Urinary Proteomics Profiles Are Useful for Detection of Cancer Biomarkers and Changes Induced by Therapeutic Procedures
Abstract
:1. Introduction
2. Results
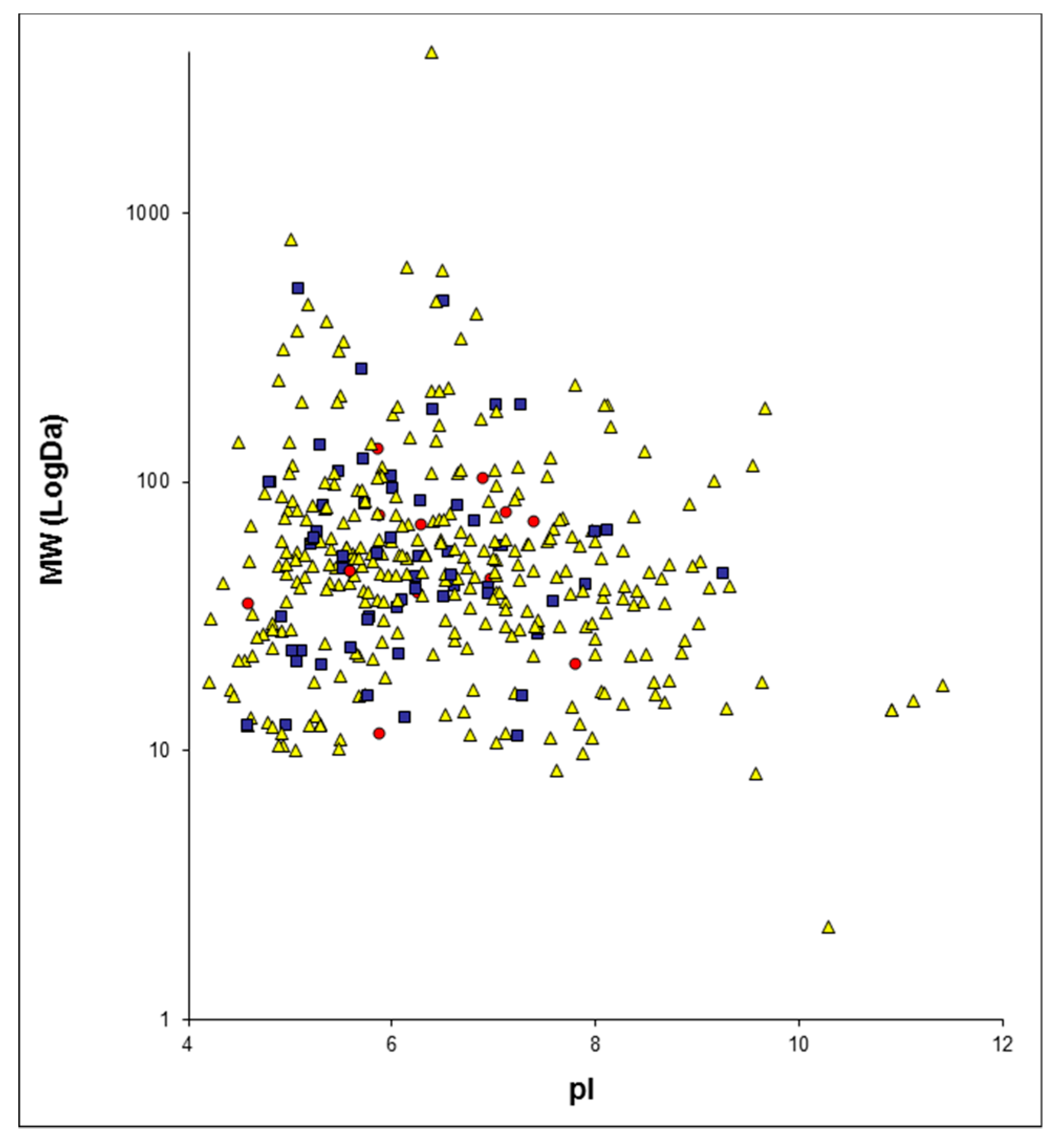
2.1. Proteome Profiles of Urine
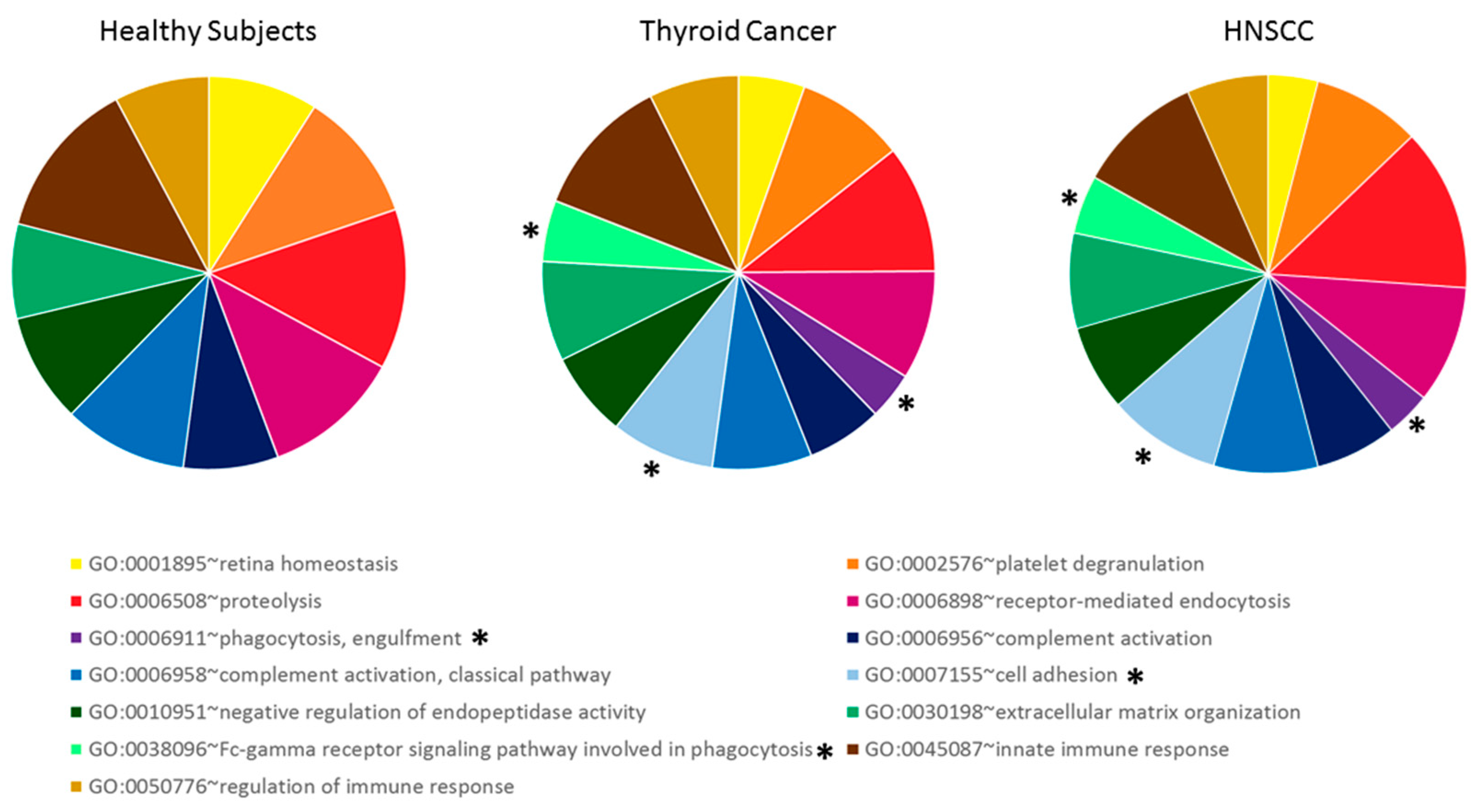
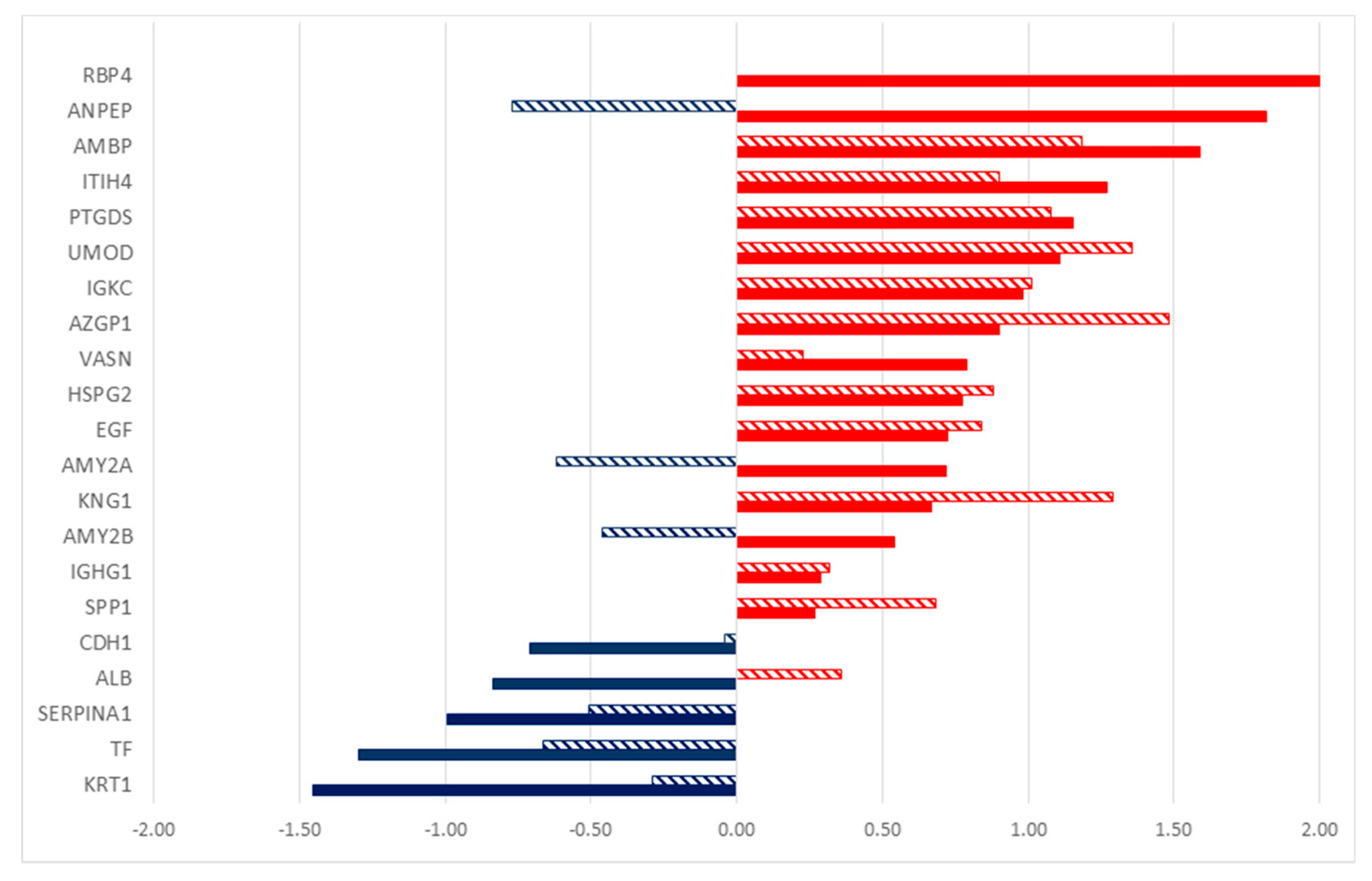
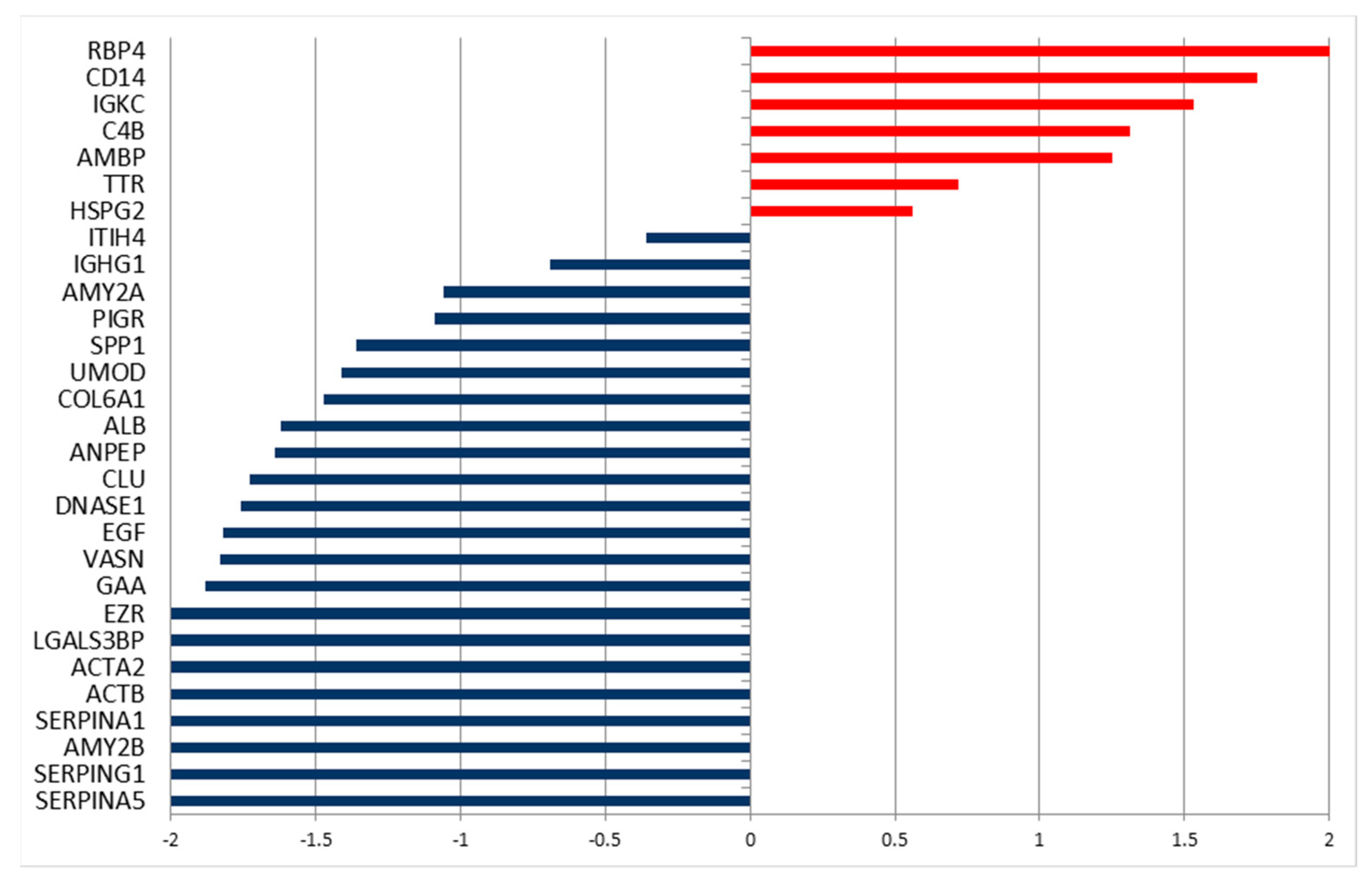
2.2. Differentially Secreted Proteins in Urine from Tumor Patients
2.3. Proteome Profiles in Urine to Monitor Effects of Interventions
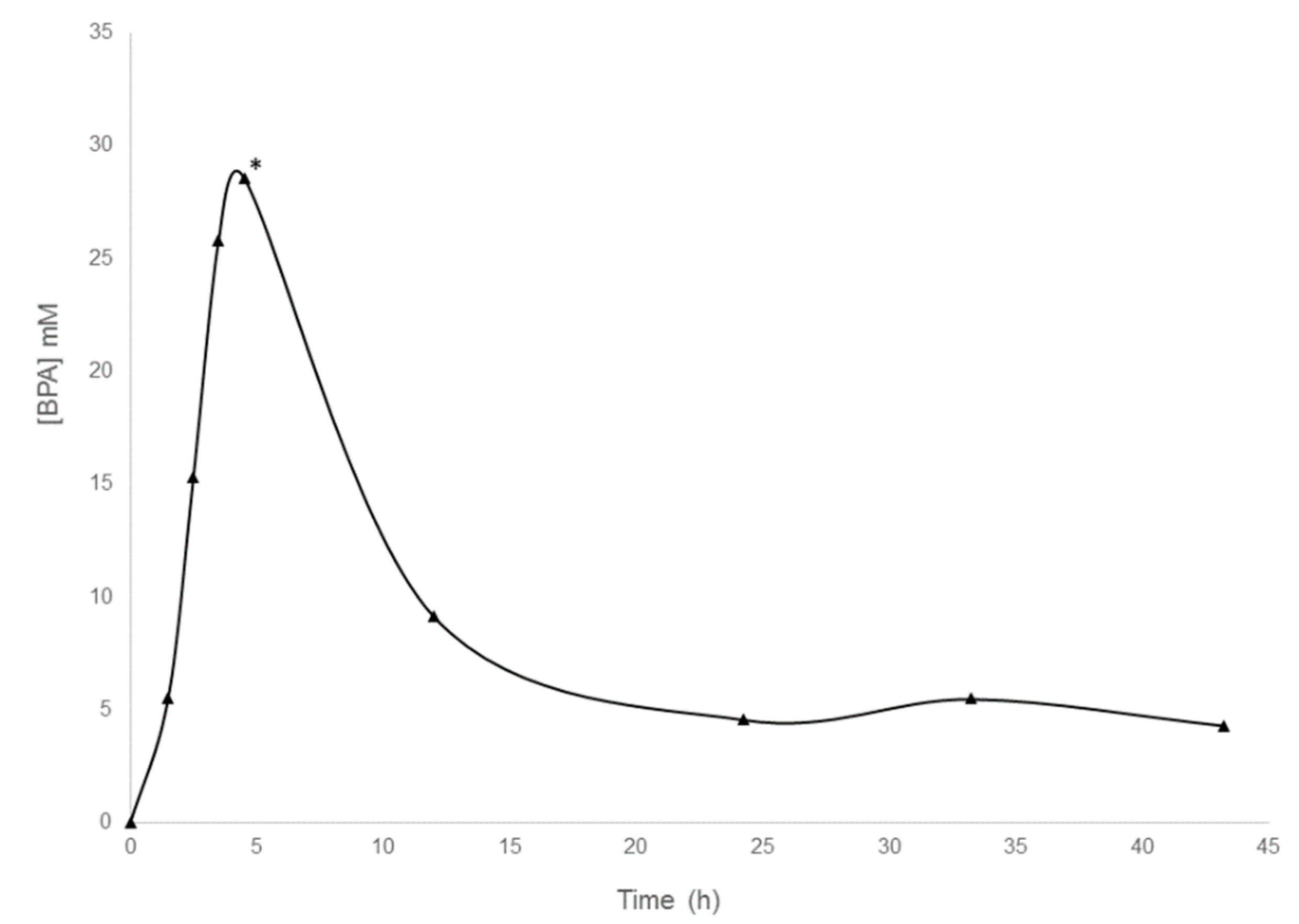
2.3.1. Infusion of a HNSCC Patient with BPA
2.3.2. Infusion of a Thyroid Cancer Patient with BSH
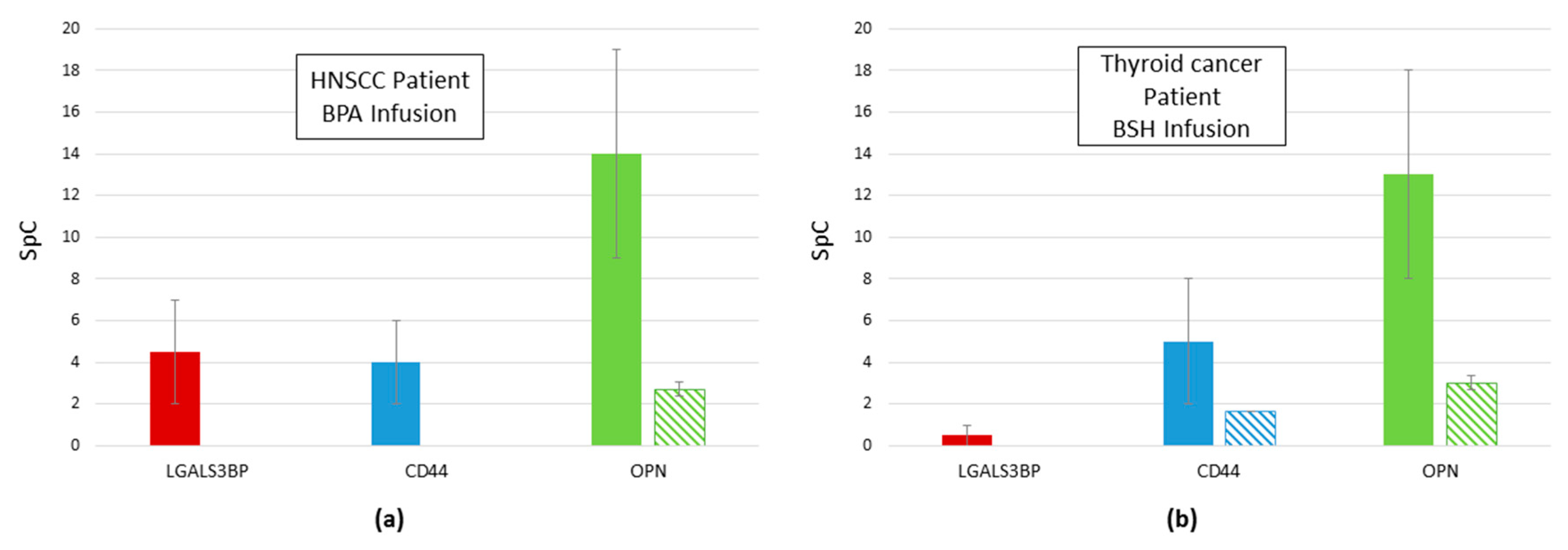
2.4. Reduction of Proteins Related to Inflammation after Infusion: galectin 3 Binding Protein, CD44, and OPN
3. Discussion
4. Materials and Methods
4.1. Patients Recruitment and Urine Collection
4.2. Boron Compounds and Urine Collection
4.3. Urine Preparation for Proteomics Analysis
4.4. Enzymatic Digestion of Protein Samples
4.5. Two-Dimensional Capillary Chromatography-Tandem Mass Spectrometry (2DC-MS/MS) Analysis
4.6. Data Processing of MS Results
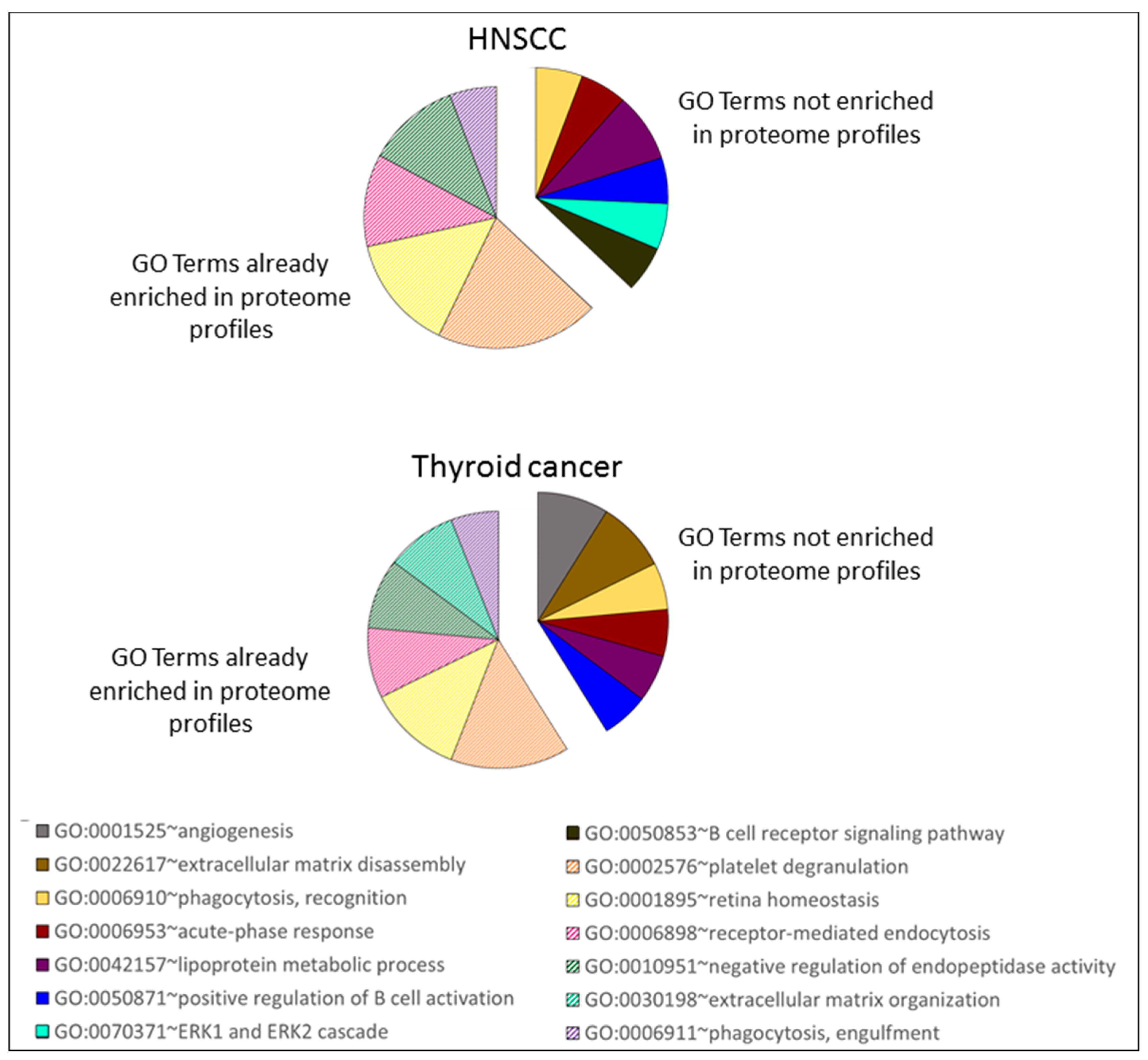
4.7. GO Terms Enrichment
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Moss, R.L. Critical review, with an optimistic outlook, on Boron Neutron Capture Therapy (BNCT). Appl. Radiat. Isot. 2014, 88, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Miyatake, S.-I.; Kawabata, S.; Hiramatsu, R.; Kuroiwa, T.; Suzuki, M.; Kondo, N.; Ono, K. Boron Neutron Capture Therapy for Malignant Brain Tumors. Neurol. Med. Chir. (Tokyo) 2016, 56, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Sauerwein, W.; Zurlo, A. The EORTC Boron Neutron Capture Therapy (BNCT) Group: achievements and future projects. Eur. J. Cancer 2002, 38, 31–34. [Google Scholar] [CrossRef]
- Kankaanranta, L.; Seppälä, T.; Koivunoro, H.; Saarilahti, K.; Atula, T.; Collan, J.; Salli, E.; Kortesniemi, M.; Uusi-Simola, J.; Välimäki, P.; et al. Boron neutron capture therapy in the treatment of locally recurred head-and-neck cancer: final analysis of a phase I/II trial. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e67–e75. