The Limitations of Collagen/CPP Hybrid Peptides as Carriers for Cancer Drugs to FaDu Cells
Abstract
:1. Introduction
2. Results
2.1. Hybrid Peptides
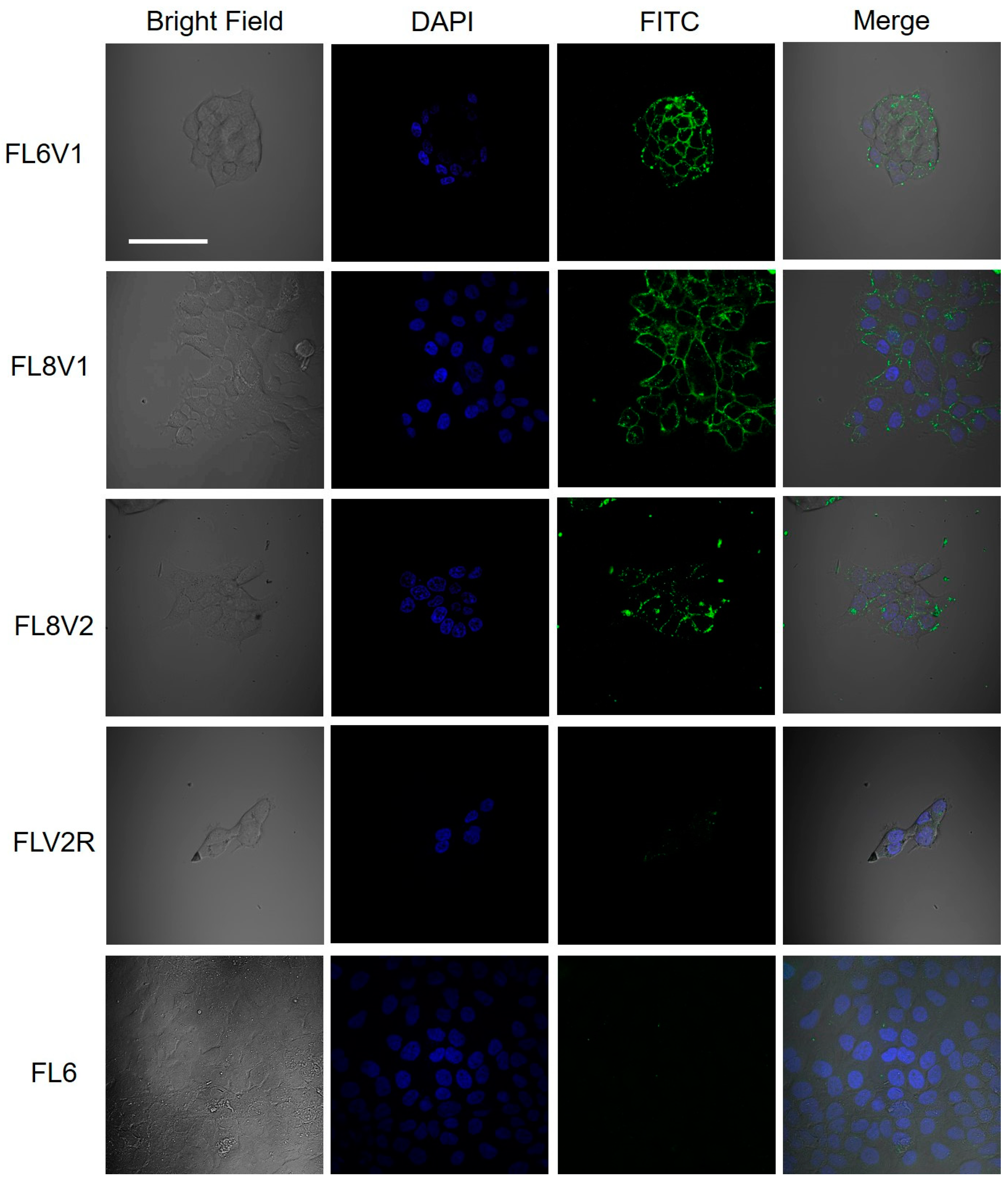
2.2. Hybrid Peptide as A Carrier of Model Drugs to FaDu Cells
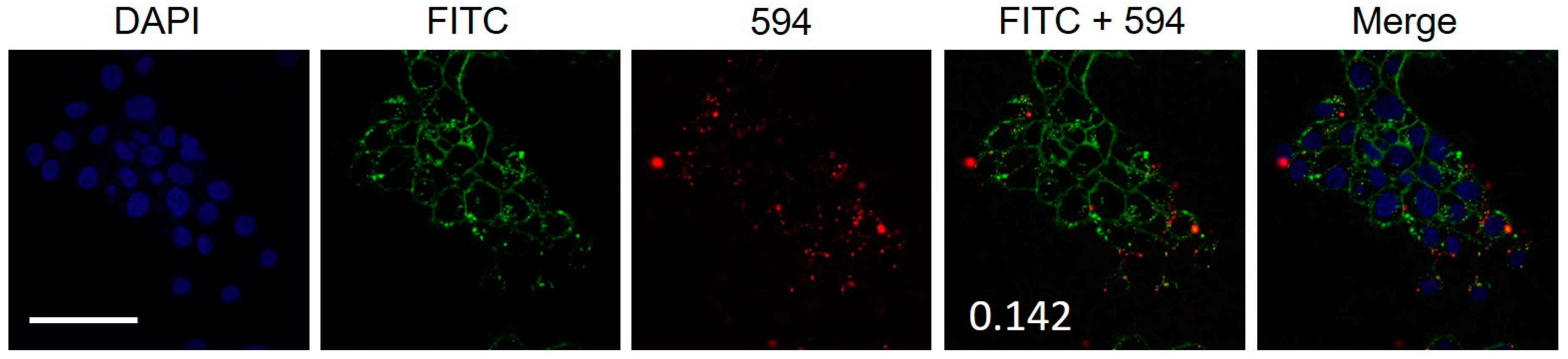
2.3. Colocalization of Hybrid Peptide Carrier and JAM-A Protein
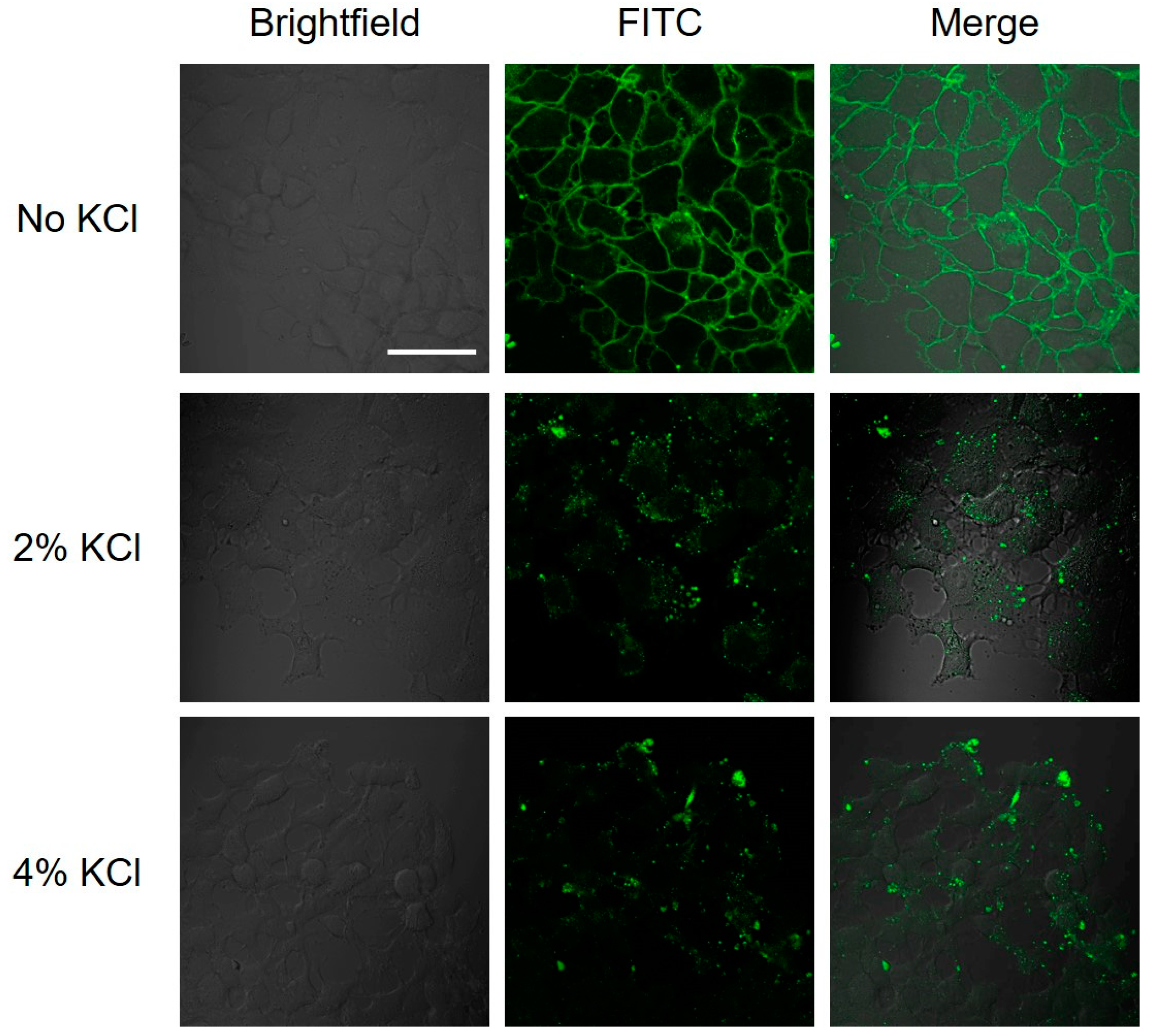
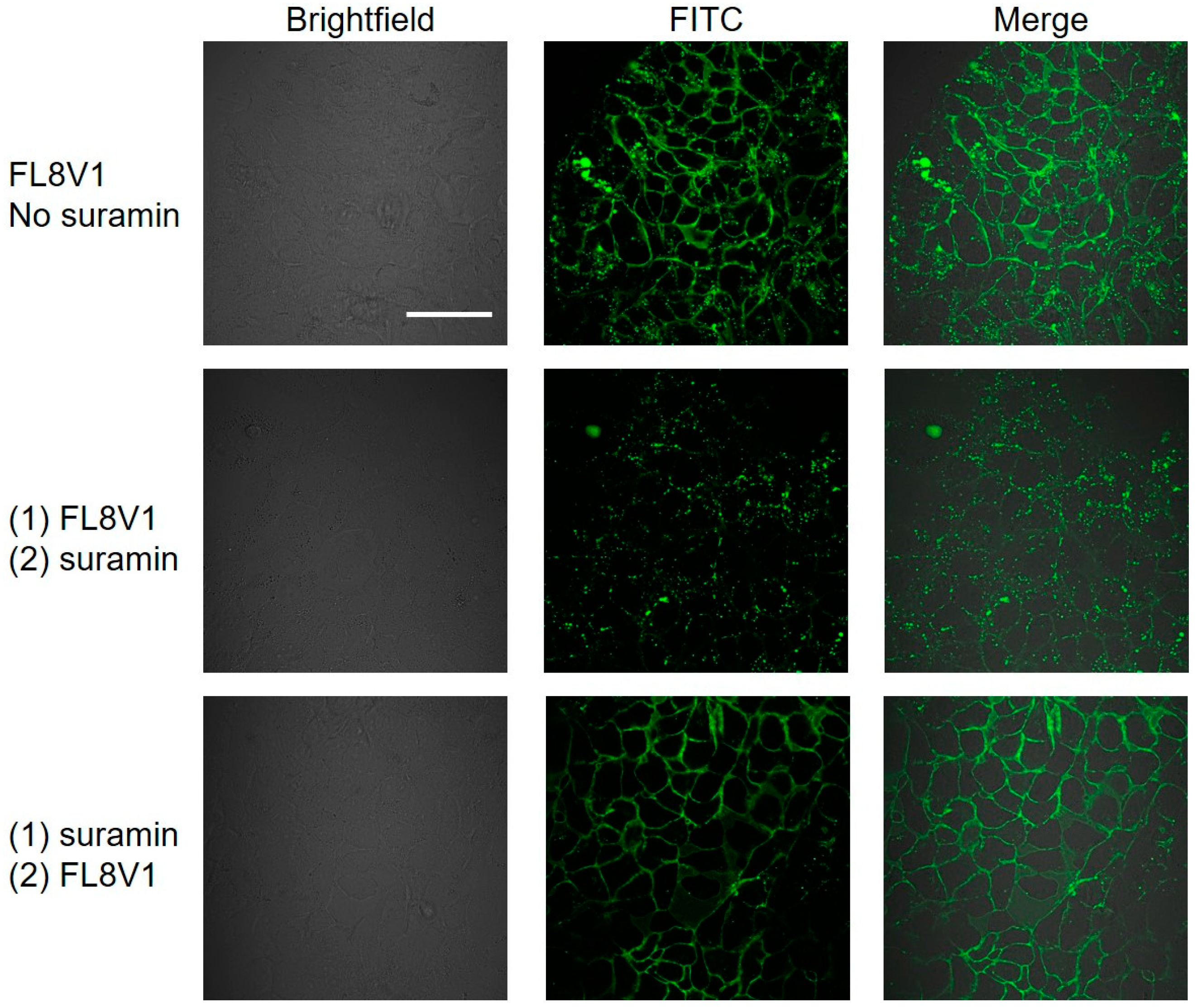
2.4. Interaction of Hybrid Peptide Carrier with FaDu Cell Surface
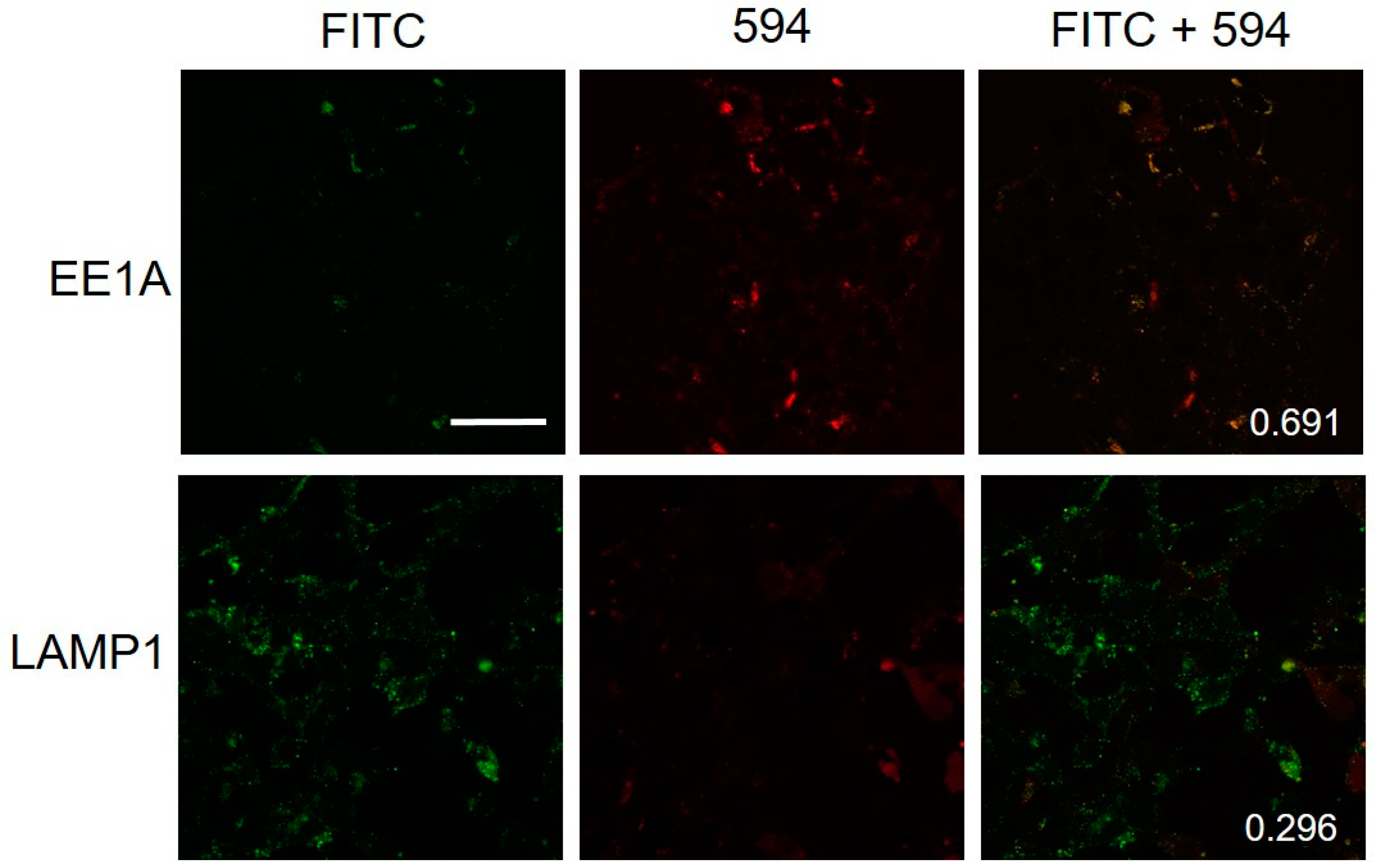
2.5. Colocalization of Hybrid Peptide Carrier and Endosomal Markers
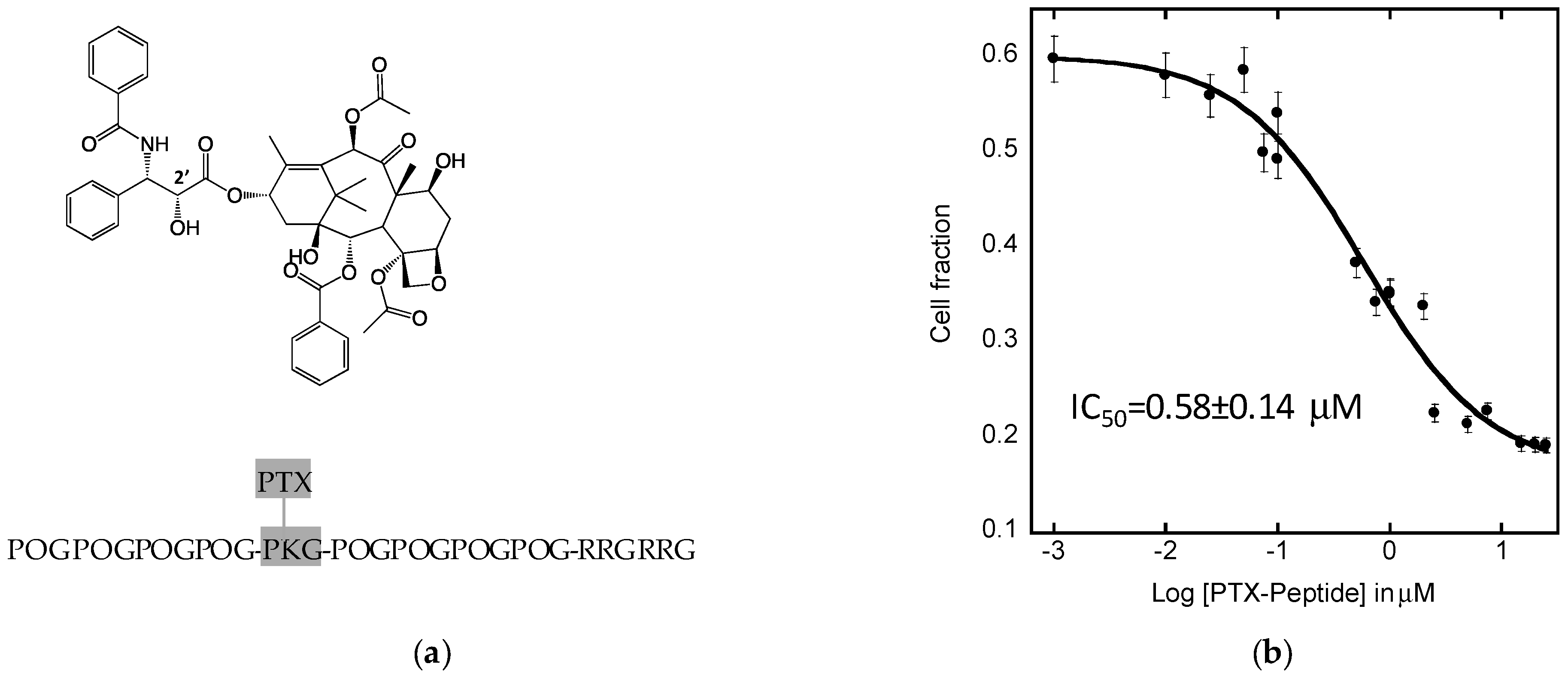
2.6. Efficacy of PTX8V1 in FaDu Cells
3. Discussion
4. Materials and Methods
4.1. Peptides
4.2. Cell Culture
4.3. Confocal Microscopy
4.4. Colocalization of JAM-A and FL8V1
4.5. Colocalization of EEA1 or LAMP1 and FL8V1
4.6. Salt Wash
4.7. Suramin Wash
4.8. IC50 Assay
4.9. Heparin Binding
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weinberg, R.A. The Biology of Cancer, 1st ed.; Garland Science: New York, NY, USA, 2007. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- National Cancer Institute. Head and Neck Cancers. Available online: https://www.cancer.gov/types/headand-neck/head-neck-fact-sheet/ (accessed on June 2018).
