 361–373. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, J. Predicting drug-target interactions using restricted boltzmann machines. Bioinformatics 2013, 29, i126–i134. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Z. A semi-supervised method for drug-target interaction prediction with consistency in networks. PLoS ONE 2013, 8, e62975. [Google Scholar] [CrossRef] [PubMed]
- Tanoli, Z.; Alam, Z.; Ianevski, A.; Wennerberg, K.; Vähä-Koskela, M.; Aittokallio, T. Interactive visual analysis of drug–target interaction networks using drug target profiler, with applications to precision medicine and drug repurposing. Brief. Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, A.; Wu, M.; Li, X.; Kwoh, C.-K. Computational prediction of drug-target interactions via ensemble learning. In Computational Methods for Drug Repurposing; Springer: Berlin, Germany, 2019; pp. 239–254. [Google Scholar]
- Zhao, Q.; Yu, H.; Ji, M.; Zhao, Y.; Chen, X. Computational model development of drug-target interaction prediction: A review. Curr. Protein Pept. Sci. 2019. [Google Scholar] [CrossRef]
- Peng, L.; Zhu, W.; Liao, B.; Duan, Y.; Chen, M.; Chen, Y.; Yang, J. Screening drug-target interactions with positive-unlabeled learning. Sci. Rep. 2017, 7, 8087. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Liao, B.; Zhu, W.; Li, Z.; Li, K. Predicting drug–target interactions with multi-information fusion. IEEE J. Biomed. Health Inform. 2017, 21, 561–572. [Google Scholar] [CrossRef]
- Ezzat, A.; Wu, M.; Li, X.-L.; Kwoh, C.-K. Computational prediction of drug–target interactions using chemogenomic approaches: An empirical survey. Brief. Bioinform. 2018. [Google Scholar] [CrossRef]
- Chen, R.; Liu, X.; Jin, S.; Lin, J.; Liu, J. Machine learning for drug-target interaction prediction. Molecules 2018, 23, 2208. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, A.; Zhao, P.; Wu, M.; Li, X.-L.; Kwoh, C.-K. Drug-target interaction prediction with graph regularized matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Peng, M.; Liao, B.; Huang, G.; Li, W.; Xie, D. The advances and challenges of deep learning application in biological big data processing. Curr. Bioinform. 2018, 13, 352–359. [Google Scholar] [CrossRef]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef]
- Sansone, E.; Natale, F.G.D.; Zhou, Z.-H. Efficient training for positive unlabeled learning. IEEE Trans. Pattern Anal. Mach. Intell. 2018. [Google Scholar] [CrossRef]
- Jain, S.; White, M.; Radivojac, P. Recovering true classifier performance in positive-unlabeled learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–10 February 2017; pp. 2066–2072. [Google Scholar]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding rna networks in cancer. Nat. Rev. Cancer 2018, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Corey, D.R. Non-coding rnas as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167. [Google Scholar] [CrossRef]
- Sampson, V.B.; Yoo, S.; Kumar, A.; Vetter, N.S.; Kolb, E.A. Micrornas and potential targets in osteosarcoma. Front. Pediatr. 2015, 3, 69. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Masoudifar, A.; Sahebkar, A.; Zare, N.; Nahand, J.S.; Rashidi, B.; Mehrabian, E.; Mohammadi, M.; Mirzaei, H.R.; Jaafari, M.R. Microrna: A novel target of curcumin in cancer therapy. J. Cell. Physiol. 2018, 233, 3004–3015. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shi, Y.-Y. A bipartite network and resource transfer-based approach to infer lncrna-environmental factor associations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Chen, G.; Cui, Q. Towards the understanding of microrna and environmental factor interactions and their relationships to human diseases. Sci. Rep. 2012, 2, 318. [Google Scholar] [CrossRef]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef]