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kato, I.; Aihara, T.; Hiratsuka, J.; Yoshimura, K.; Niimi, M.; Kimura, Y.; Ariyoshi, Y.; Haginomori, S.; Sakurai, Y.; et al. Boron neutron capture therapy outcomes for advanced or recurrent head and neck cancer. J. Radiat. Res. (Tokyo) 2014, 55, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Aihara, T.; Morita, N.; Kamitani, N.; Kumada, H.; Ono, K.; Hiratsuka, J.; Harada, T. BNCT for advanced or recurrent head and neck cancer. Appl. Radiat. Isot. 2014, 88, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Quah, D.S.; Leech, M.; Marignol, L. Clinical potential of boron neutron capture therapy for locally recurrent inoperable previously irradiated head and neck cancer. Appl. Radiat. Isot. 2015, 106, 237–241. [Google Scholar] [CrossRef]
- Kato, I.; Fujita, Y.; Maruhashi, A.; Kumada, H.; Ohmae, M.; Kirihata, M.; Imahori, Y.; Suzuki, M.; Sakrai, Y.; Sumi, T.; et al. Effectiveness of boron neutron capture therapy for recurrent head and neck malignancies. Appl. Radiat. Isot. Data Instrum. Methods Use Agric. Ind. Med. 2009, 67, S37–S42. [Google Scholar] [CrossRef]
- Wittig, A.; Collette, L.; Moss, R.; Sauerwein, W.A. Early clinical trial concept for boron neutron capture therapy: a critical assessment of the EORTC trial 11001. Appl. Radiat. Isot. Data Instrum. Methods Use Agric. Ind. Med. 2009, 67, S59–S62. [Google Scholar] [CrossRef]
- Dagrosa, A.; Carpano, M.; Perona, M.; Thomasz, L.; Nievas, S.; Cabrini, R.; Juvenal, G.; Pisarev, M. Studies for the application of boron neutron capture therapy to the treatment of differentiated thyroid cancer. Appl. Radiat. Isot. 2011, 69, 1752–1755. [Google Scholar] [CrossRef]
- Yoshida, F.; Matsumura, A.; Shibata, Y.; Yamamoto, T.; Nakauchi, H.; Okumura, M.; Nose, T. Cell cycle dependence of boron uptake from two boron compounds used for clinical neutron capture therapy. Cancer Lett. 2002, 187, 135–141. [Google Scholar] [CrossRef]
