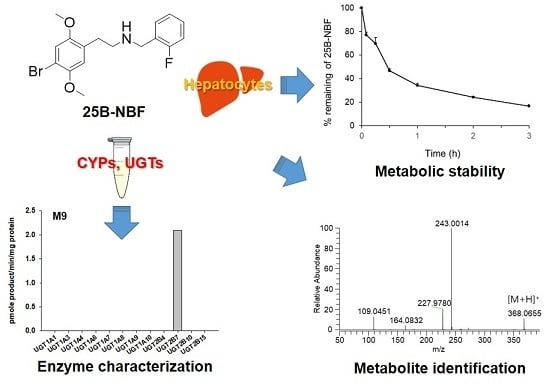
In Vitro Metabolism of 25B-NBF, 2-(4-Bromo-2,5-Dimethoxyphenyl)-N-(2-Fluorobenzyl)ethanamine, in Human Hepatocytes Using Liquid Chromatography–Mass Spectrometry
Abstract
:1. Introduction
2. Results
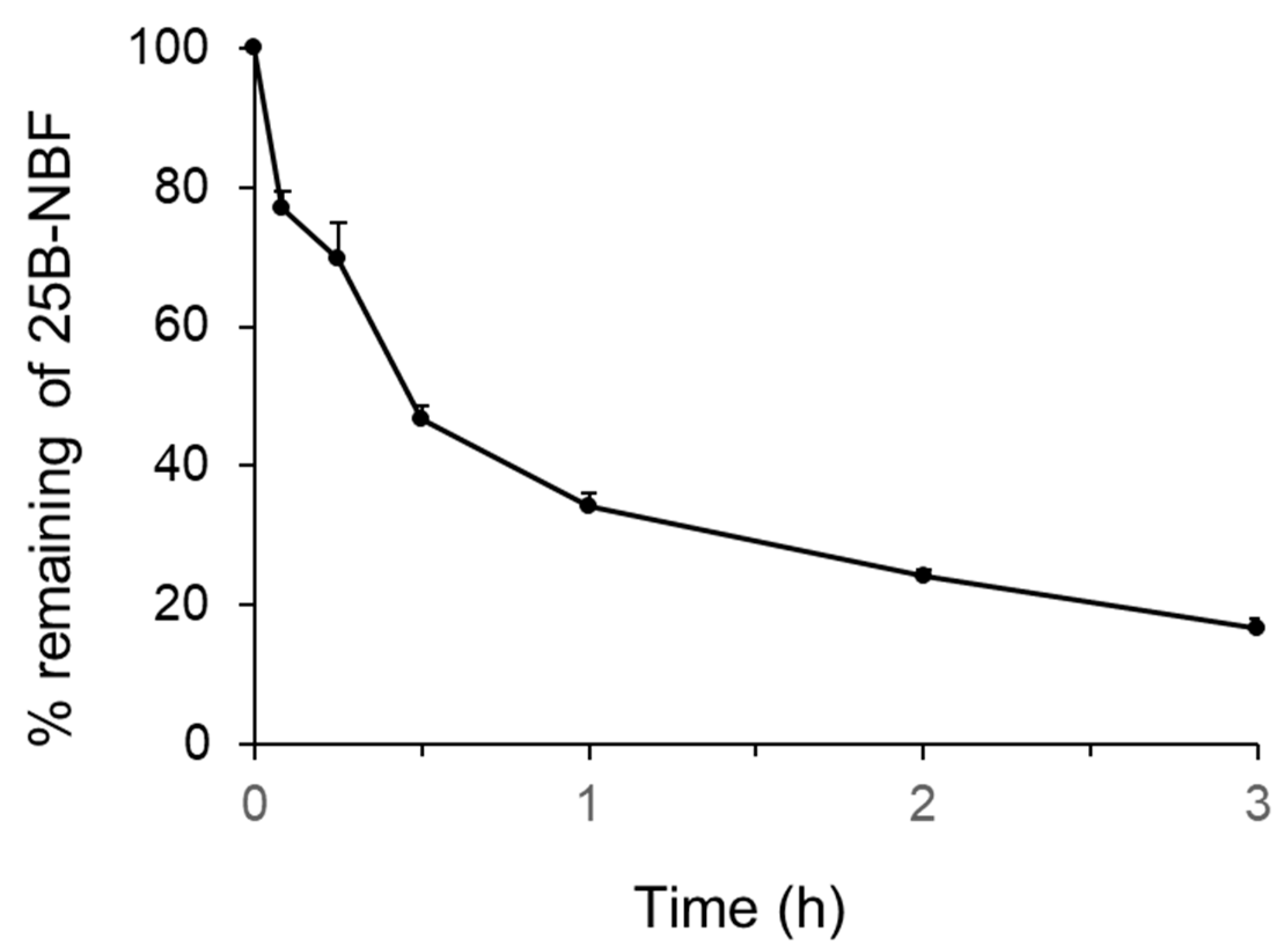
2.1. Metabolic Stability of 25B-NBF and Prediction of Its Hepatic Clearance
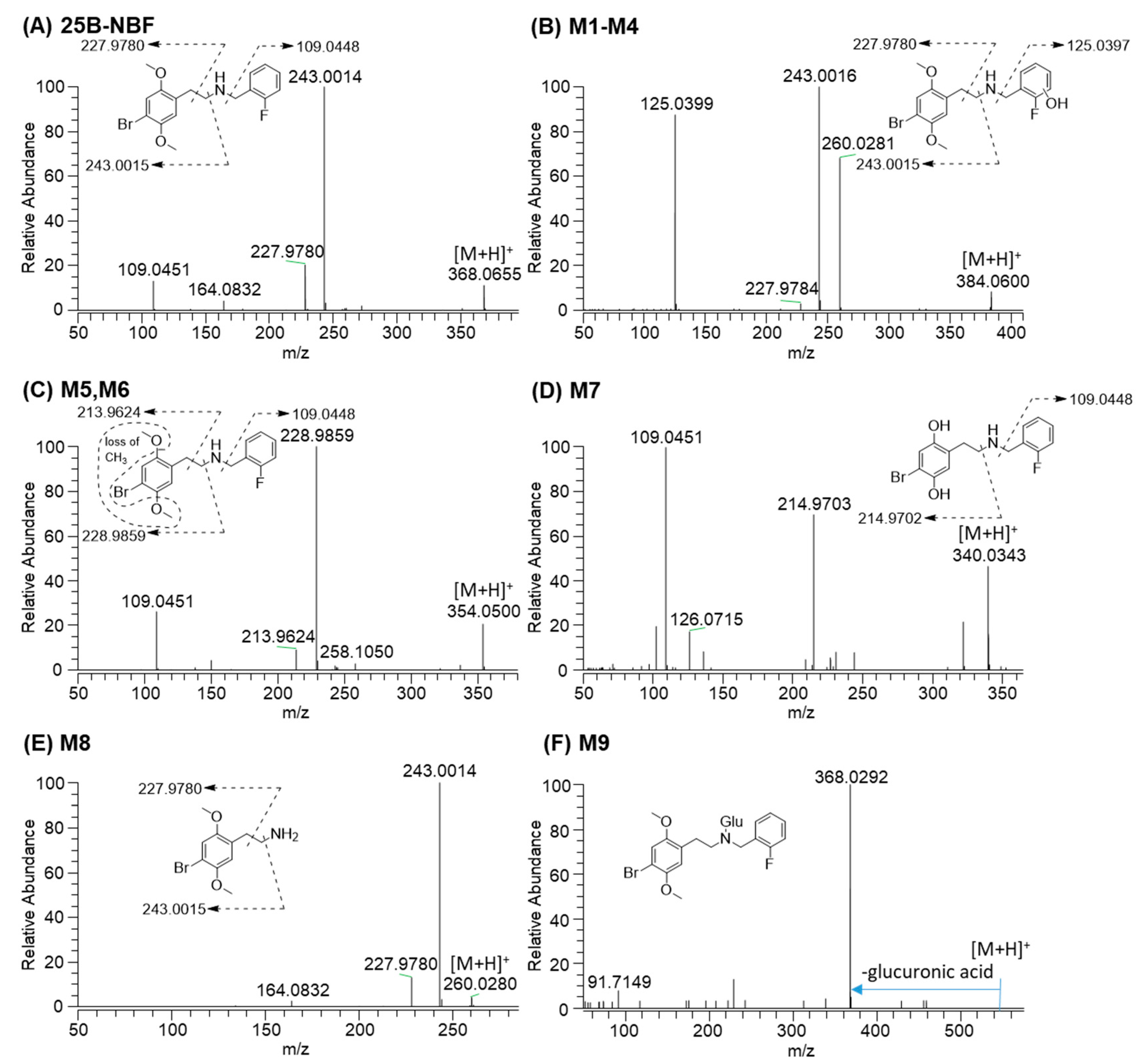
2.2. Metabolite Identification of 25B-NBF
2.3. Screening of CYPs and UGTs Responsible for the Metabolism of 25B-NBF
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Metabolic Stability of 25B-NBF in Human Hepatocytes
4.3. Metabolite Identification in Human Hepatocytes
4.4. Metabolism of 25B-NBF in Human cDNA-Expressed CYPs and UGTs
4.5. LC–MS Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- United Nations Office on Drugs and Crime: World Drug Report 2017. Available online: https://www.unodc.org/wdr2017/index.html (accessed on 19 February 2019).
- European Monitoring Centre for Drugs and Drug Addiction, European Drug Report 2017: Trends and Developments. Available online: http://www.emcdda.europa.eu/publications/edr/trends-developments/2017_en (accessed on 19 February 2019).
