Impact of Small Molecules on Intermolecular G-Quadruplex Formation
Abstract
1. Introduction
2. Results and Discussion
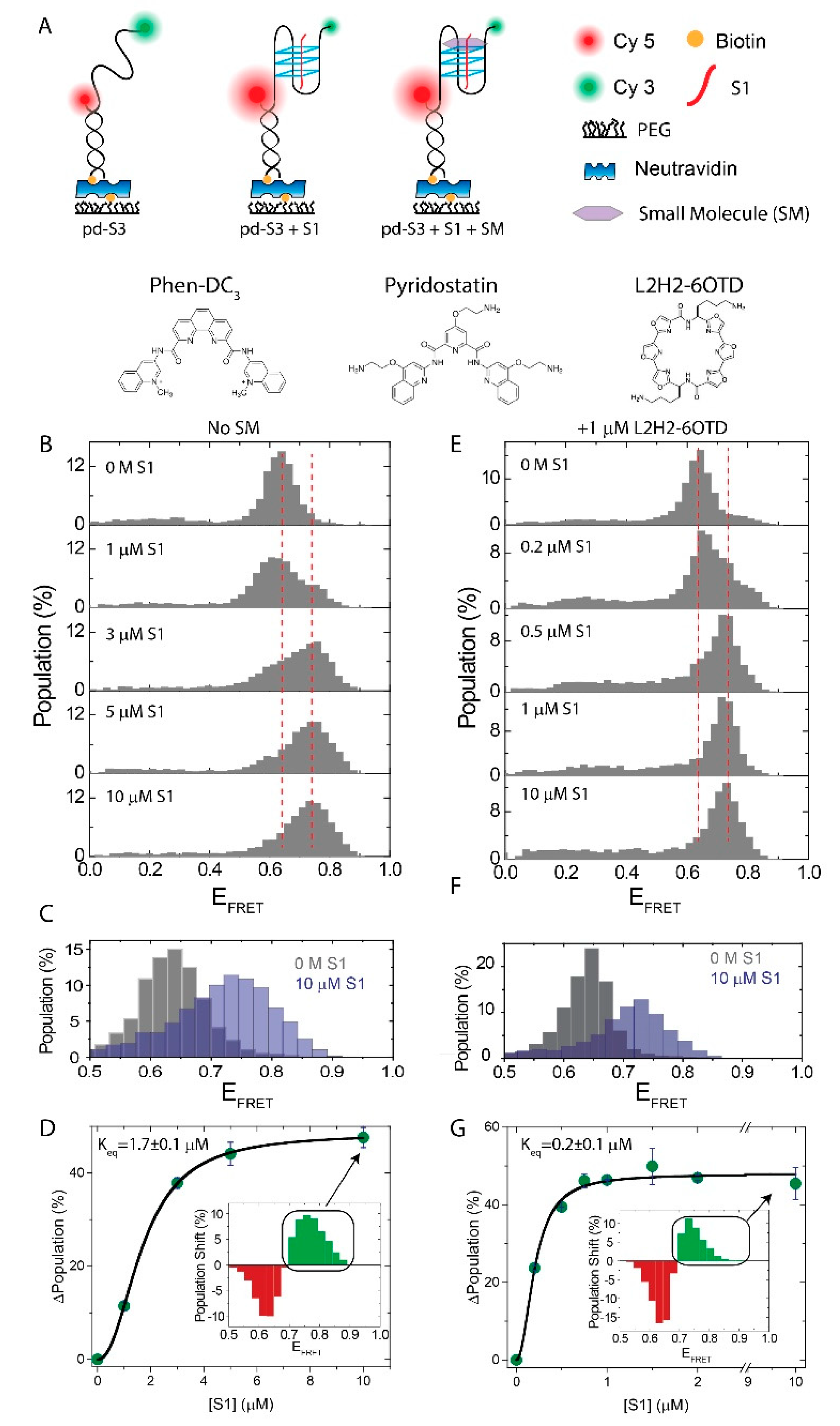
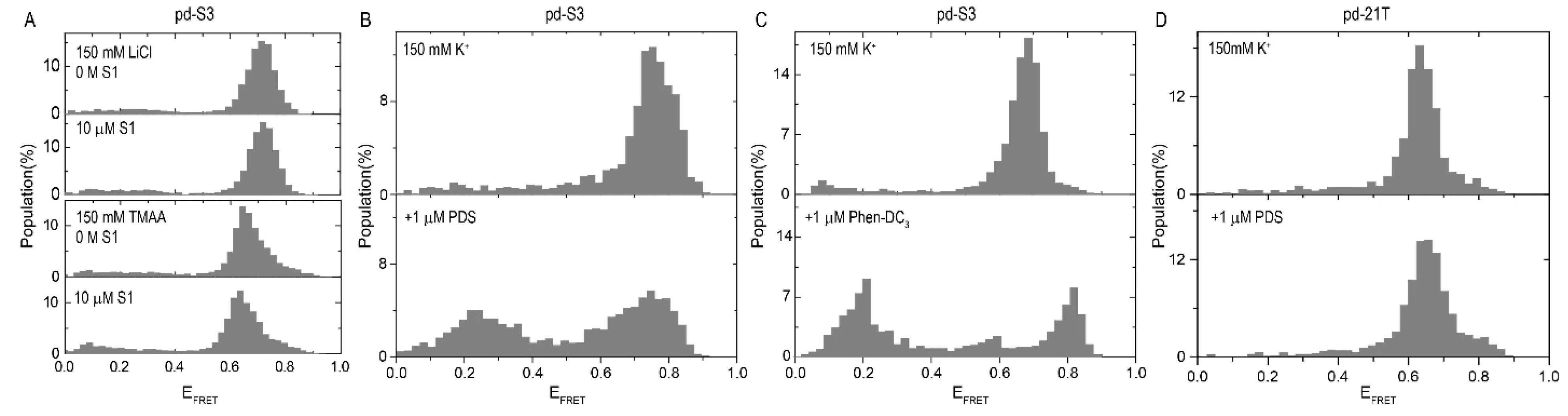
2.1. Influence of SMs on the i-GQ Formation in 3+1 GGG-Repeat Configuration
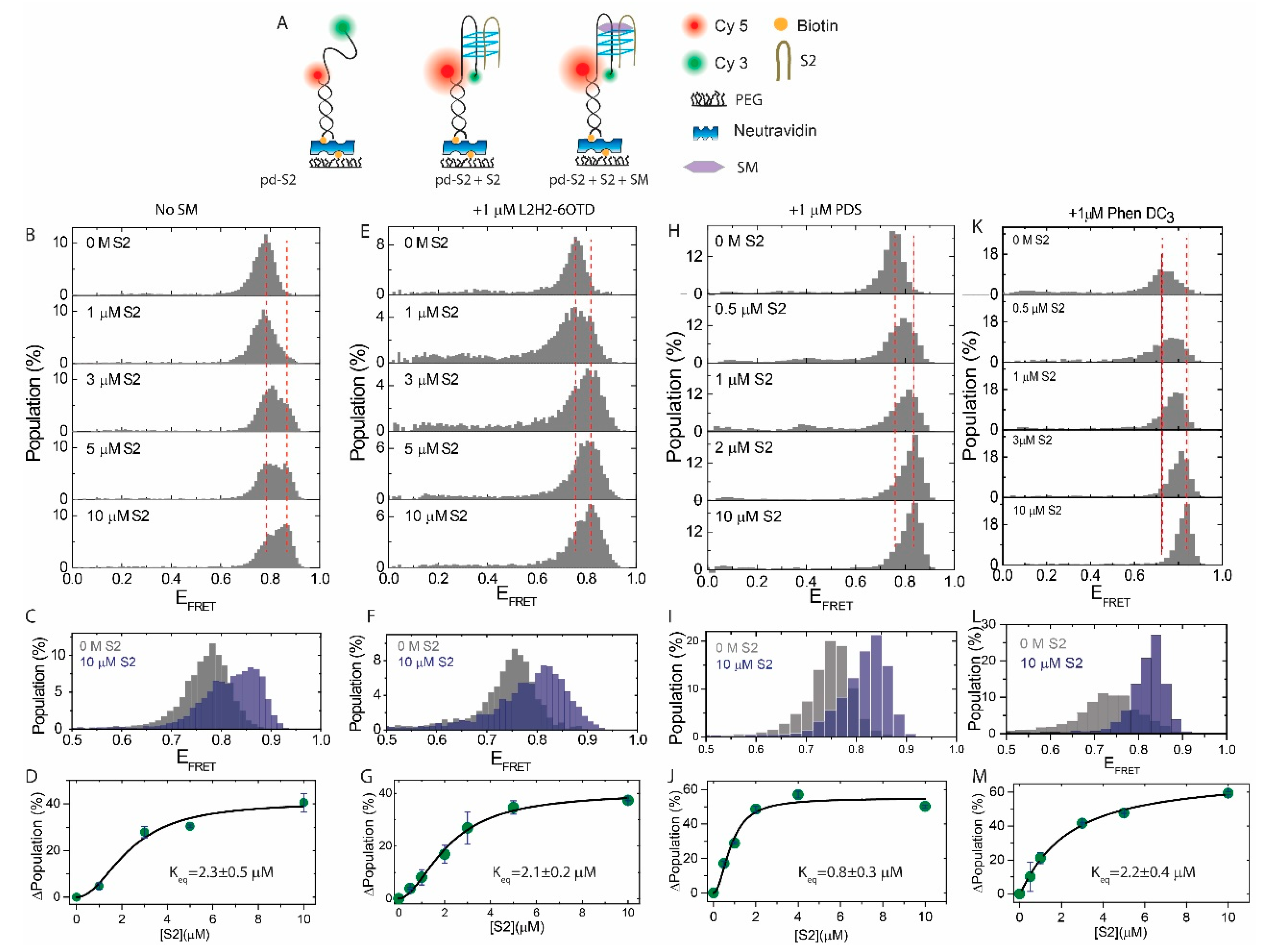
2.2. Influence of SMs on the i-GQ Formation in the 2+2 GGG-Repeat Configuration
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- McLuckie, K.I.; Di Antonio, M.; Zecchini, H.; Xian, J.; Caldas, C.; Krippendorff, B.F.; Tannahill, D.; Lowe, C.; Balasubramanian, S. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J. Am. Chem. Soc. 2013, 135, 9640–9643. [Google Scholar] [CrossRef] [PubMed]
- Merle, P.; Gueugneau, M.; Teulade-Fichou, M.P.; Muller-Barthelemy, M.; Amiard, S.; Chautard, E.; Guetta, C.; Dedieu, V.; Communal, Y.; Mergny, J.L.; et al. Highly efficient radiosensitization of human glioblastoma and lung cancer cells by a g-quadruplex DNA binding compound. Sci. Rep. 2015, 5, 16255. [Google Scholar] [CrossRef] [PubMed]
- Fernando, H.; Reszka, A.P.; Huppert, J.; Ladame, S.; Rankin, S.; Venkitaraman, A.R.; Neidle, S.; Balasubramanian, S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 2006, 45, 7854–7860. [Google Scholar] [CrossRef]
- Fernando, H.; Sewitz, S.; Darot, J.; Tavare, S.; Huppert, J.L.; Balasubramanian, S. Genome-wide analysis of a g-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009, 37, 6716–6722. [Google Scholar] [CrossRef]
- Mitchell, T.; Ramos-Montoya, A.; Di Antonio, M.; Murat, P.; Ohnmacht, S.; Micco, M.; Jurmeister, S.; Fryer, L.; Balasubramanian, S.; Neidle, S.; et al. Downregulation of androgen receptor transcription by promoter g-quadruplex stabilization as a potential alternative treatment for castrate-resistant prostate cancer. Biochemistry 2013, 52, 1429–1436. [Google Scholar] [CrossRef]
