Phenylalkanoid Glycosides (Non-Salicinoids) from Wood Chips of Salix triandra × dasyclados Hybrid Willow
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material and Metabolite Extraction
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skvortsov, A.K. Willows of Russia and Adjacent Countries: Taxonomical and Geographical Revision; University of Joensuu: Joensuu, Finland, 1999; ISBN 9517087667. [Google Scholar]
- Hedner, T.; Everts, B. The early clinical history of salicylates in rheumatology and pain. Clin. Rheumatol. 1998, 17, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Boeckler, G.A.; Gershenzon, J.; Unsicker, S.B. Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 2011, 72, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Lavola, A.; Maukonen, M.; Julkunen-Tiitto, R. Variability in the composition of phenolic compounds in winter-dormant Salix pyrolifolia in relation to plant part and age. Phytochemistry 2018, 153, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Brereton, N.J.B.; Berthod, N.; Lafleur, B.; Pedneault, K.; Pitre, F.E.; Labrecque, M. Extractable phenolic yield variation in five cultivars of mature short rotation coppice willow from four plantations in Quebec. Ind. Crops Prod. 2017, 97, 525–535. [Google Scholar] [CrossRef]
- Jaggi, J.; Haslam, E. Phenols in Salix species. Phytochemistry 1969, 8, 635–636. [Google Scholar] [CrossRef]
- Thieme, H. Zur Konstitution des Salidrosids, eines Phenolglykosids aus Salix triandra L. Naturwissenshaften 1960, 51, 360. [Google Scholar] [CrossRef]
- Wu, Y.; Dobermann, D.; Beale, M.H.; Ward, J.L. Acutifoliside, a novel benzoic acid glycoside from Salix acutifolia. Nat. Prod. Res. 2016, 30, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Noleto-Dias, C.; Ward, J.L.; Bellisai, A.; Lomax, C.; Beale, M.H. Salicin-7-sulfate: A new salicinoid from willow and implications for herbal medicine. Fitoterapia 2018, 127, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Corol, D.I.; Harflett, C.; Beale, M.H.; Ward, J.L. An efficient high throughput metabotyping platform for screening of biomass willows. Metabolites 2014, 4, 946–976. