The Polymorphisms of Oligonucleotide Probes in Wheat Cultivars Determined by ND-FISH
Abstract
1. Introduction
2. Results
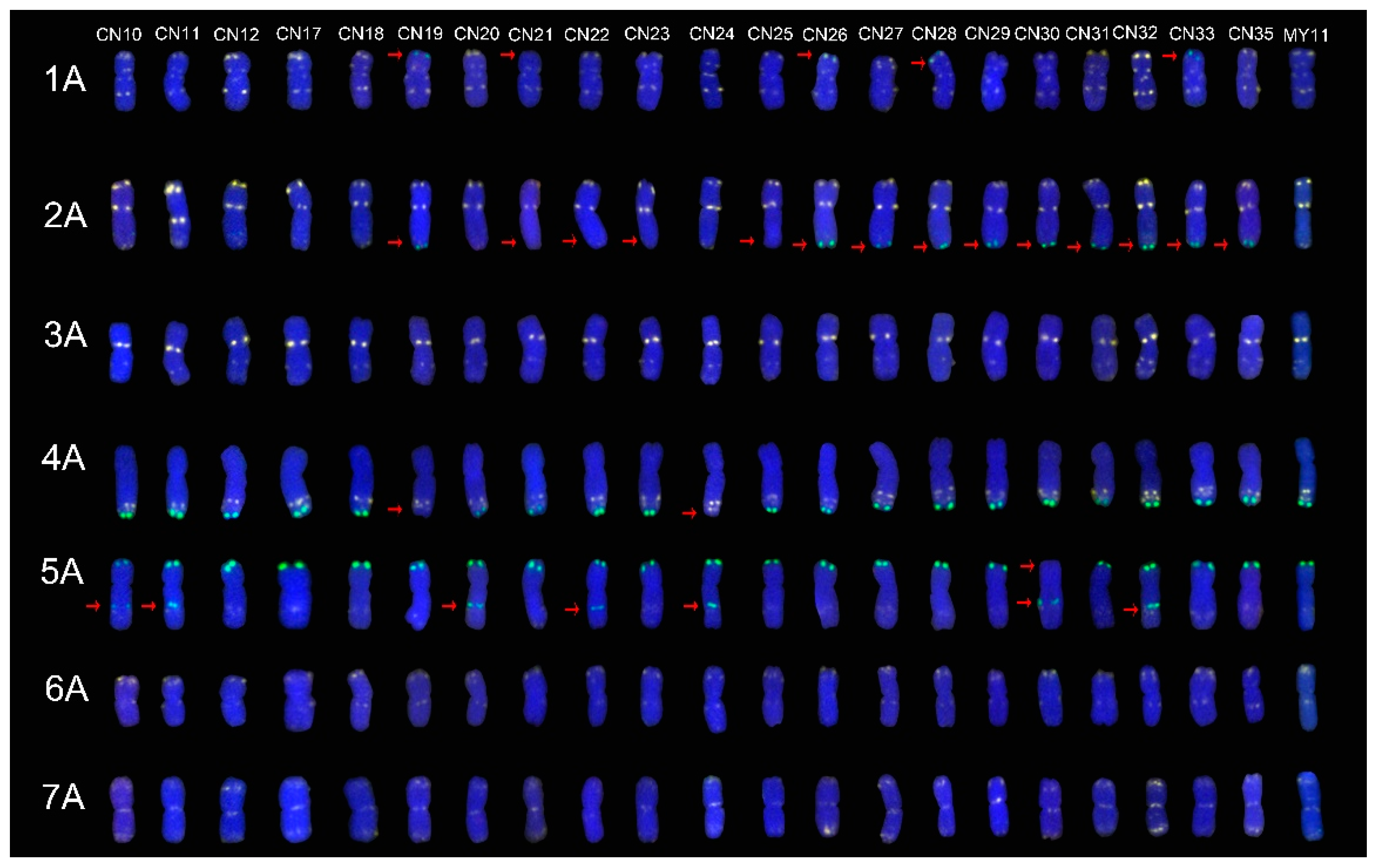
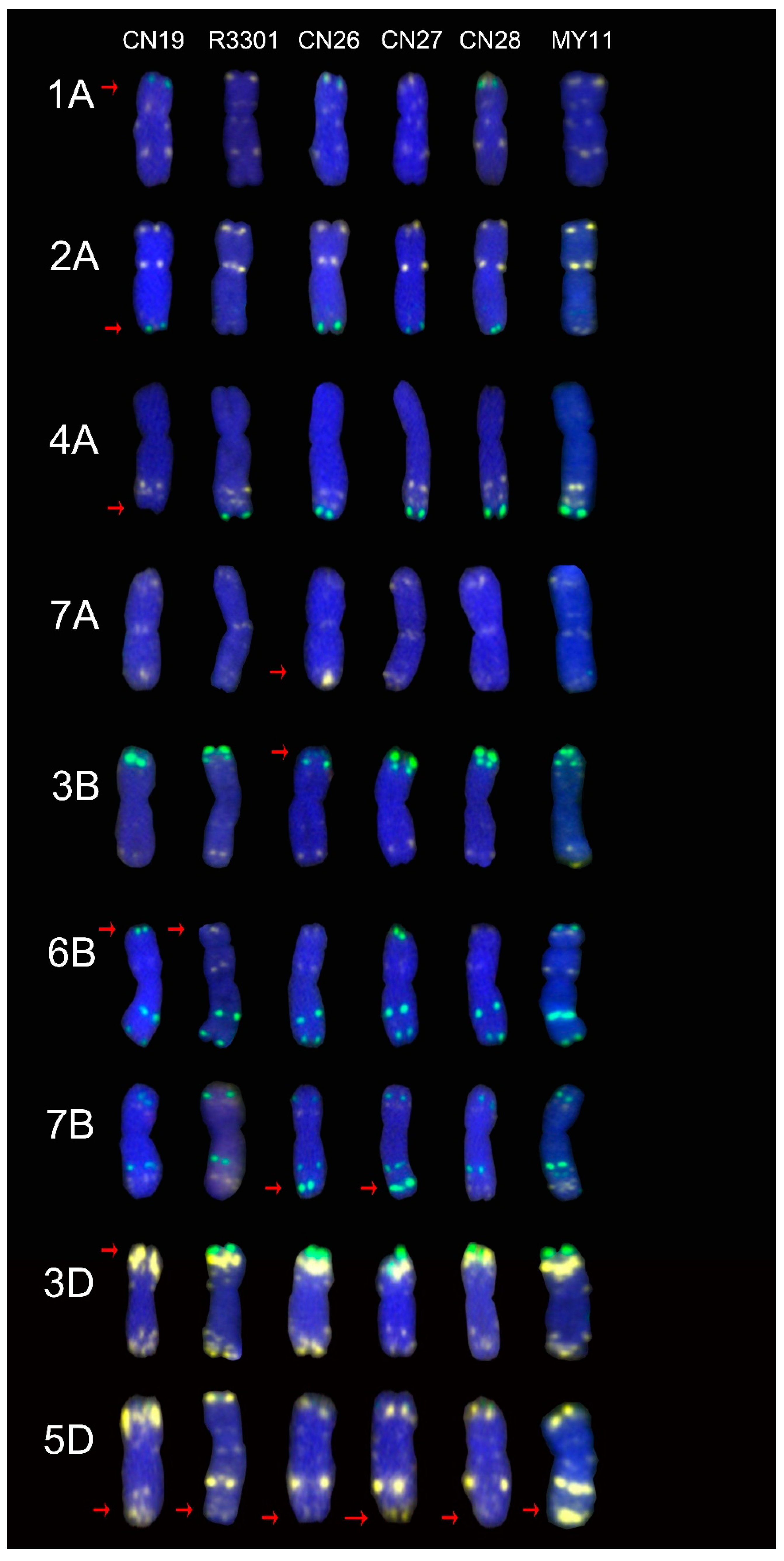
2.1. The ND-FISH Signals Indicate Polymorphisms in A-Genome Chromosomes
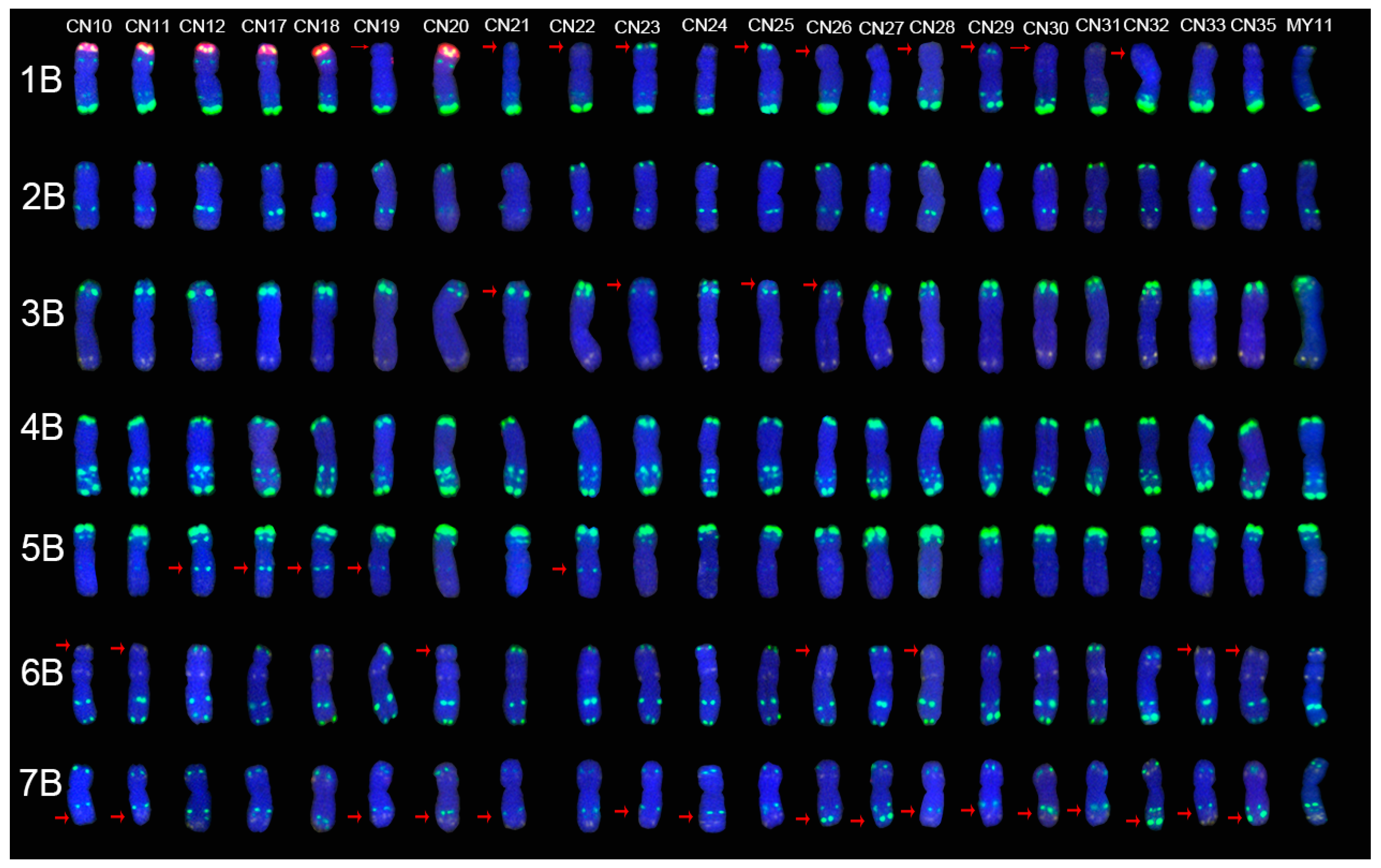
2.2. The ND-FISH Signals Indicate Polymorphisms in B-Genome Chromosomes
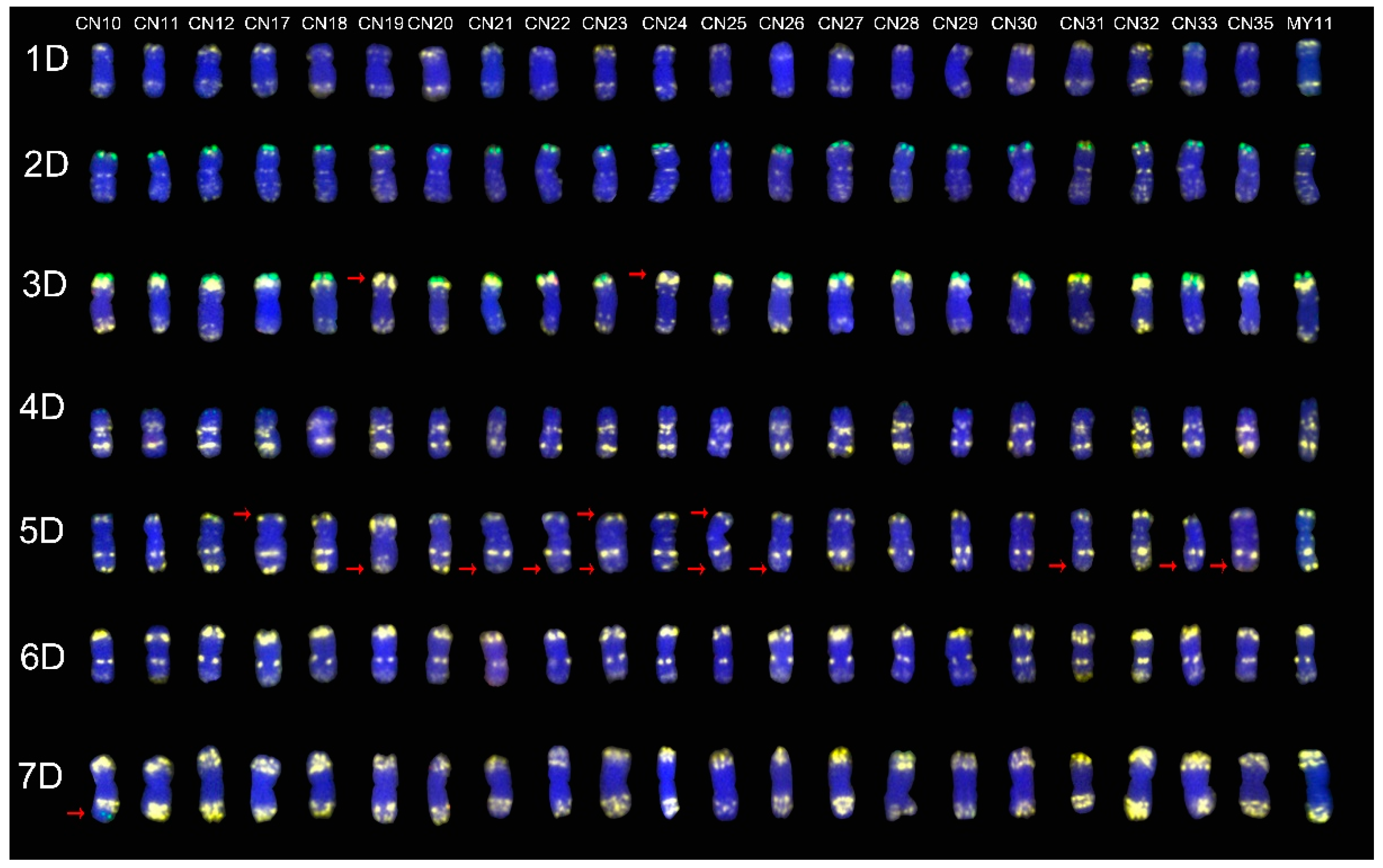
2.3. The ND-FISH Signals Indicate Polymorphisms in D-Genome Chromosomes
2.4. The Mutant Rate of Signal Patterns of ND-FISH
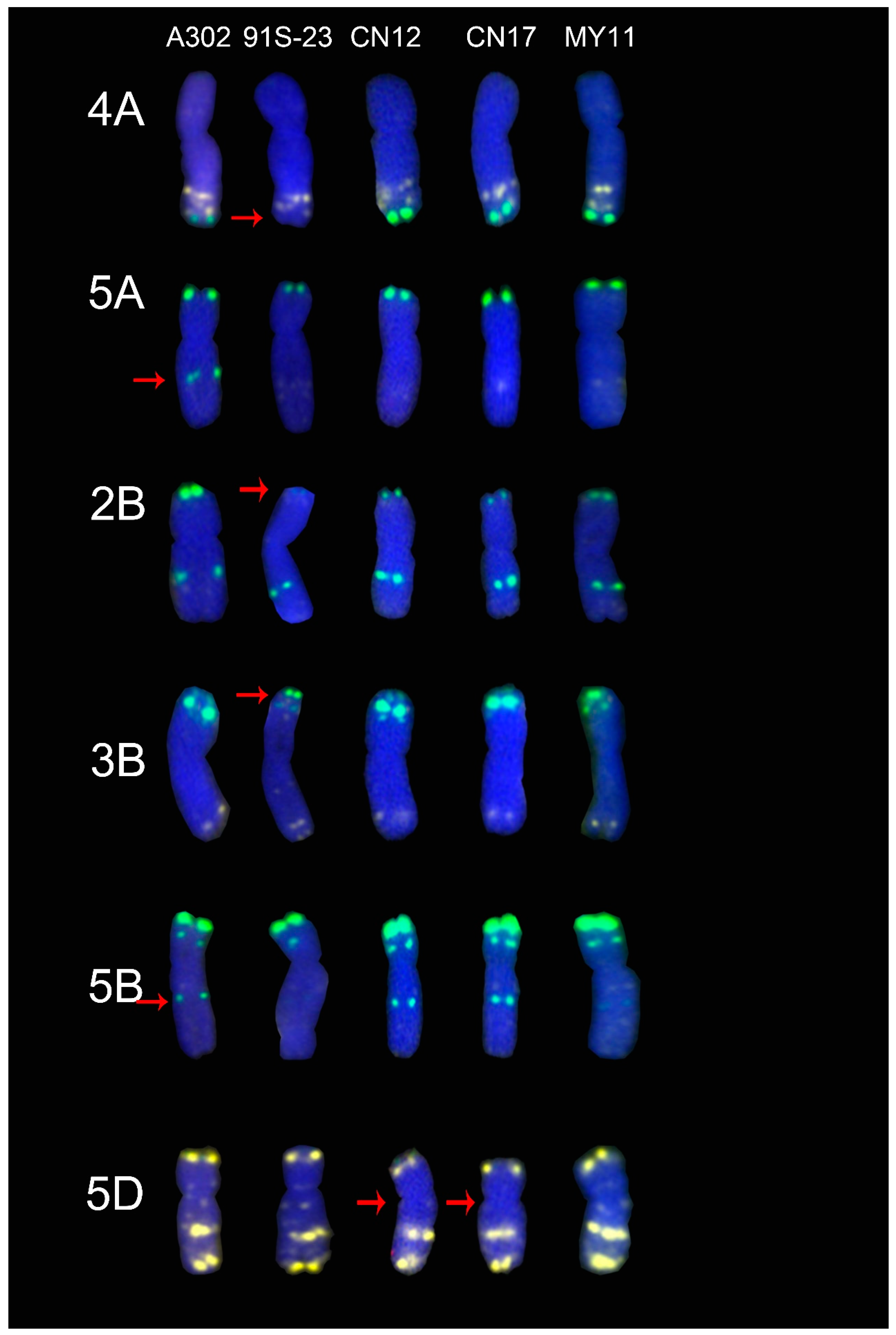
2.5. ND-FISH Signals of the Tested Cultivars Compared with Those of Their Parents
3. Discussion
3.1. The Chromosome Structure of “Chuan-nong” Series Cultivars
3.2. The Oligonucleotide Probes Can Replace the Use of Repetitive Sequence Probes
3.3. The Polymorphism Shown by ND-FISH Signal Patterns
3.4. The Sources of the ND-FISH Signal Pattern Polymorphisms
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of Chromosomes
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ekong, R.; Wolfe, J. Advances in fluorescent in situ hybridisation. Curr. Opin. Biotechnol. 1998, 9, 19–24. [Google Scholar] [CrossRef]
- Raap, A.K. Advances in fluorescence in situ hybridization. Mutat. Res. 1998, 400, 287–298. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Schwarzacher, T. The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 1998, 107, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, Á.; Jouve, N. Evolutionary trends of different repetitive DNA sequences during speciation in the genus secale. J. Hered. 