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Ma, L.; Lu, S.; Yu, L.; Tian, J.; Li, J.; Wang, H.; Xu, W. FaDu cell characteristics inducted by multidrug resistance. Oncol. Rep. 2011, 26, 1189–1195. [Google Scholar] [PubMed]
- Takes, R.; Strojan, P.; Silver, C.; Bradley, P.; Haigentz, M.; Wolf, G.; Shaha, A.; Hartl, D.; Olofsson, J.; Langendijk, J.; et al. Current trends in initial management of hypopharyngeal cancer: The declining use of open surgery. Head Neck 2010, 34, 270–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, B.A. How Taxol/paclitaxel kills cancer cells. Mol. Bio. Cell 2014, 25, 2677–2681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safavy, A. Recent developments in taxane drug delivery. Curr. Drug Deliv. 2008, 5, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Slichenmyer, W.J.; Von Hoff, D.D. New natural products in cancer chemotherapy. J. Clin. Pharmacol. 1990, 30, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Watanabe, J.; Ishihara, K. Enhanced solubility of paclitaxel using water-soluble and biocompatible 2-methacryloyloxyethyl phosphorylcholine polymers. J. Biomed. Mater. Res. 2003, 65, 209–214. [Google Scholar] [CrossRef]
- Ayalew, L.; Acuna, J.; Urfano, S.F.; Morfin, C.; Sablan, A.; Oh, M.; Gamboa, A.; Slowinska, K. Conjugation of paclitaxel to hybrid peptide carrier and biological evaluation in jurkat and A549 cancer cell lines. ACS Med. Chem. Lett. 2017, 8, 814–819. [Google Scholar] [CrossRef]
- Kalafatovic, D.; Giralt, E. Cell-penetrating peptides: Design strategies beyond primary structure and amphipathicity. Molecules 2017, 22, 1929. [Google Scholar] [CrossRef]
- Kauffman, W.B.; Fuselier, T.; He, J.; Wimley, W.C. Mechanism matters: A taxonomy of cell penetrating peptides. Trends Biochem. Sci. 2015, 40, 749–764. [Google Scholar] [CrossRef] [PubMed]
- Reissmann, S. Cell penetration: Scope and limitations by the application of cell-penetrating peptides. J. Pept. Sci. 2014, 20, 760–784. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wanga, Y.; Zhanga, X.; Zhanga, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Palm-Apergi, C.; Lorents, A.; Padari, K.; Pooga, M.; Hallbrink, M. The membrane repair response