- Morris, M.J.; Negishi, Y.; Pazsint, C.; Schonhoft, J.D.; Basu, S. An rna g-quadruplex is essential for cap-independent translation initiation in human vegf ires. J. Am. Chem. Soc. 2010, 132, 17831–17839. [Google Scholar] [CrossRef]
- Mergny, J.L.; Sen, D. DNA quadruple helices in nanotechnology. Chem. Rev. 2019. [Google Scholar] [CrossRef]
- Nasiri, H.R.; Bell, N.M.; McLuckie, K.I.; Husby, J.; Abell, C.; Neidle, S.; Balasubramanian, S. Targeting a c-myc g-quadruplex DNA with a fragment library. Chem. Commun. (Camb.) 2014, 50, 1704–1707. [Google Scholar] [CrossRef]
- Neidle, S. A personal history of quadruplex-small molecule targeting. Chem. Rec. 2015. [Google Scholar] [CrossRef]
- Ohnmacht, S.A.; Varavipour, E.; Nanjunda, R.; Pazitna, I.; Di Vita, G.; Gunaratnam, M.; Kumar, A.; Ismail, M.A.; Boykin, D.W.; Wilson, W.D.; et al. Discovery of new g-quadruplex binding chemotypes. Chem. Commun. (Camb.) 2014, 50, 960–963. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.M.; Tizkova, K.; Reszka, A.P.; Neidle, S.; Thurston, D.E. Identification of novel telomeric g-quadruplex-targeting chemical scaffolds through screening of three nci libraries. Bioorg. Med. Chem. Lett. 2012, 22, 3006–3010. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.J.; Heddi, B.; Hamon, F.; Teulade-Fichou, M.P.; Phan, A.T. Solution structure of a g-quadruplex bound to the bisquinolinium compound phen-dc(3). Angew Chem. Int. Ed. Engl. 2014, 53, 999–1002. [Google Scholar] [CrossRef]
- De Rache, A.; Mergny, J.L. Assessment of selectivity of g-quadruplex ligands via an optimised fret melting assay. Biochimie 2015, 115, 194–202. [Google Scholar] [CrossRef]