2002, 93, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.H.; Tang, Z.X.; Fu, S.L.; Yan, B.J.; Tan, F.Q.; Ren, Z.L.; Li, Z. Molecular Cytogenetic Characterization of Novel Wheat-rye T1RS.1BL Translocation Lines with High Resistance to Diseases and Great Agronomic Traits. Front. Plant Sci. 2017, 8, 799. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Flavell, R.B. The relatedness and evolution of repeated nucleotide sequences in the genomes of some Gramineae species. Biochem. Genet. 1974, 12, 243–256. [Google Scholar] [CrossRef]
- Komissarov, A.S.; Gavrilova, E.V.; Demin, S.J.; Ishov, A.M.; Podgornaya, O.I. Tandemly repeated DNA families in the mouse genome. BMC Genom. 2011, 12, 531. [Google Scholar] [CrossRef]
- Sepsi, A.; Molnár, I.; Szalay, D.; Molnár-Láng, M. Characterization of a leaf rust-resistant wheat—Thinopyrum ponticum partial amphiploid BE-1, using sequential multicolor GISH and FISH. Theor. Appl. Genet. 2008, 116, 825–834. [Google Scholar] [CrossRef]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef]
- Ren, T.H.; Li, Z.; Yan, B.J.; Tan, F.Q.; Tang, Z.X.; Fu, S.L.; Yang, M.Y.; Ren, Z.L. De novo balanced complex chromosome rearrangements involving chromosomes 1B and 3B of wheat and 1R of rye. Genome 2016, 59, 1076–1084. [Google Scholar] [CrossRef]
- Cuadrado, A.; Golczyk, H.; Jouve, N. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosom. Res. 2009, 17, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, Á.; Jouve, N. Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 2010, 119, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.L.; Chen, L.; Wang, Y.Y.; Li, M.; Yang, Z.J.; Qiu, L.; Yan, B.J.; Ren, Z.L.; Tang, Z.X. Oligonucleotide Probes for ND-FISH Analysis to Identify Rye and Wheat Chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ren, Z.L.; Tan, F.Q.; Tang, Z.X.; Fu, S.L.; Yan, B.J.; Ren, T.H. Molecular Cytogenetic Characterization of New Wheat-Rye 1R (1B) Substitution and Translocation Lines from a Chinese Secale cereal L. Aigan with Resistance to Stripe Rust. PLoS ONE 2016, 11, e0163642. [Google Scholar] [CrossRef]
- Xiao, Z.Q.; Tang, S.Y.; Qiu, L.; Tang, Z.X.; Fu, S.L. Oligonucleotides and ND-FISH Displaying Different Arrangements of Tandem Repeats and Identification of Dasypyrum villosum Chromosomes in Wheat Backgrounds. Molecules 2017, 22, 973. [Google Scholar] [CrossRef]
- Tang, S.Y.; Tang, Z.X.; Qiu, L.; Yang, Z.J.; Li, G.R.; Lang, T.; Zhu, W.Q.; Zhang, J.H.; Fu, S.L. Developing New Oligo Probes to Distinguish Specific Chromosomal Segments and the A, B, D Genomes of Wheat (Triticum aestivum L.) Using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef]
- Contento, A.; Heslop-Harrison, J.S.; Schwarzacher, T. Diversity of a major repetitive DNA sequence in diploid and polyploidy Triticeae. Cytogenet. Genome Res. 2005, 109, 34–42. [Google Scholar] [CrossRef]
- Liu, C.; Li, G.R.; Gong, W.P.; Li, G.Y.; Han, R.; Li, H.S.; Song, J.M.; Liu, A.F.; Cao, X.Y.; Chu, X.S.; et al. Molecular and cytogenetic characterization of a powdery mildew-resistant wheat-aegilops mutica partial amphiploid and addition line. Cytogenet. Genome Res. 2016, 147, 186–194. [Google Scholar] [CrossRef]
