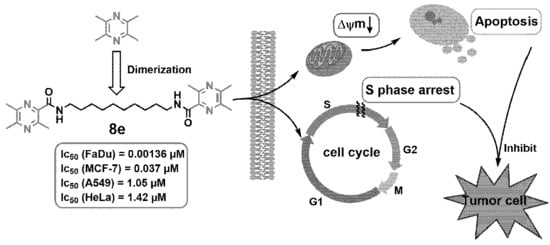
Novel Homo-Bivalent and Polyvalent Compounds Based on Ligustrazine and Heterocyclic Ring as Anticancer Agents
Abstract
1. Introduction
2. Results and Discussion
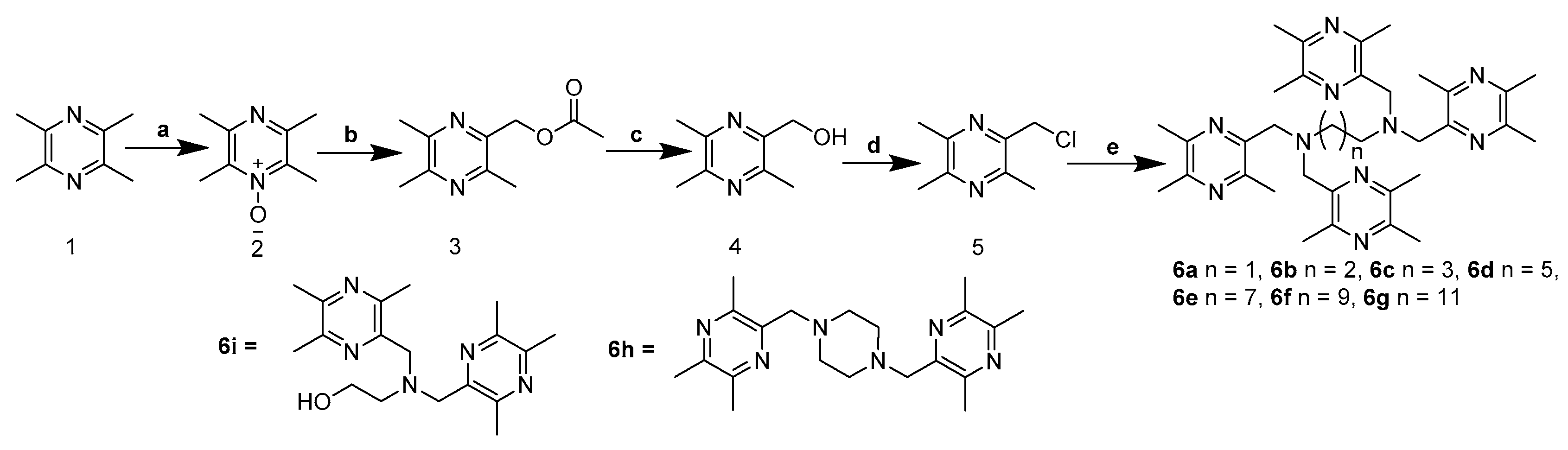