- Iida, K.; Nagasawa, K. Macrocyclic polyoxazoles as g-quadruplex ligands. Chem. Rec. 2013, 13, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Le, D.D.; Di Antonio, M.; Chan, L.K.; Balasubramanian, S. G-quadruplex ligands exhibit differential g-tetrad selectivity. Chem. Commun. (Camb.) 2015, 51, 8048–8050. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Delemos, E.; Mergny, J.L.; Teulade-Fichou, M.P.; Monchaud, D. Highly efficient g-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Majima, S.; Nakamura, T.; Seimiya, H.; Nagasawa, K. Evaluation of the interaction between long telomeric DNA and macrocyclic hexaoxazole (6otd) dimer of a g-quadruplex ligand. Molecules 2013, 18, 4328–4341. [Google Scholar] [CrossRef]
- Rodriguez, R.; Muller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.F.; Balasubramanian, S. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef]
- Kudlicki, A.S. G-quadruplexes involving both strands of genomic DNA are highly abundant and colocalize with functional sites in the human genome. PLoS ONE 2016, 11, e0146174. [Google Scholar] [CrossRef]
- Bose, K.; Lech, C.J.; Heddi, B.; Phan, A.T. High-resolution afm structure of DNA g-wires in aqueous solution. Nat. Commun. 2018, 9, 1959. [Google Scholar] [CrossRef]
- Marsh, T.C.; Henderson, E. G-wires: Self-assembly of a telomeric oligonucleotide, d(ggggttgggg), into large superstructures. Biochemistry 1994, 33, 10718–10724. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. DNA origami nanomachines. Molecules 2018, 23, 1766. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Hidaka, K.; Tran, P.L.; Mergny, J.L.; Sugiyama, H. Controlling the stoichiometry and strand polarity of a tetramolecular g-quadruplex structure by using a DNA origami frame. Nucleic Acids Res. 2013, 41, 8738–8747. [Google Scholar] [CrossRef] [PubMed]
- Yeo, Q.Y.; Loh, I.Y.; Tee, S.R.; Chiang, Y.H.; Cheng, J.; Liu, M.H.; Wang, Z.S. A DNA bipedal nanowalker with a piston-like expulsion stroke. Nanoscale 2017, 9, 12142–12149. [Google Scholar] [CrossRef]