- Halberstadt, A.L.; Geyer, M.A. Effects of the hallucinogen 2,5-dimethoxy-4-iodophenethylamine (2C-I) and superpotent N-benzyl derivatives on the head twitch response. Neuropharmacology 2014, 77, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Phonekeo, K.; Paine, J.S.; Leth-Petersen, S.; Begtrup, M.; Bräuner-Osborne, H.; Kristensen, J.L. Synthesis and Structure–Activity Relationships of N-Benzyl Phenethylamines as 5-HT2A/2C Agonists. ACS Chem. Neurosci. 2014, 5, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Braden, M.R.; Parrish, J.C.; Naylor, J.C.; Nichols, D.E. Molecular Interaction of Serotonin 5-HT2A Receptor Residues Phe339(6.51) and Phe340(6.52) with Superpotent N-Benzyl Phenethylamine Agonists. Mol. Pharmacol. 2006, 70, 1956–1964. [Google Scholar] [CrossRef] [PubMed]
- HSLF-FS 2015:35. Available online: https://en.wikipedia.org/wiki/25B-NBF (accessed on 19 February 2019).
- The Misuse of Drugs Act 1971 (Ketamine etc.) (Amendment) Order 2014. Available online: https://www.legislation.gov.uk/ukdsi/2014/9780111110904 (accessed on 19 February 2019).
- Ninnemann, A.; Stuart, G.L. The NBOMe series: A novel, dangerous group of hallucinogenic drugs. J. Stud. Alcohol Drugs 2013, 74, 977–978. [Google Scholar] [CrossRef] [PubMed]
- Poklis, J.L.; Nanco, C.R.; Troendle, M.M.; Wolf, C.E.; Poklis, A. Determination of 4-bromo-2, 5-dimethoxy-N-[(2-methoxyphenyl) methyl]-benzeneethanamine (25B-NBOMe) in serum and urine by high performance liquid chromatography with tandem mass spectrometry in a case of severe intoxication. Drug Test. Anal. 2014, 6, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Halberstadt, A.L. Pharmacology and toxicology of N-benzylphenethylamine (“NBOMe”) hallucinogens. In Neuropharmacology of New Psychoactive Substances (NPS): The Science Behind the Headlines; Baumann, M.H., Glennon, R.A., Wiley, J.L., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 283–311. [Google Scholar]