- Carpano, M.; Perona, M.; Rodriguez, C.; Nievas, S.; Olivera, M.; Santa Cruz, G.A.; Brandizzi, D.; Cabrini, R.; Pisarev, M.; Juvenal, G.J.; et al. Experimental Studies of Boronophenylalanine ((10)BPA) Biodistribution for the Individual Application of Boron Neutron Capture Therapy (BNCT) for Malignant Melanoma Treatment. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Koivunoro, H.; Hippeläinen, E.; Auterinen, I.; Kankaanranta, L.; Kulvik, M.; Laakso, J.; Seppälä, T.; Savolainen, S.; Joensuu, H. Biokinetic analysis of tissue boron (10B) concentrations of glioma patients treated with BNCT in Finland. Appl. Radiat. Isot. 2015, 106, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Svantesson, E.; Capala, J.; Markides, K.E.; Pettersson, J. Determination of boron-containing compounds in urine and blood plasma from boron neutron capture therapy patients. The importance of using coupled techniques. Anal. Chem. 2002, 74, 5358–5363. [Google Scholar] [CrossRef] [PubMed]
- Kulvik, M.; Kallio, M.; Laakso, J.; Vähätalo, J.; Hermans, R.; Järviluoma, E.; Paetau, A.; Rasilainen, M.; Ruokonen, I.; Seppälä, M.; et al. Biodistribution of boron after intravenous 4-dihydroxyborylphenylalanine-fructose (BPA-F) infusion in meningioma and schwannoma patients: A feasibility study for boron neutron capture therapy. Appl. Radiat. Isot. Data Instrum. Methods Use Agric. Ind. Med. 2015, 106, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Basilico, F.; Sauerwein, W.; Pozzi, F.; Wittig, A.; Moss, R.; Mauri, P.L. Analysis of 10B antitumoral compounds by means of flow-injection into ESI-MS/MS. J. Mass Spectrom. JMS 2005, 40, 1546–1549. [Google Scholar] [CrossRef] [PubMed]
- Bendel, P.; Wittig, A.; Basilico, F.; Mauri, P.L.; Sauerwein, W. Metabolism of borono-phenylalanine-fructose complex (BPA-fr) and borocaptate sodium (BSH) in cancer patients--results from EORTC trial 11001. J. Pharm. Biomed. Anal. 2010, 51, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Masutani, M.; Baiseitov, D.; Itoh, T.; Hirai, T.; Berikkhanova, K.; Murakami, Y.; Zhumadilov, Z.; Imahori, Y.; Hoshi, M.; Itami, J. Histological and biochemical analysis of DNA damage after BNCT in rat model. Appl. Radiat. Isot. 2014, 88, 104–108. [Google Scholar] [CrossRef]
- Mauri, P.L.; Basilico, F. Proteomic Investigations for Boron Neutron Capture Therapy. In Neutron Capture Therapy: Principles and Applications; Sauerwein, W., Wittig, A., Moss, R., Nakagawa, Y., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2012; pp. 189–200. ISBN 978-3-642-31334-9. [Google Scholar]