- Kosman, J.; Juskowiak, B. Peroxidase-mimicking dnazymes for biosensing applications: A review. Anal. Chim. Acta 2011, 707, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Poon, L.C. Rna and DNA complexes with hemin [fe(iii) heme] are efficient peroxidases and peroxygenases: How do they do it and what does it mean? Crit. Rev. Biochem. Mol. Biol. 2011, 46, 478–492. [Google Scholar] [CrossRef]
- Shimron, S.; Wang, F.; Orbach, R.; Willner, I. Amplified detection of DNA through the enzyme-free autonomous assembly of hemin/g-quadruplex dnazyme nanowires. Anal. Chem. 2012, 84, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wu, Z.S.; Shen, G.L.; Yu, R.Q. Intermolecular g-quadruplex structure-based fluorescent DNA detection system. Biosens. Bioelectron. 2013, 41, 262–267. [Google Scholar] [CrossRef]
- He, H.Z.; Chan, D.S.; Leung, C.H.; Ma, D.L. A highly selective g-quadruplex-based luminescent switch-on probe for the detection of gene deletion. Chem. Commun. (Camb.) 2012, 48, 9462–9464. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; He, B.; Lu, L.; Leung, C.H.; Mergny, J.L.; Ma, D.L. Label-free luminescent detection of lmp1 gene deletion using an intermolecular g-quadruplex-based switch-on probe. Biosens. Bioelectron. 2015, 70, 338–344. [Google Scholar] [CrossRef]
- Freeman, R.; Liu, X.; Willner, I. Chemiluminescent and chemiluminescence resonance energy transfer (cret) detection of DNA, metal ions, and aptamer-substrate complexes using hemin/g-quadruplexes and cdse/zns quantum dots. J. Am. Chem. Soc. 2011, 133, 11597–11604. [Google Scholar] [CrossRef]
- Ma, D.L.; Wang, M.; He, B.; Yang, C.; Wang, W.; Leung, C.H. A luminescent cocaine detection platform using a split g-quadruplex-selective iridium(iii) complex and a three-way DNA junction architecture. ACS Appl. Mater. Interfaces 2015, 7, 19060–19067. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, A.H.; Wurm, J.P.; Immer, C.; Weickhmann, A.K.; Wohnert, J. An intermolecular g-quadruplex as the basis for gtp recognition in the class v-gtp aptamer. RNA 2016, 22, 1750–1759. [Google Scholar] [CrossRef]