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.; Zhang, X.; Wang, L.; Zheng, Y.; Yuan, C.; Sun, G. Isolation and characterization of phenolic compounds from the leaves of Salix matsudana. Molecules 2008, 13, 1530–1537. [Google Scholar] [CrossRef]
- Sulima, P.; Krauze-Baranowska, M.; Przyborowski, J.A. Variations in the chemical composition and content of salicylic glycosides in the bark of Salix purpurea from natural locations and their significance for breeding. Fitoterapia 2017, 118, 118–125. [Google Scholar] [CrossRef]
- Binns, W.W.; Blunden, G. Effects of hybridization on leaf constituents in the genus Salix. Phytochemistry 1969, 8, 1235–1239. [Google Scholar] [CrossRef]
- Baderschneider, B.; Winterhalter, P. Isolation and characterization of novel benzoates, cinnamates, flavonoids, and lignans from Riesling wine and screening for antioxidant activity. J. Agric. Food Chem. 2001, 49, 2788–2798. [Google Scholar] [CrossRef] [PubMed]
- Zapesochnaya, G.G.; Kurkin, V.A.; Braslavskii, V.B.; Filatova, N.V. Phenolic compounds of Salix acutifolia bark. Chem. Nat. Compd. 2002, 38, 314–318. [Google Scholar] [CrossRef]
- Corradi, E.; Schmidt, N.; Räber, N.; De Mieri, M.; Hamburger, M.; Butterweck, V.; Potterat, O. Metabolite profile and antiproliferative effects in HaCaT cells of a Salix reticulata Extract. Planta Med. 2017, 83, 1149–1158. [Google Scholar] [CrossRef]
- Gao, L.; Xu, X.; Yang, J. Chemical constituents of the roots of Rheum officinale. Chem. Nat. Compd. 2013, 49, 603–605. [Google Scholar] [CrossRef]
- Coen, M.; Engel, R.; Nahrstedt, A. Chavicol β-D-glucoside, a phenylpropanoid heteroside, benzyl-β-D-glucoside and glycosidically bound volatiles from subspecies of Cedronella canariensis. Phytochemistry 1995, 40, 149–155. [Google Scholar] [CrossRef]
- Ly, T.N.; Yamauchi, R.; Shimoyamada, M.; Kato, K. Isolation and structural elucidation of some glycosides from the rhizomes of smaller Galanga (Alpinia officinarum Hance). J. Agric. Food Chem. 2002, 50, 4919–4924. [Google Scholar] [CrossRef] [PubMed]