- Sato, A.; Itoh, T.; Imamichi, S.; Kikuhara, S.; Fujimori, H.; Hirai, T.; Saito, S.; Sakurai, Y.; Tanaka, H.; Nakamura, H.; et al. Proteomic analysis of cellular response induced by boron neutron capture reaction in human squamous cell carcinoma SAS cells. Appl. Radiat. Isot. Data Instrum. Methods Use Agric. Ind. Med. 2015, 106, 213–219. [Google Scholar] [CrossRef]
- Mauri, P.; Scigelova, M. Multidimensional protein identification technology for clinical proteomic analysis. Clin. Chem. Lab. Med. 2009, 47, 636–646. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Ponta, H. Perspectives of CD44 targeting therapies. Arch. Toxicol. 2015, 89, 3–14. [Google Scholar] [CrossRef]
- Castello, L.M.; Raineri, D.; Salmi, L.; Clemente, N.; Vaschetto, R.; Quaglia, M.; Garzaro, M.; Gentilli, S.; Navalesi, P.; Cantaluppi, V.; et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Mediators Inflamm. 2017, 2017, 4049098. [Google Scholar] [CrossRef] [PubMed]
- Loimaranta, V.; Hepojoki, J.; Laaksoaho, O.; Pulliainen, A.T. Galectin-3-binding protein: A multitask glycoprotein with innate immunity functions in viral and bacterial infections. J. Leukoc. Biol. 2018, 104, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Devuyst, O.; Olinger, E.; Rampoldi, L. Uromodulin: from physiology to rare and complex kidney disorders. Nat. Rev. Nephrol. 2017, 13, 525–544. [Google Scholar] [CrossRef]
- El-Achkar, T.M.; Wu, X.-R. Uromodulin in kidney injury: an instigator, bystander, or protector? Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2012, 59, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, R.S.; Nayak, A.R.; Deshpande, P.S.; Kabra, D.; Purohit, H.J.; Taori, G.M.; Daginawala, H.F. Inter-alpha-trypsin inhibitor heavy chain 4 is a novel marker of acute ischemic stroke. Clin. Chim. Acta Int. J. Clin. Chem. 2009, 402, 160–163. [Google Scholar] [CrossRef]