- Merkina, E.E.; Fox, K.R. Kinetic stability of intermolecular DNA quadruplexes. Biophys. J. 2005, 89, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Diveshkumar, K.V.; Sakrikar, S.; Harikrishna, S.; Dhamodharan, V.; Pradeepkumar, P.I. Targeting promoter g-quadruplex dnas by indenopyrimidine-based ligands. ChemMedChem 2014, 9, 2754–2765. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Laxmi-Reddy, K.; Jena, P.V.; Baptiste, B.; Dong, Z.; Godde, F.; Ha, T.; Rodriguez, R.; Balasubramanian, S.; Huc, I. Targeting DNA g-quadruplexes with helical small molecules. Chembiochem 2014, 15, 2563–2570. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, S.A.; Neidle, S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Nakamura, T.; Yoshida, W.; Tera, M.; Nakabayashi, K.; Hata, K.; Ikebukuro, K.; Nagasawa, K. Fluorescent-ligand-mediated screening of g-quadruplex structures using a DNA microarray. Angew Chem. Int. Ed. Engl. 2013, 52, 12052–12055. [Google Scholar] [CrossRef]
- Largy, E.; Granzhan, A.; Hamon, F.; Verga, D.; Teulade-Fichou, M.P. Visualizing the quadruplex: From fluorescent ligands to light-up probes. Top. Curr. Chem. 2013, 330, 111–177. [Google Scholar] [PubMed]
- Verga, D.; Hamon, F.; Poyer, F.; Bombard, S.; Teulade-Fichou, M.P. Photo-cross-linking probes for trapping g-quadruplex DNA. Angew Chem. Int. Ed. Engl. 2014, 53, 994–998. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Tera, M.; Hirokawa, T.; Shin-ya, K.; Nagasawa, K. G-quadruplex recognition by macrocyclic hexaoxazole (6otd) dimer: Greater selectivity than monomer. Chem. Commun. (Camb.) 2009, 6481–6483. [Google Scholar] [CrossRef] [PubMed]