- Gee, P.; Schep, L.J.; Jensen, B.P.; Moore, G.; Barrington, S. Case series: Toxicity from 25B-NBOMe–a cluster of N-bomb cases. Clin. Toxicol. 2016, 54, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.H.Y.; Ching, C.K.; Tsui, M.S.H.; Chu, F.K.C.; Mak, T.W.L. Two cases of severe intoxication associated with analytically confirmed use of the novel psychoactive substances 25B-NBOMe and 25C-NBOMe. Clin. Toxicol. 2014, 52, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Caspar, A.T.; Brandt, S.D.; Stoever, A.E.; Meyer, M.R.; Maurer, H.H. Metabolic fate and detectability of the new psychoactive substances 2-(4-bromo-2,5-dimethoxyphenyl)-N-[(2-methoxyphenyl)-methyl]ethanamine (25B-NBOMe) and 2-(4-chloro-2,5-dimethoxyphenyl)-N-[(2-methoxy-phenyl)methyl]ethanamine (25C-NBOMe) in human and rat urine by GC–MS, LC–MSn, and LC–HR–MS/MS approaches. J. Pharm. Biomed. Anal. 2017, 134, 158–169. [Google Scholar] [PubMed]
- Boumrah, Y.; Humbert, L.; Phanithavong, M.; Khimeche, K.; Dahmani, A.; Allorge, D. In vitro characterization of potential CYP- and UGT-derived metabolites of the psychoactive drug 25B-NBOMe using LC-high resolution MS. Drug Test. Anal. 2016, 8, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Wohlfarth, A.; Roman, M.; Andersson, M.; Kugelberg, F.C.; Diao, X.; Carlier, J.; Eriksson, C.; Wu, X.; Konradsson, P.; Josefsson, M.; et al. 25C-NBOMe and 25I-NBOMe metabolite studies in human hepatocytes, in vivo mouse and human urine with high-resolution mass spectrometry. Drug Test. Anal. 2017, 9, 680–698. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, H.S.; Kong, T.Y.; Lee, J.Y.; Kim, J.Y.; In, M.K.; Lee, H.S. In vitro metabolism of a novel synthetic cannabinoid, EAM-2201, in human liver microsomes and human recombinant cytochrome P450s. J. Pharm. Biomed. Anal. 2016, 119, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kong, T.Y.; Moon, J.Y.; Choi, K.H.; Cho, Y.Y.; Kang, H.C.; Lee, J.Y.; Lee, H.S. Targeted and non-targeted metabolite identification of MAM-2201 in human, mouse, and rat hepatocytes. Drug Test. Anal. 2018, 10, 1328–1335. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.Y.; Kim, J.H.; Kim, D.K.; Lee, H.S. Synthetic cannabinoids are substrates and inhibitors of multiple drug-metabolizing enzymes. Arch. Pharm. Res. 2018, 41, 691–710. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.U.; Kim, J.H.; Kong, T.Y.; Choi, W.G.; Lee, H.S. Comparative metabolism of honokiol in mouse, rat, dog, monkey, and human hepatocytes. Arch. Pharm. Res. 2016, 39, 516–530. [Google Scholar] [CrossRef] [PubMed]