- Opstal-van Winden, A.W.J.; Krop, E.J.M.; Kåredal, M.H.; Gast, M.-C.W.; Lindh, C.H.; Jeppsson, M.C.; Jönsson, B.A.G.; Grobbee, D.E.; Peeters, P.H.M.; Beijnen, J.H.; et al. Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer 2011, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- Davalieva, K.; Kiprijanovska, S.; Komina, S.; Petrusevska, G.; Zografska, N.C.; Polenakovic, M. Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer. Proteome Sci. 2015, 13, 2. [Google Scholar] [CrossRef]
- Li, X.; Li, B.; Li, B.; Guo, T.; Sun, Z.; Li, X.; Chen, L.; Chen, W.; Chen, P.; Mao, Y.; et al. ITIH4: Effective Serum Marker, Early Warning and Diagnosis, Hepatocellular Carcinoma. Pathol. Oncol. Res. POR 2018, 24, 663–670. [Google Scholar] [CrossRef]
- Icer, M.A.; Gezmen-Karadag, M. The multiple functions and mechanisms of osteopontin. Clin. Biochem. 2018, 59, 17–24. [Google Scholar] [CrossRef]
- Wolff, D.; Greinix, H.; Lee, S.J.; Gooley, T.; Paczesny, S.; Pavletic, S.; Hakim, F.; Malard, F.; Jagasia, M.; Lawitschka, A.; et al. Biomarkers in chronic graft-versus-host disease: quo vadis? Bone Marrow Transplant. 2018, 53, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Wickström, M.; Larsson, R.; Nygren, P.; Gullbo, J. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011, 102, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Pérez, I.; Varona, A.; Blanco, L.; Gil, J.; Santaolalla, F.; Zabala, A.; Ibarguen, A.M.; Irazusta, J.; Larrinaga, G. Increased APN/CD13 and acid aminopeptidase activities in head and neck squamous cell carcinoma. Head Neck 2009, 31, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, H.; Yang, X.; Wang, W.; Huang, A.; Li, J.; Qin, X.; Li, F.; Lu, G.; Ding, H.; et al. Vasorin is a potential serum biomarker and drug target of hepatocarcinoma screened by subtractive-EMSA-SELEX to clinic patient serum. Oncotarget 2015, 6, 10045–10059. [Google Scholar] [CrossRef] [PubMed]
- Davalieva, K.; Kiprijanovska, S.; Maleva Kostovska, I.; Stavridis, S.; Stankov, O.; Komina, S.; Petrusevska, G.; Polenakovic, M. Comparative Proteomics Analysis of Urine Reveals Down-Regulation of Acute Phase Response Signaling and LXR/RXR Activation Pathways in Prostate Cancer. Proteomes 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Vidotto, A.; Henrique, T.; Raposo, L.S.; Maniglia, J.V.; Tajara, E.H. Salivary and serum proteomics in head and neck carcinomas: before and after surgery and radiotherapy. Cancer Biomark. Sect. Dis. Markers 2010, 8, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Zhao, X.; Jin, S.; Meng, Q.; Zhou, H.; Zhang, M. Discovery of potential bladder cancer biomarkers by comparative urine proteomics and analysis. Clin. Genitourin. Cancer 2013, 11, 56–62. [Google Scholar] [CrossRef]