- Mshvildadze, V.; Legault, J.; Lavoie, S.; Gauthier, C.; Pichette, A. Anticancer diarylheptanoid glycosides from the inner bark of Betula papyrifera. Phytochemistry 2007, 68, 2531–2536. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.K. NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Inagaki, J.; Watanabe, N.; Moon, J.-H.; Yagi, A.; Sakata, K.; Ina, K.; Luo, S. Glycosidic aroma precursors of 2–phenylethyl and benzyl alcohols from Jasminum sambac flowers. Biosci. Biotechnol. Biochem. 1995, 59, 738–739. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, K.H.; Lee, I.K.; Lee, K.H.; Choi, S.U.; Lee, K.R. A new flavonol glycoside from Hylomecon vernalis. Arch. Pharm. Res. 2012, 35, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Marchev, A.S.; Aneva, I.Y.; Koycheva, I.K.; Georgiev, M.I. Phytochemical variations of Rhodiola rosea L. wild-grown in Bulgaria. Phytochem. Lett. 2017, 20, 386–390. [Google Scholar] [CrossRef]
- Jossang, A.; Jossang, P.; Bodo, B. Cinnamrutinoses A and B, glycosides of Populus tremula. Phytochemistry 1994, 35, 547–549. [Google Scholar] [CrossRef]
- Nakanishi, T.; Iida, N.; Inatomi, Y.; Murata, H.; Inada, A.; Murata, J.; Lang, F.A.; Iinuma, M.; Tanaka, T. New neolignan and phenylpropanoid glycosides in Juniperus communis var. depressa. Heterocycles 2004, 63, 2573. [Google Scholar] [CrossRef]
- Jimenez, C.; Riguera, R. Phenylethanoid glycosides in plants: Structure and biological activity. Nat. Prod. Rep. 1994, 11, 591–606. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
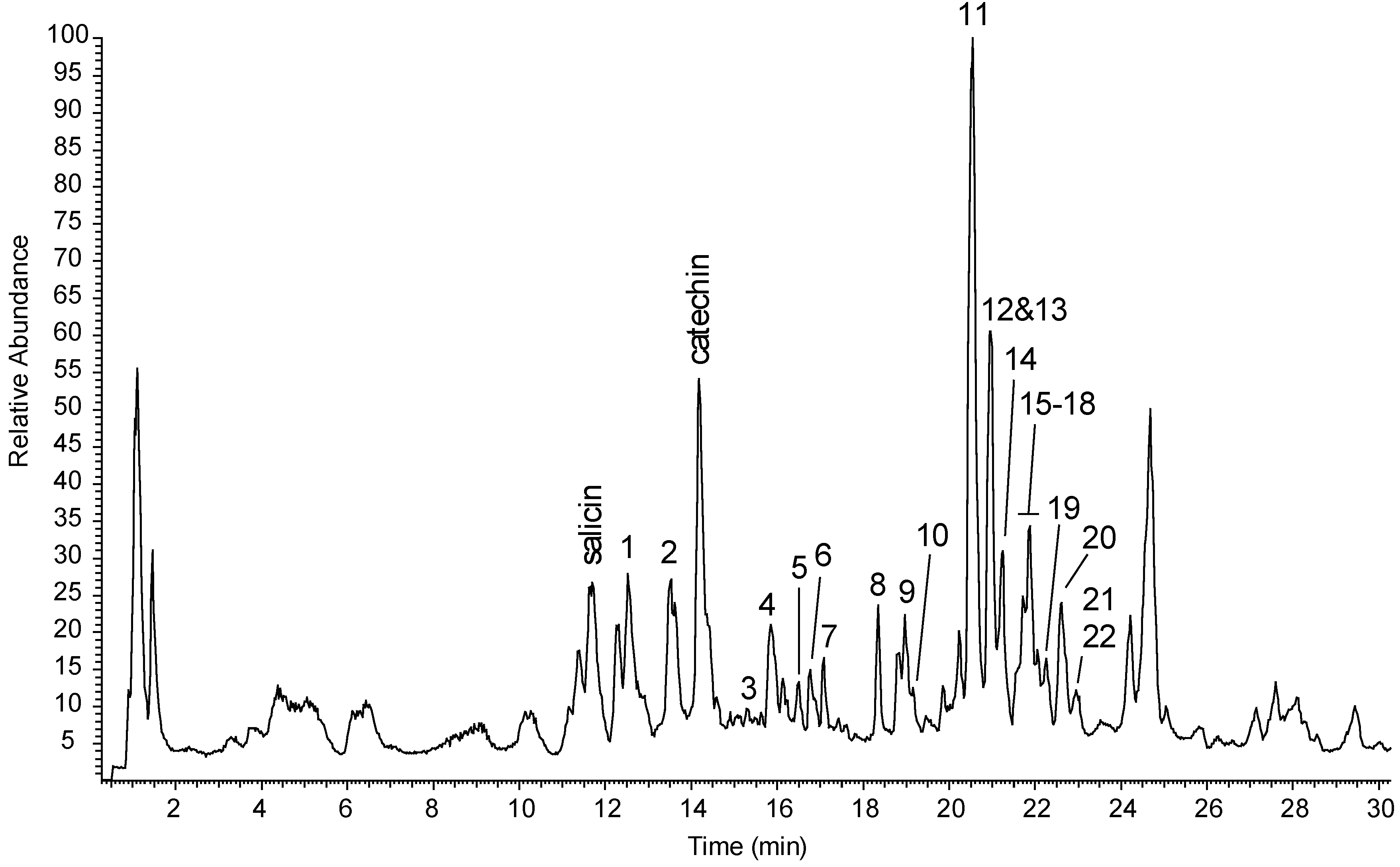
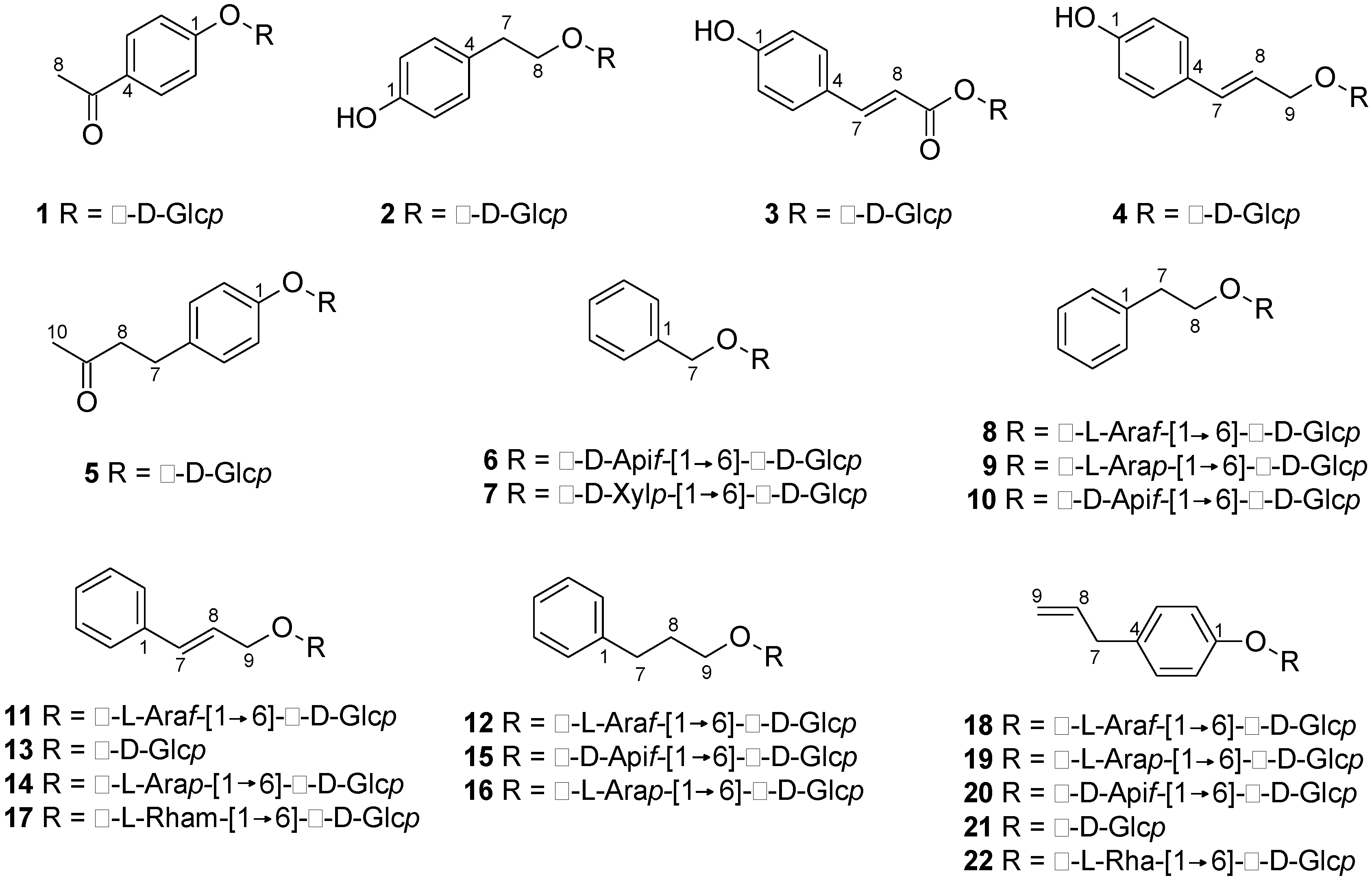
| No. | [M − H]− (m/z) | tR (min) | Formula | Δ (ppm) | MS/MS Ions (m/z) | Compound |
|---|---|---|---|---|---|---|
| 1 | 297.0981 | 12.6 | C14H18O7 | +0.55 | 135, 181 | Picein a |
| 2 | 299.1136 | 13.4 | C14H20O7 | −0.03 | 119, 137, 179 | Salidroside a |
| 3 | 325.0930 | 15.2 | C15H18O8 | +0.29 | 89, 119, 145, 163, 193 | p-Coumaroyl-β-d-glcpb |