- Delgado, A.; Carvalho, A.; Martín, A.C.; Martín, A.; Lima-Brito, J. Use of the synthetic oligo-pTa535 and oligo-pAs1 probes for identification of Hordeum chilense-origin chromosomes in hexaploid tritordeum. Genet. Resour. Crop Evol. 2016, 63, 945–951. [Google Scholar] [CrossRef]
- Ren, T.H.; Ren, Z.L.; Yang, M.Y.; Yan, B.J.; Tan, F.Q.; Fu, S.L.; Tang, Z.X.; Li, Z. Novel source of 1RS from Baili rye conferred high resistance to diseases and enhanced yield traits to common wheat. Mol. Breed. 2018, 38, 101. [Google Scholar] [CrossRef]
- Tang, S.Y.; Qiu, L.; Xiao, Z.Q.; Fu, S.L.; Tang, Z.X. New Oligonucleotide Probes for ND-FISH Analysis to Identify Barley Chromosomes and to Investigate Polymorphisms of Wheat Chromosomes. Genes 2016, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A. Origin and evolution of wheat and related triticeae species. In Alien Introgression in Wheat; Springer International Publishing: Berlin, Germany, 2015; pp. 21–76. [Google Scholar]
- Ren, T.H.; Yang, Z.J.; Yan, B.J.; Zhang, H.Q.; Fu, S.L.; Ren, Z.L. Development and characterization of a new 1BL.1RS translocation line with resistance to stripe rust and powdery mildew of wheat. Euphytica 2009, 169, 207–213. [Google Scholar] [CrossRef]
- Ren, T.H.; Chen, F.; Zhang, H.Y.; Yan, B.J.; Ren, Z.L. Application of 1RS.1BL Translocation in the Breeding of “Chuannong” Series Wheat Cultivars. J. Triticeae Crops 2011, 31, 430–436. [Google Scholar]