- Fujita, K.; Kume, H.; Matsuzaki, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Uemura, M.; Miyagawa, Y.; Tomonaga, T.; Nonomura, N. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep. 2017, 7, 42961. [Google Scholar] [CrossRef]
- Saraswat, M.; Joenväärä, S.; Seppänen, H.; Mustonen, H.; Haglund, C.; Renkonen, R. Comparative proteomic profiling of the serum differentiates pancreatic cancer from chronic pancreatitis. Cancer Med. 2017, 6, 1738–1751. [Google Scholar] [CrossRef]
- Gong, Y.; Pu, W.; Jin, H.; Yang, P.; Zeng, H.; Wang, Y.; Pang, F.; Ma, X. Quantitative proteomics of CSF reveals potential predicted biomarkers for extranodal NK-/T-cell lymphoma of nasal-type with ethmoidal sinus metastasis. Life Sci. 2018, 198, 94–98. [Google Scholar] [CrossRef]
- Roy, F. van Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat. Rev. Cancer 2014, 14, 121–134. [Google Scholar] [PubMed]
- Jabbari, S.; Hedayati, M.; Yaghmaei, P.; Parivar, K. Medullary Thyroid Carcinoma--Circulating Status of Vaspin and Retinol Binding Protein-4 in Iranian Patients. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 6507–6512. [Google Scholar] [CrossRef]
- Chen, Y.; Azman, S.N.; Kerishnan, J.P.; Zain, R.B.; Chen, Y.N.; Wong, Y.-L.; Gopinath, S.C.B. Identification of host-immune response protein candidates in the sera of human oral squamous cell carcinoma patients. PloS One 2014, 9, e109012. [Google Scholar] [CrossRef] [PubMed]
- Shevde, L.A.; Samant, R.S. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. J. Int. Soc. Matrix Biol. 2014, 37, 131–141. [Google Scholar] [CrossRef]
- Karpinsky, G.; Fatyga, A.; Krawczyk, M.A.; Chamera, M.; Sande, N.; Szmyd, D.; Izycka-Swieszewska, E.; Bien, E. Osteopontin: its potential role in cancer of children and young adults. Biomark. Med. 2017, 11, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Marcondes, M.C.G.; Poling, M.; Watry, D.D.; Hall, D.; Fox, H.S. In vivo osteopontin-induced macrophage accumulation is dependent on CD44 expression. Cell. Immunol. 2008, 254, 56–62. [Google Scholar] [CrossRef]