| 4 | 311.1133 | 15.8 | C15H20O7 | −1.11 | 149, 161 | Triandrin b |
| 5 | 325.1289 | 16.3 | C16H22O7 | +0.47 | 163 | p-Hydroxybenzylacetone-β-d-glcpb |
| 6 | 401.1453 | 16.7 | C18H26O10 | +0.01 | 161, 269 | Benzyl-β-d-apif-(1→6)-β-d-glcp b |
| 7 | 401.1454 | 17.1 | C18H26O10 | +0.31 | 161, 269 | Benzyl-β-d-xylp-(1→6)-β-d-glcp b |
| 8 | 415.1611 | 18.3 | C19H28O10 | +0.31 | 179, 191, 283 | 2-phenylethyl-α-l-araf-(1→6)-β-d-glcp |
| 9 | 415.1612 | 18.9 | C19H28O10 | +0.46 | 149, 179, 191, 283 | 2-Phenylethyl-α-l-arap-(1→6)-β-d-glcp b |
| 10 | 415.1610 | 19.0 | C19H28O10 | +0.07 | 89, 149, 191, 283 | 2-Phenylethyl-β-d-apif-(1→6)-β-d-glcp b |
| 11 | 427.1611 | 20.5 | C20H28O10 | +0.24 | 125, 133, 149, 161, 191, 293 | Rosarin a |
| 12 | 429.1768 | 20.9 | C20H30O10 | +0.32 | 101, 131, 161, 297 | Dihydrorosarin b |
| 13 | 341.1242 * | 21.0 | C15H20O6 | +0.04 | 133, 161 | Rosin b |
| 14 | 427.1610 | 21.2 | C20H28O10 | +0.09 | 125, 133, 149, 161, 191, 233, 293 | Rosavin a |
| 15 | 429.1766 | 21.5 | C20H30O10 | +0.03 | 101, 131, 161, 297 | Phenylpropanol-β-d-apif-(1→6)-β-d-glcp c |
| 16 | 429.1767 | 21.6 | C20H30O10 | +0.11 | 101, 131, 161, 297 | Dihydrorosavin b |
| 17 | 441.1768 | 21.8 | C21H30O10 | +0.52 | 101, 125, 163, 247, 307 | Cinnamrutinose A b |
| 18 | 427.1611 | 21.9 | C20H28O10 | +0.38 | 89, 125, 133, 191, 233, 293 | Chavicol-α-l-araf-(1→6)-β-d-glcp b |
| 19 | 427.1610 | 22.1 | C20H28O10 | +0.02 | 89, 125, 133, 149, 191, 233, 293 | Chavicol-α-l-arap-(1→6)-β-d-glcp c |
| 20 | 427.1610 | 22.6 | C20H28O10 | +0.09 | 89, 125, 133, 149, 191, 233, 293 | Chavicol-β-d-apif-(1→6)-β-d-glcp b |
| 21 | 341.1243 * | 22.9 | C15H20O6 | +0.31 | 133, 161 | Chavicol-glucoside b |
| 22 | 441.1766 | 23.0 | C21H30O10 | −0.03 | 101, 125, 163, 247, 307 | Chavicol-rutinoside b |
| Position | (1) | (2) | (3) | (4) | (5) | (13) | (21) |
|---|---|---|---|---|---|---|---|
| 1 | − | − | − | − | − | − | − |
| 2 | 7.22 d (8.9) | 6.84 d (8.6) | 6.93 d (8.6) | 6.88 d (8.6) | 7.07 d (8.6) | 7.52 d (7.4) | 7.07 d (8.7) |