- Shewry, P.R.; Parmar, S.; Fulrath, N.; Kasarda, D.D.; Miller, T.E. Chromosomal locations of the structural genes for secalins in wild perennial rye (Secale montanum Guss.) and cultivated rye (S. cereal L.) determined by two-dimensional electrophoresis. Can. J. Genet. Cytol. 1986, 28, 76–83. [Google Scholar] [CrossRef]
- Moreno-Sevilla, B.; Baenziger, P.S.; Shelton, D.R.; Graybosch, R.A.; Peterson, C.J. Agronomic performance and end-use quality of 1B vs. 1BL/1RS genotypes derived from winter wheat ‘Rawhide’. Crop Sci. 1995, 35, 1607–1612. [Google Scholar] [CrossRef]
- Graybosch, R.A. Uneasy unions: Quality effects of rye chromatin transfers to wheat. J. Cereal Sci. 2001, 33, 3–16. [Google Scholar] [CrossRef]
- Schneider, A.; Linc, G.; Molnár-Láng, M.; Graner, A. Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed. 2003, 122, 396–400. [Google Scholar] [CrossRef]
- Mirzaghaderi, G.; Houben, A.; Badaeva, E.D. Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol. Cytogenet. 2014, 7, 91. [Google Scholar] [CrossRef]
- Mcintyre, C.L.; Pereira, S.; Moran, L.B.; Appels, R. New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 1990, 33, 635–640. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Vitellozzi, F.; Jouve, N.; Ceoloni, C. Fluorescence in situ hybridization with multiple repeated DNA probes applied to the analysis of wheat-rye chromosome pairing. Theor. Appl. Genet. 1997, 94, 347–355. [Google Scholar] [CrossRef]
- Yang, X.F.; Wang, C.Y.; Li, X.; Chen, C.H.; Tian, Z.R.; Wang, Y.J.; Ji, W.Q. Development and molecular cytogenetic identification of a novel wheat-leymus mollis Lm#7Ns (7D) disomic substitution line with stripe rust resistance. PLoS ONE 2015, 10, e0140227. [Google Scholar]