- Wright, J.; Yang, A.Y.; Ganesh, A.; Sastry, S.S.; Ma, Y. Robust face recognition via sparse representation. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 31, 210–227. [Google Scholar] [CrossRef]
- Wang, E.; Zou, J.; Zaman, N.; Beitel, L.K.; Trifiro, M.; Paliouras, M. Cancer systems biology in the genome sequencing era: Part 1, dissecting and modeling of tumor clones and their networks. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2013; pp. 279–285. [Google Scholar]
- Hauser, A.S.; Chavali, S.; Masuho, I.; Jahn, L.J.; Martemyanov, K.A.; Gloriam, D.E.; Babu, M.M. Pharmacogenomics of gpcr drug targets. Cell 2018, 172, 41–54. [Google Scholar] [CrossRef]
- Francia, R.D.; Monaco, A.D.; Saggese, M.; Iaccarino, G.; Crisci, S.; Frigeri, F.; Filippi, R.D.; Berretta, M.; Pinto, A. Pharmacological profile and pharmacogenomics of anti-cancer drugs used for targeted therapy. Curr. Cancer Drug Targets 2018, 18, 499–511. [Google Scholar] [CrossRef]









| Dataset | Drugs () | Targets () | Interactions |
|---|---|---|---|
| enzyme | 445 | 664 | 2926 |
| ion channel | 210 | 204 | 1476 |
| GPCRs | 223 | 95 | 635 |
| nuclear receptor | 54 | 26 | 90 |
| AUC | ||||
| Dataset | KRM | BLM | RLS | BLM-NII |
| Enzyme | 86.4 | 97.6 | 97.8 | 98.8 |
| Ion Channel | 81.9 | 97.3 | 98.4 | 99.0 |
| GPCR | 76.5 | 95.5 | 95.4 | 98.4 |
| Nuclear Receptor | 74.9 | 88.1 | 92.2 | 98.1 |
| AUPR | ||||
| Dataset | KRM | BLM | RLS | BLM-NII |
| Enzyme | 6.30 | 83.3 | 91.5 | 92.9 |
| Ion Channel | 17.2 | 78.1 | 94.3 | 95.0 |
| GPCR | 10.9 | 66.7 | 79.0 | 86.5 |
| Nuclear Receptor | 17.1 | 61.2 | 68.4 | 86.6 |
| AUC | |||||
| Dataset | NetLapRLS | BLM-NII | WNN-GIP | KBMF2K | NRLMF |
| Enzyme | 97.2 | 97.8 | 96.4 | 90.5 | 98.7 |
| Ion Channel | 96.9 | 98.1 | 95.9 | 96.1 | 98.9 |
| GPCR | 91.5 | 95.0 | 94.4 | 92.6 | 96.9 |
| Nuclear Receptor | 85.0 | 90.5 | 90.1 | 87.7 | 95.0 |
| AUPR | |||||
| Dataset | NetLapRLS | BLM-NII | WNN-GIP | KBMF2K | NRLMF |
| Enzyme | 78.9 | 75.2 | 70.6 | 65.4 | 89.2 |
| Ion Channel | 83.7 | 82.1 | 71.7 | 77.1 | 90.6 |
| GPCR | 61.6 | 52.4 | 52.0 | 57.8 | 74.9 |
| Nuclear Receptor | 46.5 | 65.9 | 58.9 | 53.4 | 72.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Li, Z.; Yang, J.; Tian, G.; Liu, F.; Wen, H.; Peng, L.; Chen, M.; Xiang, J.; Peng, L. Revealing Drug-Target Interactions with Computational Models and Algorithms. Molecules 2019, 24, 1714. https://doi.org/10.3390/molecules24091714
Zhou L, Li Z, Yang J, Tian G, Liu F, Wen H, Peng L, Chen M, Xiang J, Peng L. Revealing Drug-Target Interactions with Computational Models and Algorithms. Molecules. 2019; 24(9):1714. https://doi.org/10.3390/molecules24091714
Chicago/Turabian StyleZhou, Liqian, Zejun Li, Jialiang Yang, Geng Tian, Fuxing Liu, Hong Wen, Li Peng, Min Chen, Ju Xiang, and Lihong Peng. 2019. "Revealing Drug-Target Interactions with Computational Models and Algorithms" Molecules 24, no. 9: 1714. https://doi.org/10.3390/molecules24091714
APA StyleZhou, L., Li, Z., Yang, J., Tian, G., Liu, F., Wen, H., Peng, L., Chen, M., Xiang, J., & Peng, L. (2019). Revealing Drug-Target Interactions with Computational Models and Algorithms. Molecules, 24(9), 1714. https://doi.org/10.3390/molecules24091714