masks membrane disturbances caused by cell-penetrating peptide uptake. FASEB J. 2009, 23, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Verdurmen, W.P.; Brock, R. Biological responses towards cationic peptides and drug carriers. Trends Pharmacol. Sci. 2011, 32, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.; Feher, K.M.; Hu, C.; Slowinska, K. Peptide internalization enabled by folding: Triple helical cell-penetrating peptides. J. Peptide Sci. 2015, 21, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Snigireva, A.V.; Vrublevskaya, V.V.; Afanasyev, V.N.; Morenkov, O.S. Cell surface heparan sulfate proteoglycans are involved in the binding of Hsp90α and Hsp90β to the cell plasma membrane. Cell Adhes. Migr. 2015, 9, 460–468. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Le, W.; Wang, Y.; Li, Z.; Wang, D.; Ren, L.; Lin, L.; Cui, S.; Hu, J.J.; Hu, Y.; et al. Targeting negative surface charges of cancer cells by multifunctional nanoprobes. Theranostics 2016, 6, 1887–1898. [Google Scholar] [CrossRef] [PubMed]
- Murono, E.P.; Washburn, A.L.; Goforth, D.P.; Wu, N. Evidence that both receptor- and heparan sulfate proteoglycan-bound basic fibroblast growth factor are internalized by cultured immature Leydig cells. Molec. Cell. Endocrin. 1993, 98, 81–90. [Google Scholar] [CrossRef]
- Barash, U.; Cohen-Kaplan, V.; Dowek, I.; Sanderson, R.D.; Ilan, N.; Vlodavsky, I. Proteoglycans in health and disease: New concepts for heparanase function in tumor progression and metastasis. FEBS J. 2010, 277, 3890–3903. [Google Scholar] [CrossRef]
- Bella, J.; Eaton, M.; Brodsky, B. Crystal structure and molecular structure of a collagen like peptide at 1.9 angstrom resolution. Science 1994, 266, 74–81. [Google Scholar] [CrossRef]
- Persikov, A.V.; Ramshaw, J.A.M.; Brodsky, B. Prediction of collagen stability from amino acid sequence. J. Biol. Chem. 2005, 280, 19343–19349. [Google Scholar] [CrossRef]
- Shin, M.C.; Zhang, J.; Min, K.A.; Lee, K.; Byun, Y.; David, A.E.; He, H.; Yang, V.C. Cell-penetrating peptides: Achievements and challenges in application for cancer treatment. J. Biomed. Mater. Res. Part A 2014, 102A, 575–587. [Google Scholar] [CrossRef]