- Jiang, M.; Xiao, Z.Q.; Fu, S.L.; Tang, Z.X. FISH Karyotype of 85 Common Wheat Cultivars/Lines Displayed by ND-FISH Using Oligonucleotide Probes. Cereal Res. Commun. 2017, 45, 549–563. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A.; Fahima, T.; Korol, A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J Exp Bot. 2012, 63, 5045–5059. [Google Scholar] [CrossRef]
- Tang, Z.X.; Fu, S.X.; Ren, Z.L.; Zou, Y.T. Rapid evolution of simple sequence repeat induced by allopolyploidization. J. Mol. Evol. 2009, 69, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Szakács, E.; Molnár-Láng, M. Molecular cytogenetic evaluation of chromosome instability in Triticum aestivum-secale cereale disomic addition lines. J. Appl. Genet. 2010, 51, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.L.; Yang, M.Y.; Ren, Z.L.; Yan, B.J.; Tang, Z.X.; Gustafson, J.P. Abnormal mitosis induced by wheat-rye 1R monosomic addition lines. Genome 2014, 57, 21. [Google Scholar] [CrossRef]
- Luo, P.G.; Zhang, H.Y.; Shu, K.; Wu, X.H.; Zhang, H.Q.; Ren, Z.L. The physiological genetic effects of 1BL/1RS translocated chromosome in “stay green” wheat cultivar CN17. Can. J. Plant Sci. 2009, 89, 1–10. [Google Scholar] [CrossRef]
- Hu, Y.S.; Ren, T.H.; Li, Z.; Tang, Y.Z.; Ren, Z.L.; Yan, B.J. Molecular mapping and genetic analysis of a QTL controlling spike formation rate and tiller number in wheat. Gene 2017, 634, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; He, Z.C.; Zhang, H.Q.; Tan, F.Q.; Jiang, H.R. A New Record on Wheat Yield in Chengdu Plain with the Consonance-type Cultivar Chuan-nong17. J. Sichuan Agric. Univ. 2003, 21, 85–87. [Google Scholar]