| 3 | 8.03 d (8.9) | 7.21 d (8.6) | 7.60 d (8.6) | 7.41 d (8.6) | 7.23 d (8.6) | 7.41 d (7.6) | 7.23 d (8.7) |
| 4 | − | − | − | − | − | 7.34 d (7.5) | − |
| 5 | 8.03 d (8.9) | 7.21 d (8.6) | 7.60 d (8.6) | 7.41 d (8.6) | 7.23 d (8.6) | 7.41 d (7.6) | 7.23 d (8.7) |
| 6 | 7.22 d (8.9) | 6.84 d (8.6) | 6.93 d (8.6) | 6.88 d (8.6) | 7.07 d (8.6) | 7.52 d (7.4) | 7.07 d (8.7) |
| 7 | − | 2.88 t (7.1) | 7.84 d (15.9) | 3.68 d (15.9) | 2.86 m | 6.77 d (16.3) | 3.36 d (6.8) |
| 8 | 2.64 s | 4.08 dt (7.1, 10.1) | 6.48 d (15.9) | 6.26 dt (15.9, 6.6) | 2.88 m | 6.42 m | 6.01 ddt (17, 10.1, 6.8) |
| 3.85 dt (7.3, 10.1) | |||||||
| 9 | − | − | − | 4.51 ddd (1.2, 6.2, 12.4) | − | 4.55 m, 4.41 m | 5.08 d (17.0) |
| 4.38 ddd (1.0, 7.0, 12.4) | |||||||
| 10 | − | − | − | − | 2.19 s | − | − |
| 1′ | 5.23 d (7.5) | 4.44 d (8.0) | 5.65 d (8.0) | 4.52 d (8.0) | 5.05 d (7.7) | 4.53 d (8.0) | 5.04 d (7.7) |
| 2′ | 3.60–3.62 m | 3.23 dd (8.0, 9.3) | 3.62–3.46 m | 3.29 dd (8.0, 9.3) | 3.53 d (7.8) | 3.49–3.39 m | 3.59–3.41 m |
| 3′ | 3.60–3.62 m | 3.45 d (9.0) | 3.62–3.46 m | 3.47 d (8.9) | 3.58 d (9.0) | 3.49–3.39 m | 3.59–3.41 m |
| 4′ | 3.52 d (9.7) | 3.39 d (8.9) | 3.62–3.46 m | 3.39 d (9.6) | 3.47 d (9.0) | 3.49–3.39 m | 3.59–3.41 m |
| 5′ | 3.67 ddd (2.2, 5.7, 9.8) | 3.40 ddd (2.2, 9.8, 6.7) | 3.62–3.46 m | 3.41 ddd (2.1, 5.7, 9.8) | 3.43 m | 3.49–3.39 m | 3.59–3.41 m |
| 6′ | 3.94 dd (2.2, 12.5) | 3.89 dd (2.2, 12.3) | 3.79 dd (2.0, 12.0) | 3.90 dd (2.1, 12.3) | 3.92 dd (2.1, 12.4) | 3.91 dd (2.1, 12.3) | 3.90 dd (2.3, 12.4) |
| 3.77 dd (5.7, 12.5) | 3.70 dd (5.7, 12.3) | 3.71 dd (5.2, 12.0) | 3.72 dd (5.8, 12.3) | 3.73 dd (5.8, 12.4) | 3.72 dd (6.0, 12.2) | 3.71 dd (5.6, 12.2) |
| Position | (17) | (18) | (20) | (22) |
|---|---|---|---|---|
| 2 | 7.50 d (7.3) | 7.07 d (8.7) | 7.07 d (8.6) | 7.07 d (8.6) |
| 3 | 7.40 d (7.6) | 7.22 d (8.7) | 7.23 d (8.6) | 7.23 d (8.6) |
| 4 | 7.33 d (7.4) | − | − | − |
| 5 | 7.40 d (7.6) | 7.22 d (8.7) | 7.23 d (8.6) | 7.23 d (8.6) |