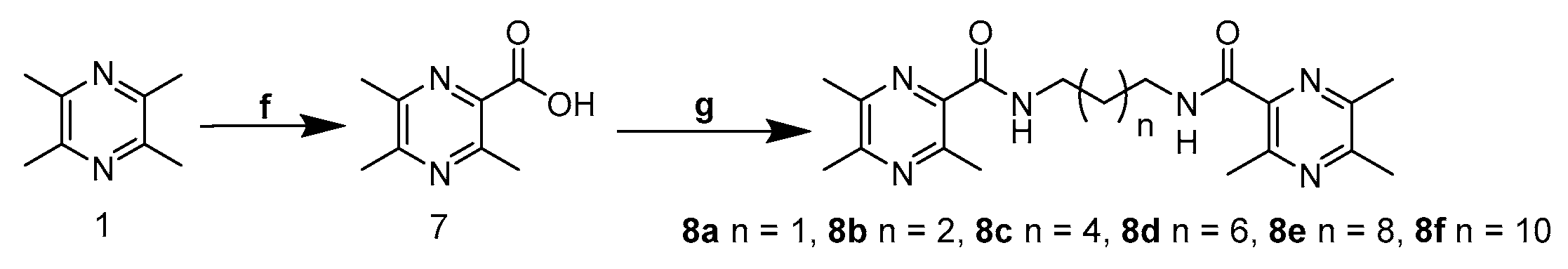
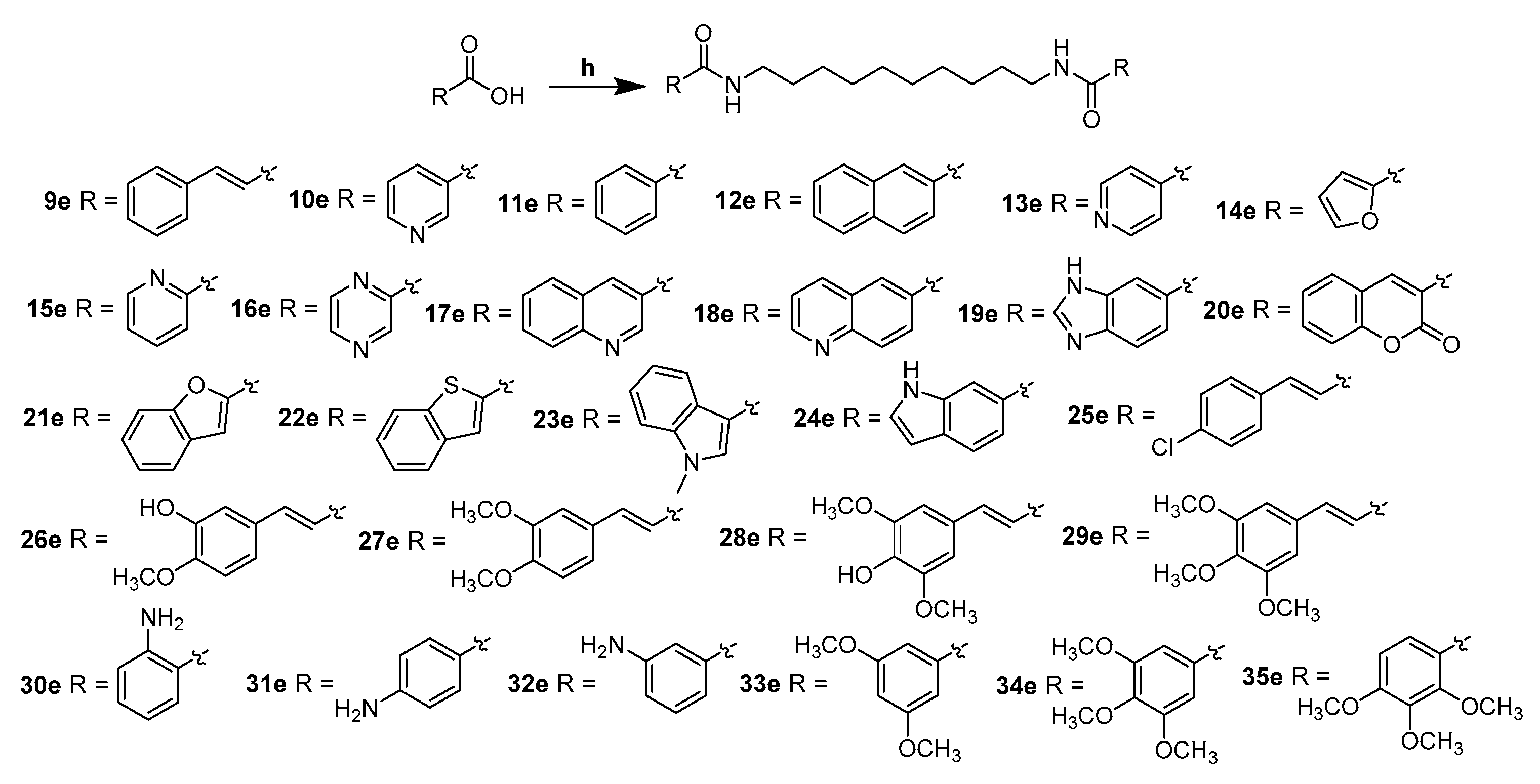
2.1. Chemistry