- Wittig, B.M.; Sabat, R.; Holzlöhner, P.; Witte-Händel, E.; Heilmann, K.; Witte, K.; Triebus, J.; Tzankov, A.; Laman, J.D.; Bokemeyer, B.; et al. Absence of specific alternatively spliced exon of CD44 in macrophages prevents colitis. Mucosal Immunol. 2018, 11, 846–860. [Google Scholar] [CrossRef]
- Nielsen, F.H. Update on human health effects of boron. J. Trace Elem. Med. Biol. Organ Soc. Miner. Trace Elem. GMS 2014, 28, 383–387. [Google Scholar] [CrossRef]
- Routray, I.; Ali, S. Boron Induces Lymphocyte Proliferation and Modulates the Priming Effects of Lipopolysaccharide on Macrophages. PloS One 2016, 11, e0150607. [Google Scholar] [CrossRef]
- Ferrari, E.; De Palma, A.; Mauri, P. Emerging MS-based platforms for the characterization of tumor-derived exosomes isolated from human biofluids: challenges and promises of MudPIT. Expert Rev. Proteomics 2017, 14, 757–767. [Google Scholar] [CrossRef]
- Wittig, A.; Malago, M.; Collette, L.; Huiskamp, R.; Bührmann, S.; Nievaart, V.; Kaiser, G.M.; Jöckel, K.-H.; Schmid, K.W.; Ortmann, U.; et al. Uptake of two 10B-compounds in liver metastases of colorectal adenocarcinoma for extracorporeal irradiation with boron neutron capture therapy (EORTC Trial 11001). Int. J. Cancer 2008, 122, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Wittig, A.; Collette, L.; Appelman, K.; Bührmann, S.; Jäckel, M.C.; Jöckel, K.-H.; Schmid, K.W.; Ortmann, U.; Moss, R.; Sauerwein, W.A.G. EORTC trial 11001: distribution of two 10B-compounds in patients with squamous cell carcinoma of head and neck, a translational research/phase 1 trial. J. Cell. Mol. Med. 2009, 13, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Mauri, P.; Dehò, G. Chapter 6 A Proteomic Approach to the Analysis of RNA Degradosome Composition in Escherichia coli. In Methods in Enzymology; RNA Turnover in Bacteria, Archaea and Organelles; Academic Press: Cambridge, MA, USA, 2008; Volume 447, pp. 99–117. [Google Scholar]