| 6 | 7.50 d (7.3) | 7.07 d (8.7) | 7.07 d (8.6) | 7.07 d (8.6) |
| 7 | 6.74 d (16.0) | 3.36 d (6.7) | 3.36 d (6.9) | 3.36 d (6.9) |
| 8 | 6.39 dt (6.5, 16.0) | 6.01 ddt (16.9, 10.1, 6.7). | 6.01 ddt (16.9, 10.1, 6.7) | 6.01 ddt (16.9, 10.1, 6.7) |
| 9 | 4.50 ddd (12.7, 6.0, 1.2) | 5.07 m | 5.08 m | 5.08 m |
| 4.4 ddd (1.1, 6.8, 12.7) | ||||
| 1′ | 4.51 d (8.0) | 5.04 d (7.7) | 5.03 d (7.6) | 5.03 d (7.6) |
| 2′ | 3.46 d (9.1) | 3.54 d (7.7) | 3.53 d (7.6) | 3.55–3.40 m |
| 3′ | 3.45 d (9.1) | 3.50 d (9.1) | 3.56 d (8.9) | 3.55–3.40 m |
| 4′ | 3.41 dd (1.8, 9.6) | 3.57 d (9.0) | 3.48 d (9.2) | 3.55–3.40 m |
| 5′ | 3.53 ddd (1.8, 5.9, 9.6) | 3.73 m | 3.70 m | 3.55–3.40 m |
| 6′ | 3.97 dd (1.8, 11.6) 3.68 dd (5.9, 11.6) | 4.04 dd (1.3, 11.1) 3.70 m | 4.02 dd (2.3, 11.4) 3.72 m | 4.12 dd (1.7, 11.9) 3.74 m |
| 1″ | 4.52 d (1.7) | 5.00 d (1.3) | 5.05 d (3.1) | 5.07 d (1.8) |
| 2″ | 3.94 dd (1.8, 3.4) | 4.06 dd (1.5, 3.3) | 3.94 d (3.1) | 4.09 dd (1.6, 3.3) |
| 3″ | 3.77 dd (3.4, 9.7) | 3.89 dd (3.3, 5.9) | − | 3.75 m |
| 4″ | 3.42 d (9.6) | 4.00 td (3.3, 5.9) | 3.83 d (10.1) 3.99 d (10.1) | 3.40 m |
| 5″ | 3.72 dd (6.3, 9.6) | 3.74 dd (3.3, 12.3) 3.64 dd (5.7, 12.3) | 3.60 s | 3.72 dd (6.2, 9.3) |
| 6″ | 1.27 d (6.3) | − | − | 1.19 d (6.2) |
| Position | (6) | (7) | (8) | (9) | (10) |
|---|---|---|---|---|---|
| 2 | 7.47 dd (1.6, 8.2) | 7.46 m | 7.40–7.37 m | 7.39–7.36 m | 7.39–7.36 m |
| 3 | 7.44 dd (7.1, 1.6) | 7.44 dd (1.5, 7.6) | 7.37–7.33 m | 7.36–7.34 m | 7.36–7.34 m |
| 4 | 7.41 d (7.0) | 7.41 dd (1.6, 8.7) | 7.29 t (7.3) | 7.29 t (7.0) | 7.29 t (7.0) |
| 5 | 7.44 dd (7.1, 1.6) | 7.44 dd (1.5, 7.6) | 7.37–7.33 m | 7.36–7.34 m | 7.36–7.34 m |
| 6 | 7.47 dd (1.6, 8.2) | 7.46 m | 7.40–7.37 m | 7.39–7.36 m | 7.39–7.36 m |
| 7 | 4.91 d (11.7) | 4.94 d (11.7) | 2.97 t (7.0) | 2.97 t (7.0) | 2.97 t (7.0) |
| 4.75 d (11.7) | 4.75 d (11.7) | ||||
| 8 | − | − | 3.91 m | 3.91 m | 3.91 m |
| 4.12 dt (6.2, 9.0) | 4.13 m | 4.13 m | |||
| 1′ | 4.52 d (8.0) | 4.52 d (7.9) | 4.45 d (7.9) | 4.45 d (8.0) | 4.45 d (8.0) |