2.2. Anti-Proliferative Activity In Vitro
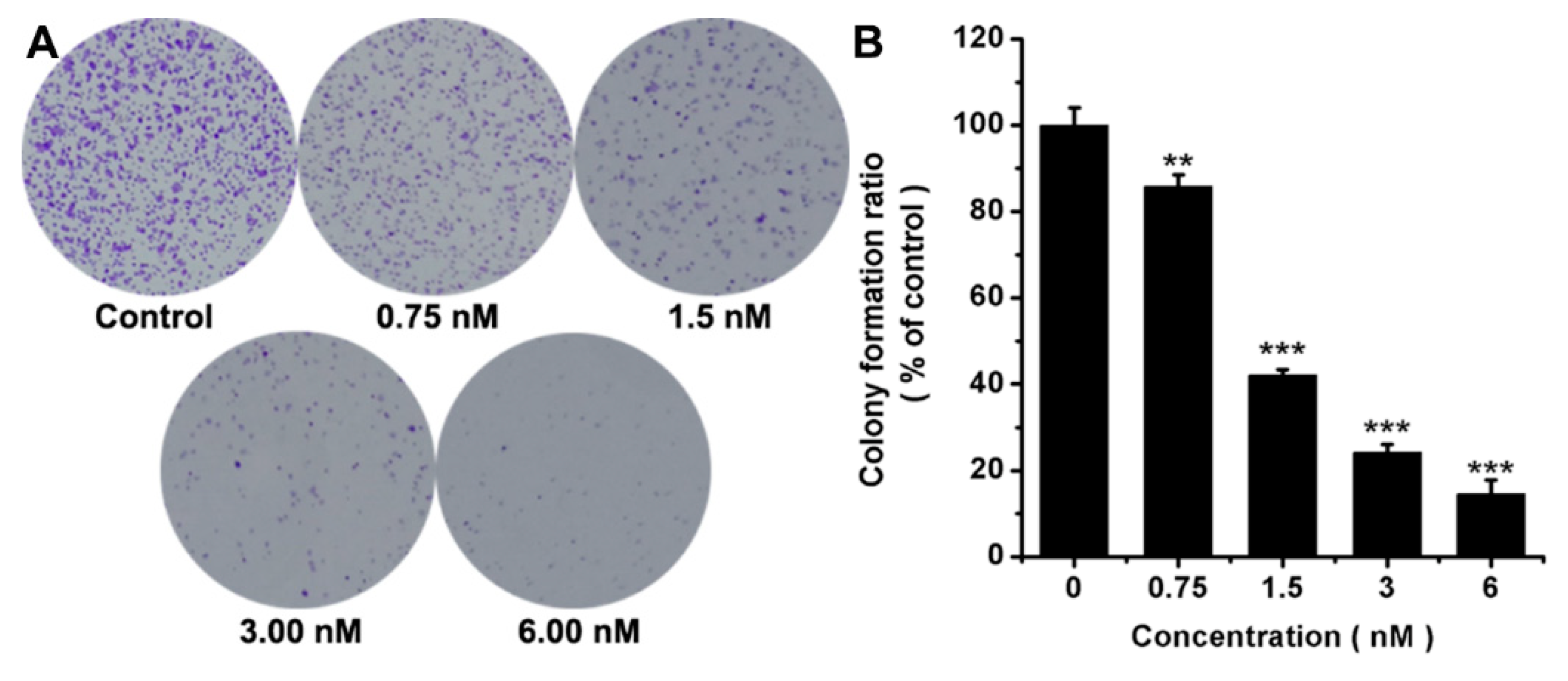
2.3. Colony Formation Assay