- Vigani, G.; Di Silvestre, D.; Agresta, A.M.; Donnini, S.; Mauri, P.; Gehl, C.; Bittner, F.; Murgia, I. Molybdenum and iron mutually impact their homeostasis in cucumber (Cucumis sativus) plants. New Phytol. 2017, 213, 1222–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; VerBerkmoes, N.C.; Langston, M.A.; Uberbacher, E.; Hettich, R.L.; Samatova, N.F. Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 2006, 5, 2909–2918. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Subject | Age | Diagnosis |
|---|---|---|
| P12 | 45.3 | Squamous cell carcinoma of the head and neck region |
| P21 | 50.6 | Squamous cell carcinoma of the head and neck region |
| P20 | 68.5 | Squamous cell carcinoma of the head and neck region |
| P22 | 53.1 | Squamous cell carcinoma of the head and neck region |
| P11 | 79.6 | Thyroid carcinoma |
| P14 | 34.6 | Papillary thyroid carcinoma |
| P16 | 34.0 | Papillary thyroid carcinoma |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrari, E.; Wittig, A.; Basilico, F.; Rossi, R.; De Palma, A.; Di Silvestre, D.; Sauerwein, W.A.G.; Mauri, P.L. Urinary Proteomics Profiles Are Useful for Detection of Cancer Biomarkers and Changes Induced by Therapeutic Procedures. Molecules 2019, 24, 794. https://doi.org/10.3390/molecules24040794
Ferrari E, Wittig A, Basilico F, Rossi R, De Palma A, Di Silvestre D, Sauerwein WAG, Mauri PL. Urinary Proteomics Profiles Are Useful for Detection of Cancer Biomarkers and Changes Induced by Therapeutic Procedures. Molecules. 2019; 24(4):794. https://doi.org/10.3390/molecules24040794
Chicago/Turabian StyleFerrari, Emanuele, Andrea Wittig, Fabrizio Basilico, Rossana Rossi, Antonella De Palma, Dario Di Silvestre, Wolfgang A.G. Sauerwein, and Pier Luigi Mauri. 2019. "Urinary Proteomics Profiles Are Useful for Detection of Cancer Biomarkers and Changes Induced by Therapeutic Procedures" Molecules 24, no. 4: 794. https://doi.org/10.3390/molecules24040794
APA StyleFerrari, E., Wittig, A., Basilico, F., Rossi, R., De Palma, A., Di Silvestre, D., Sauerwein, W. A. G., & Mauri, P. L. (2019). Urinary Proteomics Profiles Are Useful for Detection of Cancer Biomarkers and Changes Induced by Therapeutic Procedures. Molecules, 24(4), 794. https://doi.org/10.3390/molecules24040794