| 2′ | 3.28 m | 3.28 m | 3.25 d (9.0) | 3.23 d (8.0) | 3.23 d (8.0) |
| 3′ | 3.43 m | 3.43 m | 3.46 d (9.0) | 3.45 m | 3.45 m |
| 4′ | 3.46 d (9.0) | 3.46 m | 3.41 d (9.4) | 3.45 m | 3.45 m |
| 5′ | 3.53 ddd (2.0, 6.0, 9.0) | 3.59 m | 3.56 dd (2.0, 5.9) | 3.39 dd (2.0, 9.0) | 3.39 dd (2.0, 9.0) |
| 6′ | 4.02 dd (2.0, 11.6) 3.73 dd (6.0, 11.6) | 4.15 dd (1.9, 11.7), 3.85 dd (5.7, 11.7) | 3.68 dd (5.8, 12.1) 4.03 dd (2.0, 12.1) | 4.01 dd (1.9, 11.6), 3.70 dd (5.8, 11.5) | 4.01 dd (1.9, 11.6), 3.70 dd (5.8, 11.5) |
| 1″ | 5.11 d (3.2) | 4.45 d (7.8) | 5.05 d (1.4) | 4.44 d (8.0) | 5.09 d (3.2) |
| 2″ | 4.01 d (3.2) | 3.32 m | 4.10 dd (1.5, 3.3) | 3.30 m | 3.98 d (3.2) |
| 3″ | − | 3.42 m | 3.92 dd (3.3, 5.7) | 3.45 m | − |
| 4″ | 4.06 d (10.1) 3.89 d (10.1) | 3.59 m | 4.05 dd (3.3, 5.9) | 3.89 m | 4.03 d (10.1) 3.87 d (10.1) |
| 5″ | 3.66 s | 3.30 m, 3.95 dd (5.4, 11.6) | 3.80 dd (3.2, 12.2) 3.69 dd (5.0, 12.2) | 3.89 dd (2.2, 12.3) 3.69 dd (5.0, 12.3) | 3.63 d (1.7) |
| 6″ | − | − | − | − | − |
| (12) | (16) | |||||||
|---|---|---|---|---|---|---|---|---|
| Position | δC a | δH b | COSY | HMBC | δC a | δH b | COSY | HMBC |
| 1 | 145.3 | − | 145.3 | − | − | |||
| 2 | 131.6 | 7.30 d (7.0) | H-3/5, 4 | C-4 | 131.6 | 7.31 d (7.1) | H-3/5, 4 | C-3/5, 4, 7 |
| 3 | 131.5 | 7.35 d (7.5) | H-2/6, 4 | C-1 | 131.5 | 7.35 d (7.5) | H-2/6, 4 | C-1, 2/6 |
| 4 | 129.1 | 7.25 t (7.3) | H-2/6, 3/5 | C-2/6 | 129.1 | 7.25 t (7.3) | H-2/6, 3/5 | C-2/6 |
| 5 | 131.5 | 7.35 d (7.5) | H-2/6, 4 | C-1 | 131.5 | 7.35 d (7.5) | H-2/6, 4 | C-1, 2/6 |
| 6 | 131.6 | 7.30 d (7.0) | H-3/5, 4 | C-4 | 131.6 | 7.31 d (7.1) | H-3/5, 4 | C-3/5, 4, 7 |
| 7 | 34.3 | 2.71 t (7.6) | H-8 | C-1, 2/6, 9 | 34.3 | 2.71 t (7.6) | H-8 | C-1, 2/6, 8, 9 |
| 8 | 33.8 | 1.93 dt (6.7, 13.7) | H-7, 9 | C-1, 9 | 33.8 | 1.93 dt (6.7, 13.7) | H-7, 9 | C-1, 7, 9 |
| 9 | 72.8 | 3.88 m | H-8 | 72.8 | 3.89 m | H-8 | ||
| 3.65 m | 3.65 m | |||||||
| 1′ | 105.5 | 4.41 d (7.9) | H-2′ | C-9 | 105.9 | 4.42 d (7.4) | H-2′ | C-9 |