- Leth-Petersen, S.; Gabel-Jensen, C.; Gillings, N.; Lehel, S.; Hansen, H.D.; Knudsen, G.M.; Kristensen, J.L. Metabolic fate of hallucinogenic NBOMes. Chem. Res. Toxicol. 2016, 29, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Caspar, A.T.; Helfer, A.G.; Michely, J.A.; Auwärter, V.; Brandt, S.D.; Meyer, M.R.; Maurer, H.H. Studies on the metabolism and toxicological detection of the new psychoactive designer drug 2-(4-iodo-2,5-dimethoxyphenyl)-N-[(2-methoxyphenyl)methyl]ethanamine (25I-NBOMe) in human and rat urine using GC-MS, LC-MSn, and LC-HR-MS/MS. Anal. Bioanal. Chem. 2015, 407, 6697–6719. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.M.; Holm, N.B.; Leth-Petersen, S.; Kristensen, J.L.; Olsen, L.; Linnet, K. Characterization of the hepatic cytochrome P450 enzymes involved in the metabolism of 25I-NBOMe and 25I-NBOH. Drug Test. Anal. 2017, 9, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Castillo, C.; Villalobos, C.; Moya, P.R.; Sáez, P.; Cassels, B.K.; Huidobro-Toro, J.P. Differences in potency and efficacy of a series of phenylisopropylamine/phenylethylamine pairs at 5-HT(2A) and 5-HT(2C) receptors. Br. J. Pharmacol. 2002, 136, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Villalobos, C.A.; Bull, P.; Sáez, P.; Cassels, B.K.; Huidobro-Toro, J.P. 4-Bromo-2,5-dimethoxyphenethylamine (2C-B) and structurally related phenylethylamines are potent 5-HT(2A) receptor antagonists in Xenopus laevis oocytes. Br. J. Pharmacol. 2004, 141, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, T.; Nagasawa, K.; Kuwayama, K.; Tsujikawa, K.; Iwata, Y.T.; Inoue, H. Analysis of 4-bromo-2,5-dimethoxyphenethylamine abuser’s urine: Identification and quantitation of urinary metabolites. J. Forensic Sci. 2013, 58, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Theobald, D.S.; Maurer, H.H. Identification of monoamine oxidase and cytochrome P450 isoenzymes involved in the deamination of phenethylamine-derived designer drugs (2C-series). Biochem. Pharmacol. 