- Gupta, B.; Levchenko, T.S.; Torchilin, V.P. Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv. Drug Deliv. Rev. 2005, 57, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Rangan, S.R.S. A new human cell line (FaDu) from a hypopharyngeal carcinoma. Cancer 1972, 29, 117–121. [Google Scholar] [CrossRef] [Green Version]
- Kadletz, L.; Heiduschka, G.; Domayer, J.; Schmid, R.; Enzenhofer, E.; Thurnher, D. Evaluation of spheroid head and neck squamous cell carcinoma cell models in monolayer culture. Oncol. Lett. 2015, 10, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Campell, H.; Maiers, J.; DeMali, K.A. Interplay between tight junctions & adherens junctions. Experim. Cell Res. 2017, 358, 39–44. [Google Scholar]
- Salvador, E.; Burek, M.; Förster, C.Y. Tight Junctions and the Tumor Microenvironment. Curr. Pathiol. Rep. 2016, 4, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiba, H.; Osanai, M.; Murata, M.; Kojima, T.; Sawada, N. Transmembrane proteins of tight junctions. Biochim. Biophys. Acta 2008, 1778, 588–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakuki, T.; Kurose, M.; Takano, K.; Kondoh, A.; Obata, K.; Nomura, K.; Miyata, R.; Kaneko, Y.; Konno, T.; Takahashi, S.; et al. Dysregulation of junctional adhesion molecule A via p63/GATA-3 in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 33887–33900. [Google Scholar] [CrossRef] [PubMed]