| 2′ | 76.2 | 3.25 d (9.2) | H-1′, 3′ | C-3′, 4′ | 76.2 | 3.28 d (9.0) | H-1′, 3′ | C-1′, 3′, 4′ |
| 3′ | 78.9 | 3.45 m | H-2′ | C-2′, 4′ | 79 | 3.45 m | H-2′ | C-5′ |
| 4′ | 72.5 | 3.45 m | H-5′ | C-3′ | 72.5 | 3.45 m | H-5′ | C-5′ |
| 5′ | 72.7 | 3.41 dd (9.5, 2.5) | H4′ | C-2′, 3′, 4′, 6′ | 72.7 | 3.39 dd (9.0, 3.5) | H4′ | C-3′, 4′ |
| 6′ | 69.7 | 4.02 dd (11.5, 1.9) | C-4′, 5′ | 71.5 | 4.11 dd (1.9, 11.7) | C-1′ | ||
| 3.68 dd (11.5, 6.0) | 3.82 dd (5.5, 11.7) | |||||||
| 1″ | 111.2 | 5.04 d (1.3) | C-6′, 2″, C-3″ | 104.4 | 4.53 d (7.9) | C-2″ | ||
| 2″ | 87 | 4.02 dd (1.8, 3.6) | C-3″ | 72.1 | 3.58 dd (1.2, 5.4) | C-3″ | ||
| 3″ | 79.5 | 3.90 dd (6.2, 3.0) | C-4″ | 78.4 | 3.57 d (5.5) | H-4″ | C-2″ | |
| 4″ | 84 | 4.07 dd (1.6, 3.3) | C-3″ | 68.1 | 3.92 d (5.5) | H-3″, 5″ | C-2″, 3″ | |
| 5″ | 64.3 | 3.77 dd (12.3, 3.3) | C-3″ | 63.7 | 3.89 dd (12.3, 2.3) | H-4″ | C-3″ | |
| 3.66 dd (12.3, 5.0) | 3.71 dd (12.3, 5.8) | |||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noleto-Dias, C.; Wu, Y.; Bellisai, A.; Macalpine, W.; Beale, M.H.; Ward, J.L. Phenylalkanoid Glycosides (Non-Salicinoids) from Wood Chips of Salix triandra × dasyclados Hybrid Willow. Molecules 2019, 24, 1152. https://doi.org/10.3390/molecules24061152
Noleto-Dias C, Wu Y, Bellisai A, Macalpine W, Beale MH, Ward JL. Phenylalkanoid Glycosides (Non-Salicinoids) from Wood Chips of Salix triandra × dasyclados Hybrid Willow. Molecules. 2019; 24(6):1152. https://doi.org/10.3390/molecules24061152
Chicago/Turabian StyleNoleto-Dias, Clarice, Yanqi Wu, Alice Bellisai, William Macalpine, Michael H. Beale, and Jane L. Ward. 2019. "Phenylalkanoid Glycosides (Non-Salicinoids) from Wood Chips of Salix triandra × dasyclados Hybrid Willow" Molecules 24, no. 6: 1152. https://doi.org/10.3390/molecules24061152
APA StyleNoleto-Dias, C., Wu, Y., Bellisai, A., Macalpine, W., Beale, M. H., & Ward, J. L. (2019). Phenylalkanoid Glycosides (Non-Salicinoids) from Wood Chips of Salix triandra × dasyclados Hybrid Willow. Molecules, 24(6), 1152. https://doi.org/10.3390/molecules24061152