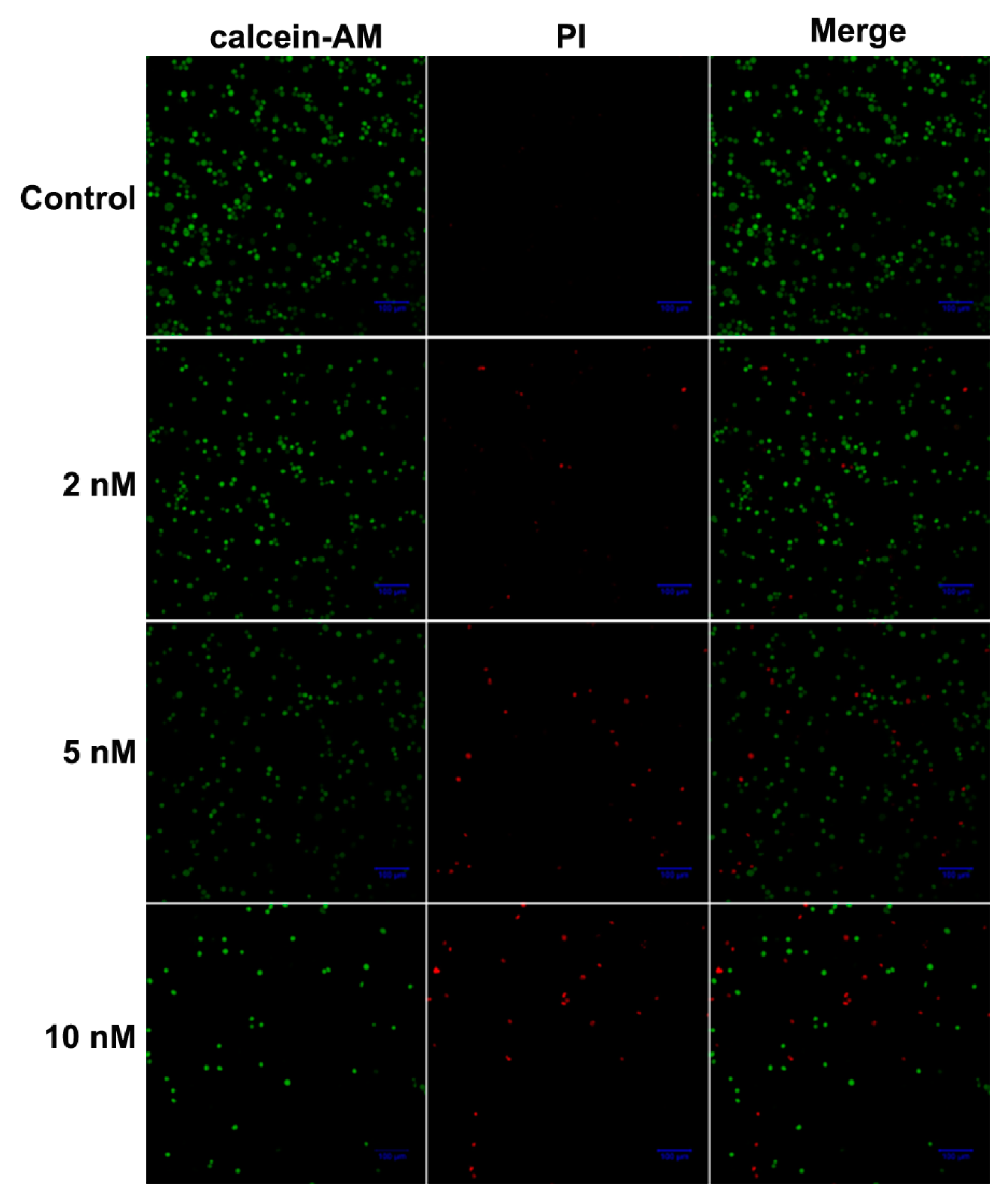
2.4. Live/Dead Staining
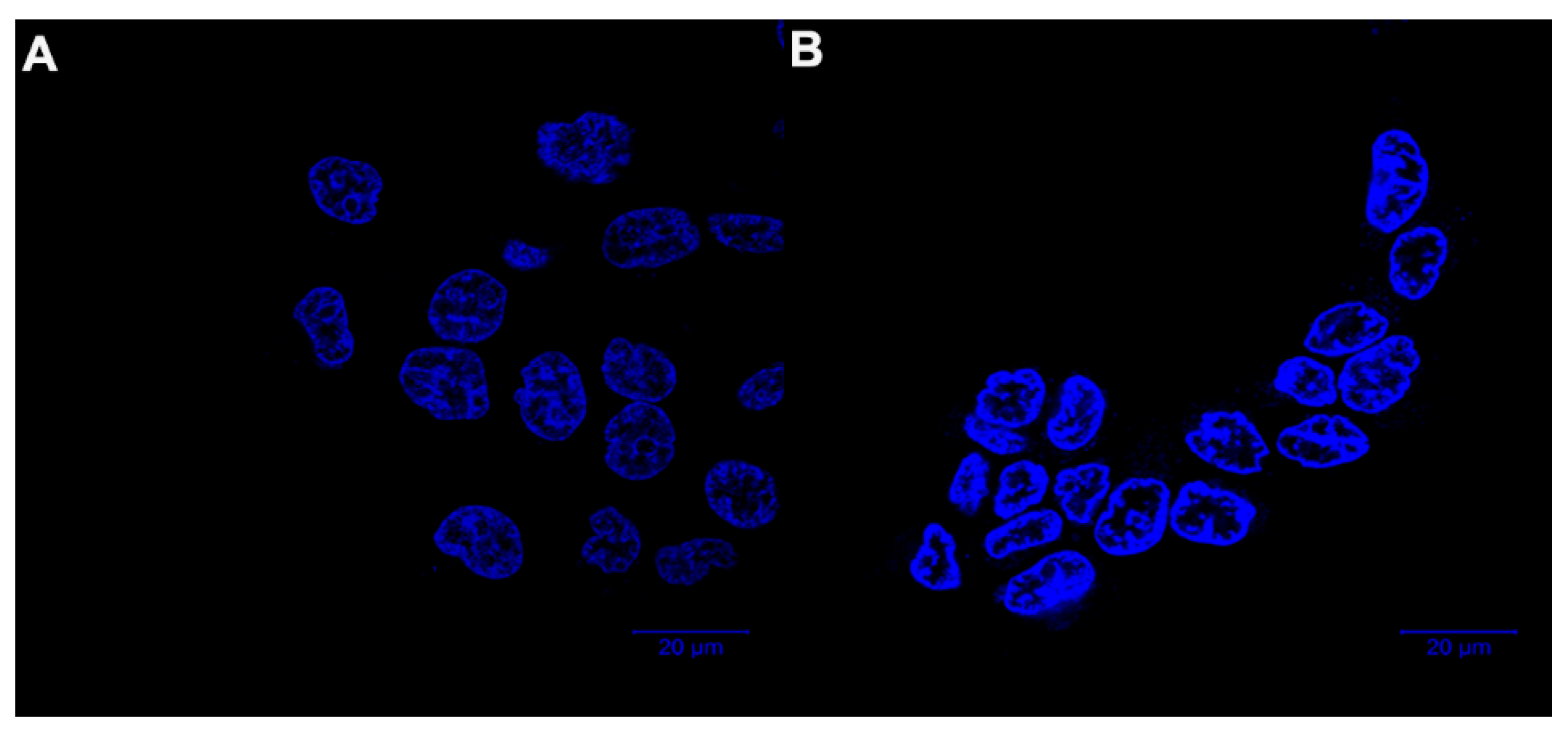
2.5. Morphological