- Luo, P.G.; Hu, X.Y.; Ren, Z.L.; Zhang, H.Y.; Yang, Z.J. Allelic analysis of stripe rust resistance genes on wheat chromosome 2BS. Genome 2008, 51, 922–927. [Google Scholar] [CrossRef]
- Luo, P.G.; Luo, H.Y.; Chang, Z.J.; Zhang, H.Y.; Zhang, M.; Ren, Z.L. Characterization and chromosomal location of Pm40 in common wheat: A new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor. Appl Genet. 2009, 118, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
| Cultivars | Cross Combinations | Released Time |
|---|---|---|
| Chuan-nong10 (CN10) | 78-5038 a × 85DH5015 | 2003 |
| Chuan-nong11 (CN11) | 78-5038 a × 85DH5015 | 2001 |
| Chuan-nong12 (CN12) | 91S-23 b × A302 | 2002 |
| Chuan-nong17 (CN17) | 91S-23 b × A302 | 2002 |
| Chuan-nong18 (CN18) | R164-1 b × A302 | 2003 |
| Chuan-nong19 (CN19) | Qian1104A × R935 b | 2003 |
| Chuan-nong20 (CN20) | 78-5038 a × 85DH5015 | 2003 |
| Chuan-nong21 (CN21) | R841 b × Qianhui3 | 2004 |
| Chuan-nong22 (CN22) | R164 b × 86-104 | 2005 |
| Chuan-nong23 (CN23) | R1685 b × MY26 | 2005 |
| Chuan-nong24 (CN24) | CN10 a × Yunfan52894-2 | 2007 |
| Chuan-nong25 (CN25) | 96 I-225 × 91S-5-4 b | 2007 |
| Chuan-nong26 (CN26) | CN19 × R3301 b | 2006 |
| Chuan-nong27 (CN27) | CN19 × R3301 b | 2009 |
| Chuan-nong28 (CN28) | CN19 × R3301 b | 2011 |
| Chuan-nong29 (CN29) | (02017 × CN19) × R131 | 2015 |
| Chuan-nong30 (CN30) | (03FR1349-1 × 54789) × CN27 | 2016 |
| Chuan-nong31 (CN31) | Not release | |
| Chuan-nong32 (CN32) | CN27 × 80978 | 2017 |