- Pembrey, R.S.; Marshall, K.C.; Schneider, R.P. Cell surface analysis techniques: What do cell preparation protocols do to cell surface properties? Appl. Envirom. Microbiol. 1999, 65, 2877–2894. [Google Scholar]
- Stein, C.A.; Khan, T.M.; Khaled, Z.; Tonkinson, J.L. Cell surface binding and cellular internalization properties of suramin, a novel antineoplastic agent. Clin. Cancer Res. 1995, 1, 509–517. [Google Scholar] [PubMed]
- Mayor, S.; Pagano, R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.T. Macropinocytosis: Searching for an endocytic identity and role in the uptake of cell penetrating peptides. J. Cell Mol. Med. 2007, 11, 670–684. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the all peptides listed in Table 1 are available from the authors. |
| Peptide | Sequence |
|---|---|
| FL6V1 | FITC-BaGG-POGPOGPOGPOGPOGPOG-RRGRRG |
| FL8V1 | FITC-BaGG-POGPOGPOGPOGPOGPOGPOGPOG-RRGRRG |
| FL8V2 | FITC-BaGG-POGPOGPOGPOGPOGPOGPOGPOG-RRRRRR |
| FLV2R | FITC-BaGG-GPPOOGPGGGPOOPGOOPGOOPGGOOPP-RRRRRR |
| FL6 | FITC-BaGG-POGPOGPOGPOGPOGPOG |
| PTX8V1 | POGPOGPOGPOG-PK[PTX]G-POGPOGPOGPOG-RRGRRG |