Observation by Hoechst 33,342 Staining
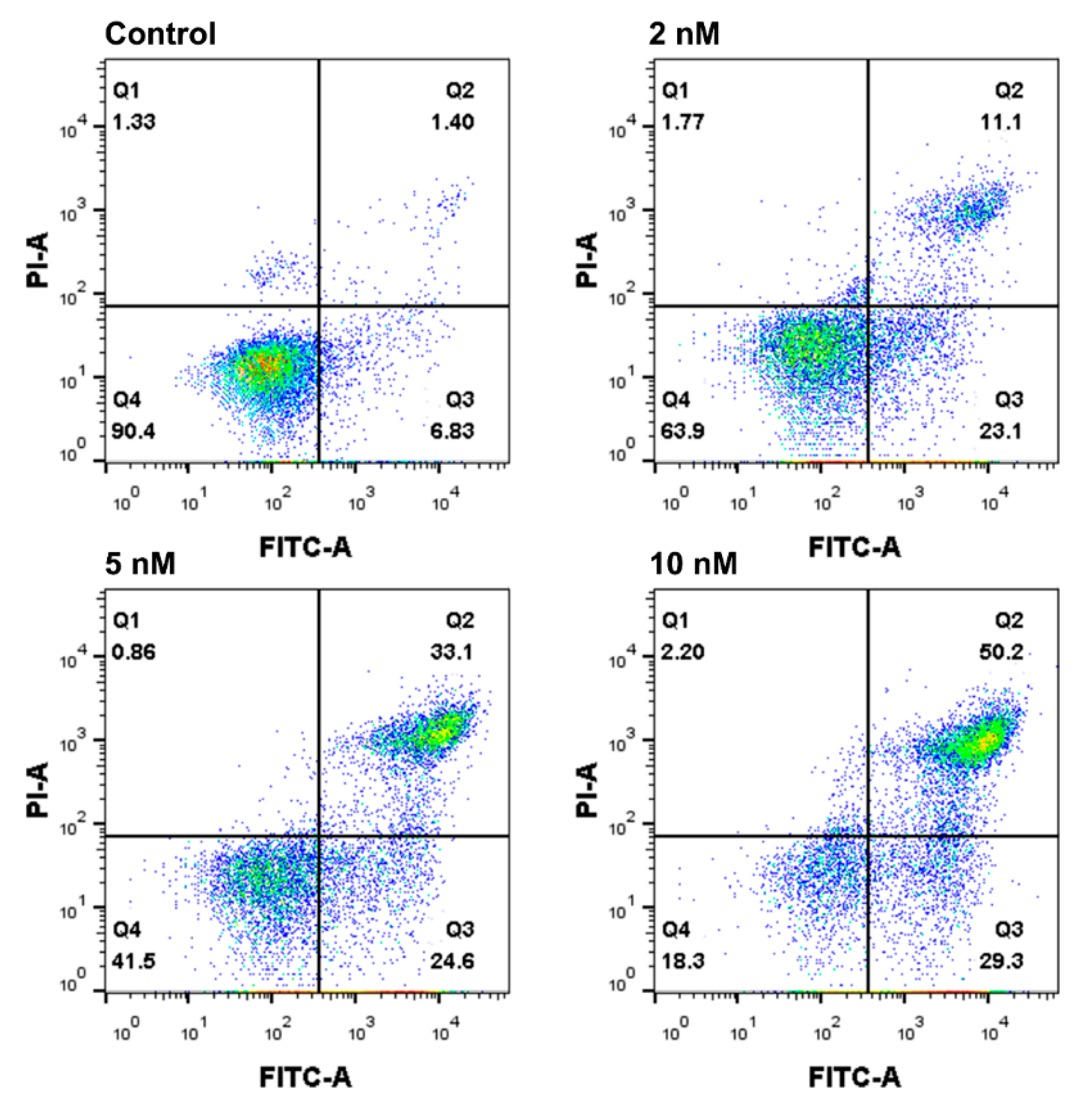
2.6. Apoptosis Analysis by Annexin