| Chuan-nong33(CN33) | Not release | |
| Chuan-nong35 (CN35) | Not release | |
| Mianyang11(MY11) | control |
| Probes | Sequences (5′-3′) |
|---|---|
| Oligo-pSc119.2-1 | CCGTTTTGTGGACTATTACTCACCGCTTTGGGGTCCCATAGCTAT |
| Oligo-pTa535-1 | AAAAACTTGACGCACGTCACGTACAAATTGGACAAACTCTTTCGGAGTATCAGGGTTTC |
| Oligo-Ku | GATCGAGACTTCTAGCAATAGGCAAAAATAGTAATGGTATCCGGGTTCG |
| Oligo-pSc200 | CTCACTTGCTTTGAGAGTCTCGATCAATTCGGACTCTAGGTTGATTTTTGTATTTTCT |
| Oligo-pSc250 | TGTGTTGTTCTTGGACAAAACAATGCATACCATCTCTTCTAC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, T.; He, M.; Sun, Z.; Tan, F.; Luo, P.; Tang, Z.; Fu, S.; Yan, B.; Ren, Z.; Li, Z. The Polymorphisms of Oligonucleotide Probes in Wheat Cultivars Determined by ND-FISH. Molecules 2019, 24, 1126. https://doi.org/10.3390/molecules24061126
Ren T, He M, Sun Z, Tan F, Luo P, Tang Z, Fu S, Yan B, Ren Z, Li Z. The Polymorphisms of Oligonucleotide Probes in Wheat Cultivars Determined by ND-FISH. Molecules. 2019; 24(6):1126. https://doi.org/10.3390/molecules24061126
Chicago/Turabian StyleRen, Tianheng, Maojie He, Zixin Sun, Feiquan Tan, Peigao Luo, Zongxiang Tang, Shulan Fu, Benju Yan, Zhenglong Ren, and Zhi Li. 2019. "The Polymorphisms of Oligonucleotide Probes in Wheat Cultivars Determined by ND-FISH" Molecules 24, no. 6: 1126. https://doi.org/10.3390/molecules24061126
APA StyleRen, T., He, M., Sun, Z., Tan, F., Luo, P., Tang, Z., Fu, S., Yan, B., Ren, Z., & Li, Z. (2019). The Polymorphisms of Oligonucleotide Probes in Wheat Cultivars Determined by ND-FISH. Molecules, 24(6), 1126. https://doi.org/10.3390/molecules24061126