| Peptide | Helix @ 37 °C | (POG)n | CPP | Cell Surface 1 |
|---|---|---|---|---|
| FL6V1 | No | Yes | Yes (1) | Yes |
| FL8V1 | Yes | Yes | Yes (1) | Yes |
| FL8V2 | Yes | Yes | Yes (2) | Yes |
| FLV2R | No | No | Yes (2) | No |
| FL6 | Yes | Yes | No | No |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, K.; Morfin, C.; Slowinska, K. The Limitations of Collagen/CPP Hybrid Peptides as Carriers for Cancer Drugs to FaDu Cells. Molecules 2019, 24, 676. https://doi.org/10.3390/molecules24040676
Ho K, Morfin C, Slowinska K. The Limitations of Collagen/CPP Hybrid Peptides as Carriers for Cancer Drugs to FaDu Cells. Molecules. 2019; 24(4):676. https://doi.org/10.3390/molecules24040676
Chicago/Turabian StyleHo, Kevin, Cristobal Morfin, and Katarzyna Slowinska. 2019. "The Limitations of Collagen/CPP Hybrid Peptides as Carriers for Cancer Drugs to FaDu Cells" Molecules 24, no. 4: 676. https://doi.org/10.3390/molecules24040676
APA StyleHo, K., Morfin, C., & Slowinska, K. (2019). The Limitations of Collagen/CPP Hybrid Peptides as Carriers for Cancer Drugs to FaDu Cells. Molecules, 24(4), 676. https://doi.org/10.3390/molecules24040676