2007, 73, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Carmo, H.; Hengstler, J.G.; Boer, D.d.; Ringel, M.; Remião, F.; Carvalho, F.; Fernandes, E.; Reys, L.A.d.; Oesch, F.; Bastos, M.d.L. Metabolic pathways of 4-bromo-2,5-dimethoxyphenethylamine (2C-B): Analysis of phase I metabolism with hepatocytes of six species including human. Toxicology 2005, 206, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.R.; Maurer, H.H. Metabolism of designer drugs of abuse: An updated review. Curr. Drug Metab. 2010, 11, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Morris, T. Physiological parameters in laboratory animals and humans. Pharm. Res. 1993, 10, 1093–1095. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, T.; Gan, L.-S. The role of drug metabolism in drug discovery. In Enzyme Inhibition in Drug Discovery and Development; Lu, C., Li, A.P., Eds.; Wiley: New York, NJ, USA, 2010; pp. 91–176. [Google Scholar]
Sample Availability: Not available |
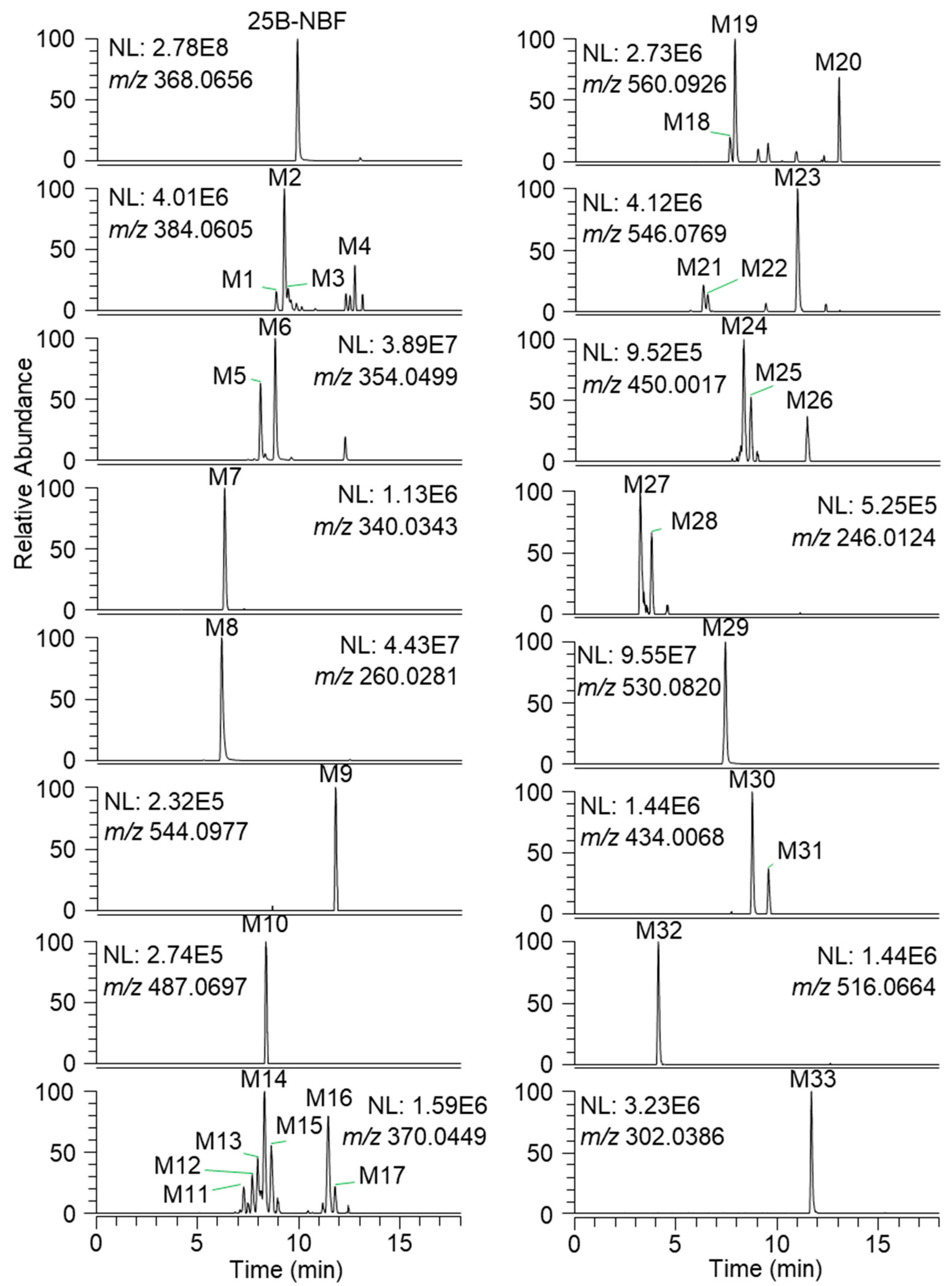
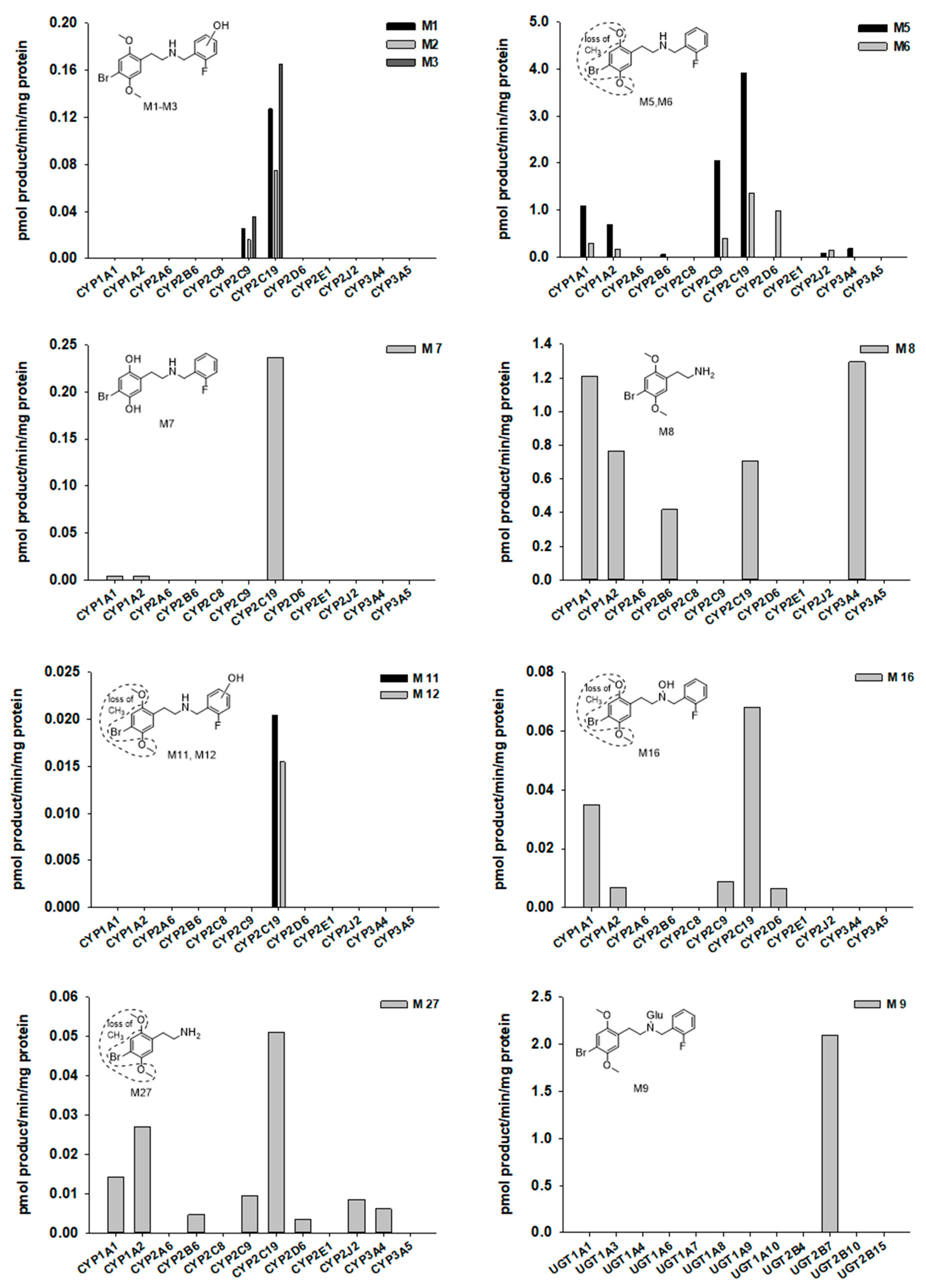
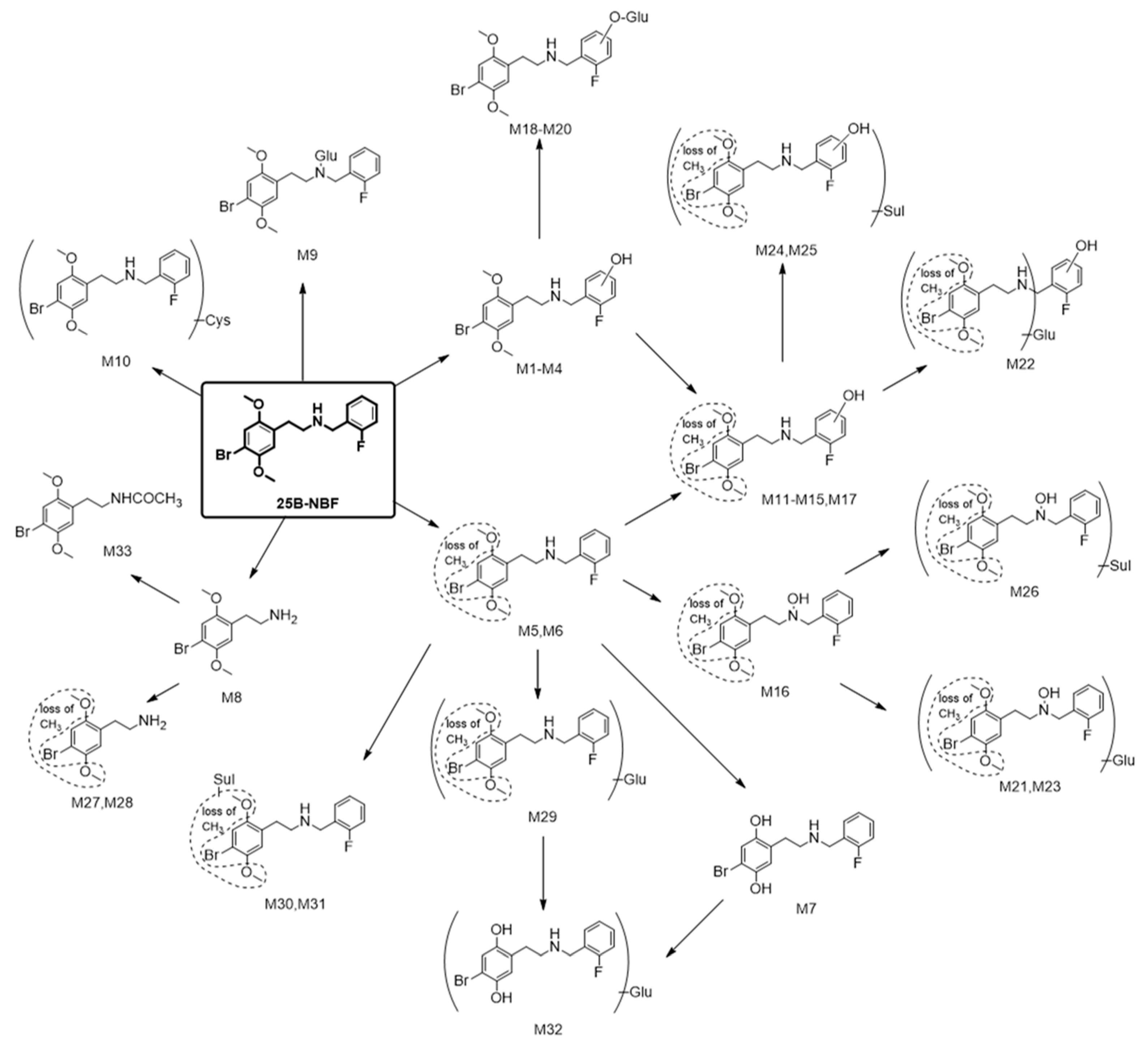
| ID | RT (min) | Formula | Exact Mass ([M + H]+) | Error (ppm) | Fragment Ions | Biotransformation |
|---|---|---|---|---|---|---|
| 25B-NBF | 9.9 | C17H19BrFNO2 | 368.0656 | −0.5 | 243.0014, 227.9780, 109.0451 | Parent |
| M1 | 8.8 | C17H19BrFNO3 | 384.0605 | 1.0 | 260.0281, 243.0015, 227.9780, 125.0399 | Monohydroxylation |
| M2 | 9.2 | C17H19BrFNO3 | 384.0605 | −0.3 | 260.0281, 243.0016, 227.9780, 125.0399 | Monohydroxylation |
| M3 | 9.4 | C17H19BrFNO3 | 384.0605 | 0.0 | 260.0280, 243.0015, 227.9780, 125.0399 | Monohydroxylation |
| M4 | 12.7 | C17H19BrFNO3 | 384.0605 | −0.8 | 260.0280, 243.0014, 227.9780, 125.0398 | Monohydroxylation |
| M5 | 8.1 | C16H17BrFNO2 | 354.0499 | -0.3 | 228.9858, 213.9626, 109.0451 | O-Demethylation |
| M6 | 8.8 | C16H17BrFNO2 | 354.0499 | 0.3 | 228.9859, 213.9624, 109.0451 | O-Demethylation |
| M7 | 6.3 | C15H15BrFNO2 | 340.0343 | −0.9 | 214.9703, 109.0451 | Bis-O-demethylation |
| M8 | 6.1 | C10H14BrNO2 | 260.0281 | -0.8 | 243.0014, 227.9780 | N-Debenzylation |