- Marchand, A.; Gabelica, V. Native electrospray mass spectrometry of DNA g-quadruplexes in potassium solution. J. Am. Soc. Mass. Spectrom. 2014, 25, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, S.; Aznauryan, M.; Haustrup, E.K.; Schiott, B.; Birkedal, V.; Corry, B. Dynamics of fluorescent dyes attached to g-quadruplex DNA and their effect on fret experiments. Chemphyschem 2015. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available either commercially (PDS and Phen-DC3) or (L2H2-OTD) from the authors. |
| Construct | Sequence (5′–3′) |
|---|---|
| Stem | Cy5-GCCTCGCTGCCGTCGCCA-Biotin |
| pd-S3 | TGGCGACGGCAGCGAGG AGGGTTAGGGTTAGGGTTAG-Cy3 + Stem |
| pd-S2 | TGG CGA CGG CAG CGA GGC AGGGTTAGGGTTAG-Cy3 + Stem |
| pd-21T | TGG CGA CGG CAG CGA GGC TTTTTTTTTTTTTTTTTTTTT-Cy3 + Stem |
| S1 | AGGGT |
| S2 | GGGTTAGGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gyawali, P.; GC, K.; Ma, Y.; Abeysirigunawardena, S.; Nagasawa, K.; Balci, H. Impact of Small Molecules on Intermolecular G-Quadruplex Formation. Molecules 2019, 24, 1570. https://doi.org/10.3390/molecules24081570
Gyawali P, GC K, Ma Y, Abeysirigunawardena S, Nagasawa K, Balci H. Impact of Small Molecules on Intermolecular G-Quadruplex Formation. Molecules. 2019; 24(8):1570. https://doi.org/10.3390/molecules24081570
Chicago/Turabian StyleGyawali, Prabesh, Keshav GC, Yue Ma, Sanjaya Abeysirigunawardena, Kazuo Nagasawa, and Hamza Balci. 2019. "Impact of Small Molecules on Intermolecular G-Quadruplex Formation" Molecules 24, no. 8: 1570. https://doi.org/10.3390/molecules24081570
APA StyleGyawali, P., GC, K., Ma, Y., Abeysirigunawardena, S., Nagasawa, K., & Balci, H. (2019). Impact of Small Molecules on Intermolecular G-Quadruplex Formation. Molecules, 24(8), 1570. https://doi.org/10.3390/molecules24081570