| M9 | 11.8 | C23H27BrFNO8 | 544.0977 | −1.8 | 368.0292 | Glucuronidation |
| M10 | 8.4 | C20H24BrFN2O4S | 487.0697 | 0.4 | 243.0015, 227.9780 | Cystein conjugation |
| M11 | 7.3 | C16H17BrFNO3 | 370.0449 | −1.4 | 245.0046, 228.9859, 125.0399 | Monohydroxylation + O-Demethylation |
| M12 | 7.7 | C16H17BrFNO3 | 370.0449 | −0.8 | 245.0046, 228.9859, 125.0398 | Monohydroxylation + O-Demethylation |
| M13 | 8.0 | C16H17BrFNO3 | 370.0449 | −0.3 | 245.0046, 228.9858, 125.0399 | Monohydroxylation + O-Demethylation |
| M14 | 8.3 | C16H17BrFNO3 | 370.0449 | −0.5 | 245.0046, 228.9858, 125.0398 | Monohydroxylation + O-Demethylation |
| M15 | 8.6 | C16H17BrFNO3 | 370.0449 | −0.8 | 245.0046, 228.9859, 125.0399 | Monohydroxylation + O-Demethylation |
| M16 | 11.5 | C16H17BrFNO3 | 370.0449 | −0.8 | 228.9859, 214.9702, 109.0451 | Monohydroxylation + O-Demethylation |
| M17 | 11.8 | C16H17BrFNO3 | 370.0449 | 0.0 | 245.0046, 228.9858, 125.0399 | Monohydroxylation + O-Demethylation |
| M18 | 7.6 | C23H27BrFNO9 | 560.0926 | 0.7 | 384.0605, 243.0013, 125.0398 | Monohydroxylation + Glucuronidation |
| M19 | 7.9 | C23H27BrFNO9 | 560.0926 | 0.4 | 384.0606, 260.0280, 125.0399 | Monohydroxylation + Glucuronidation |
| M20 | 13.0 | C23H27BrFNO9 | 560.0926 | 0.4 | 384.0601, 243.0013, 125.0399 | Monohydroxylation + Glucuronidation |
| M21 | 6.3 | C22H25BrFNO9 | 546.0769 | 0.4 | 370.0447, 352.0345, 228.9859, 109.0450 | Monohydroxylation + O-Demethylation + Glucuronidation |
| M22 | 6.5 | C22H25BrFNO9 | 546.0769 | 0.4 | 370.0447, 246.0124, 228.9861, 125.0399 | Monohydroxylation + O-Demethylation + Glucuronidation |
| M23 | 11.0 | C22H25BrFNO9 | 546.0769 | 1.1 | 370.0451, 228.9859, 214.9702, 109.0450 | Monohydroxylation + O-Demethylation + Glucuronidation |
| M24 | 8.3 | C16H17BrFNO6S | 450.0017 | 0.9 | 370.0449, 228.9857, 125.0398 | Monohydroxylation + O-Demethylation + Sulfation |
| M25 | 8.7 | C16H17BrFNO6S | 450.0017 | 0.2 | 370.0449, 228.9857, 125.0399 | Monohydroxylation + O-Demethylation + Sulfation |
| M26 | 11.4 | C16H17BrFNO6S | 450.0017 | −0.7 | 370.0439, 228.9857, 109.0451 | Monohydroxylation + O-Demethylation + Sulfation |
| M27 | 3.2 | C9H12BrNO2 | 246.0124 | −1.2 | 228.9859 | O-Demethylation + N-Debenzylation |
| M28 | 3.8 | C9H12BrNO2 | 246.0124 | 0.4 | 228.9858 | O-Demethylation + N-Debenzylation |
| M29 | 7.4 | C22H25BrFNO8 | 530.0820 | 0.6 | 354.0498, 228.9859 | O-Demethylation + Glucuronidation |
| M30 | 8.8 | C16H17BrFNO5S | 434.0068 | −0.2 | 354.0499, 308.9427, 228.9859, 109.0451 | O-Demethylation + Sulfation |
| M31 | 9.6 | C16H17BrFNO5S | 434.0068 | −0.2 | 354.0501, 308.9427, 228.9859, 109.0451 | O-Demethylation + Sulfation |
| M32 | 4.1 | C21H23BrFNO8 | 516.0664 | 0.2 | 340.0343, 214.9701, 109.0450 | Bis-O-demethylation + Glucuronidation |
| M33 | 11.7 | C12H16BrNO3 | 302.0386 | −0.3 | 260.0280, 243.0015, 227.9781 | N-Dearylation + Acetylation |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H.; Kim, S.; Lee, J.; In, S.; Cho, Y.-Y.; Kang, H.C.; Lee, J.Y.; Lee, H.S. In Vitro Metabolism of 25B-NBF, 2-(4-Bromo-2,5-Dimethoxyphenyl)-N-(2-Fluorobenzyl)ethanamine, in Human Hepatocytes Using Liquid Chromatography–Mass Spectrometry. Molecules 2019, 24, 818. https://doi.org/10.3390/molecules24040818
Kim J-H, Kim S, Lee J, In S, Cho Y-Y, Kang HC, Lee JY, Lee HS. In Vitro Metabolism of 25B-NBF, 2-(4-Bromo-2,5-Dimethoxyphenyl)-N-(2-Fluorobenzyl)ethanamine, in Human Hepatocytes Using Liquid Chromatography–Mass Spectrometry. Molecules. 2019; 24(4):818. https://doi.org/10.3390/molecules24040818
Chicago/Turabian StyleKim, Ju-Hyun, Sunjoo Kim, Jaesin Lee, Sangwhan In, Yong-Yeon Cho, Han Chang Kang, Joo Young Lee, and Hye Suk Lee. 2019. "In Vitro Metabolism of 25B-NBF, 2-(4-Bromo-2,5-Dimethoxyphenyl)-N-(2-Fluorobenzyl)ethanamine, in Human Hepatocytes Using Liquid Chromatography–Mass Spectrometry" Molecules 24, no. 4: 818. https://doi.org/10.3390/molecules24040818
APA StyleKim, J.-H., Kim, S., Lee, J., In, S., Cho, Y.-Y., Kang, H. C., Lee, J. Y., & Lee, H. S. (2019). In Vitro Metabolism of 25B-NBF, 2-(4-Bromo-2,5-Dimethoxyphenyl)-N-(2-Fluorobenzyl)ethanamine, in Human Hepatocytes Using Liquid Chromatography–Mass Spectrometry. Molecules, 24(4), 818. https://doi.org/10.3390/molecules24040818