V-FITC/PI Staining
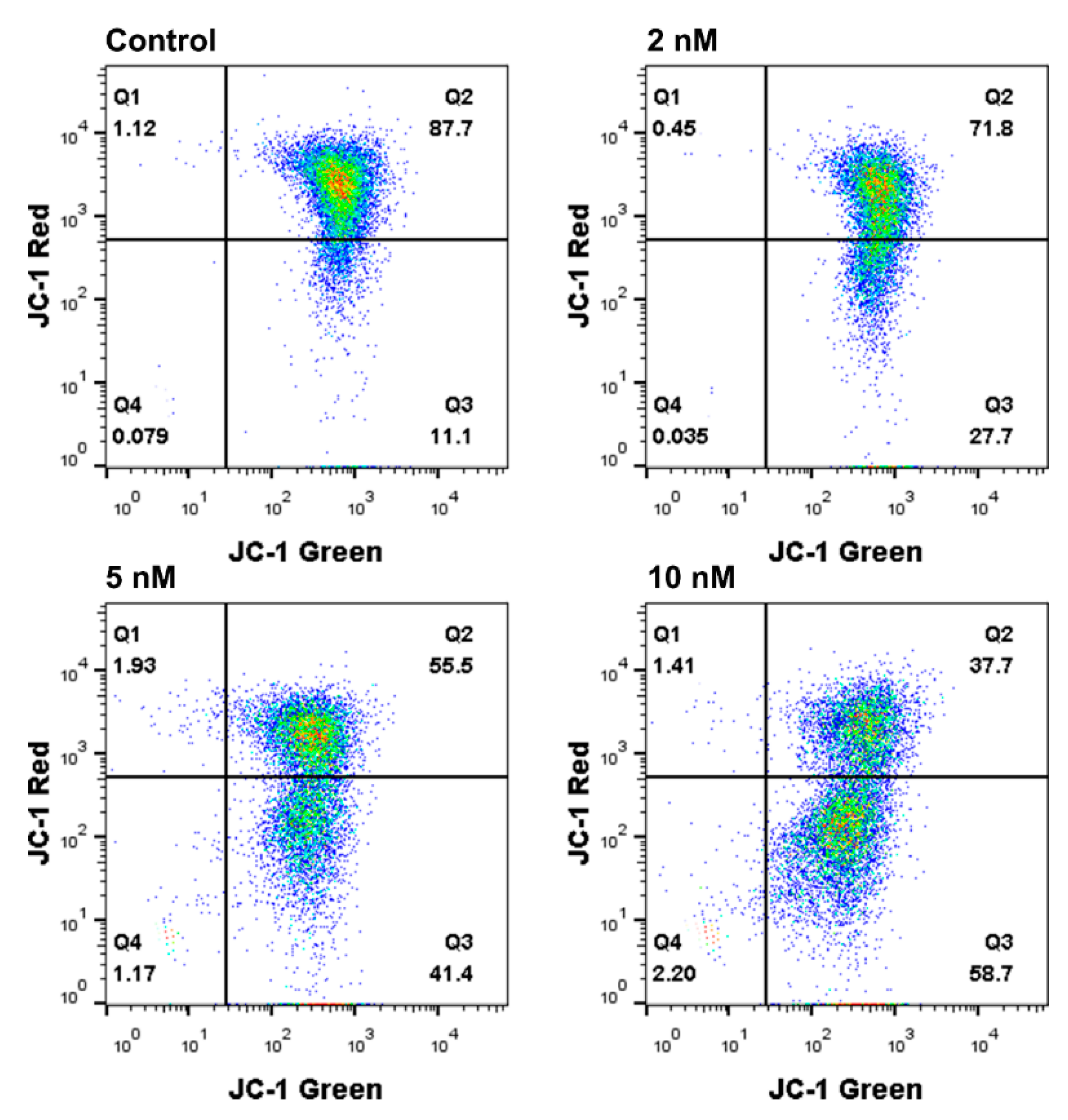
2.7. Mitochondrial Membrane Potential (ΔΨm) Analysis
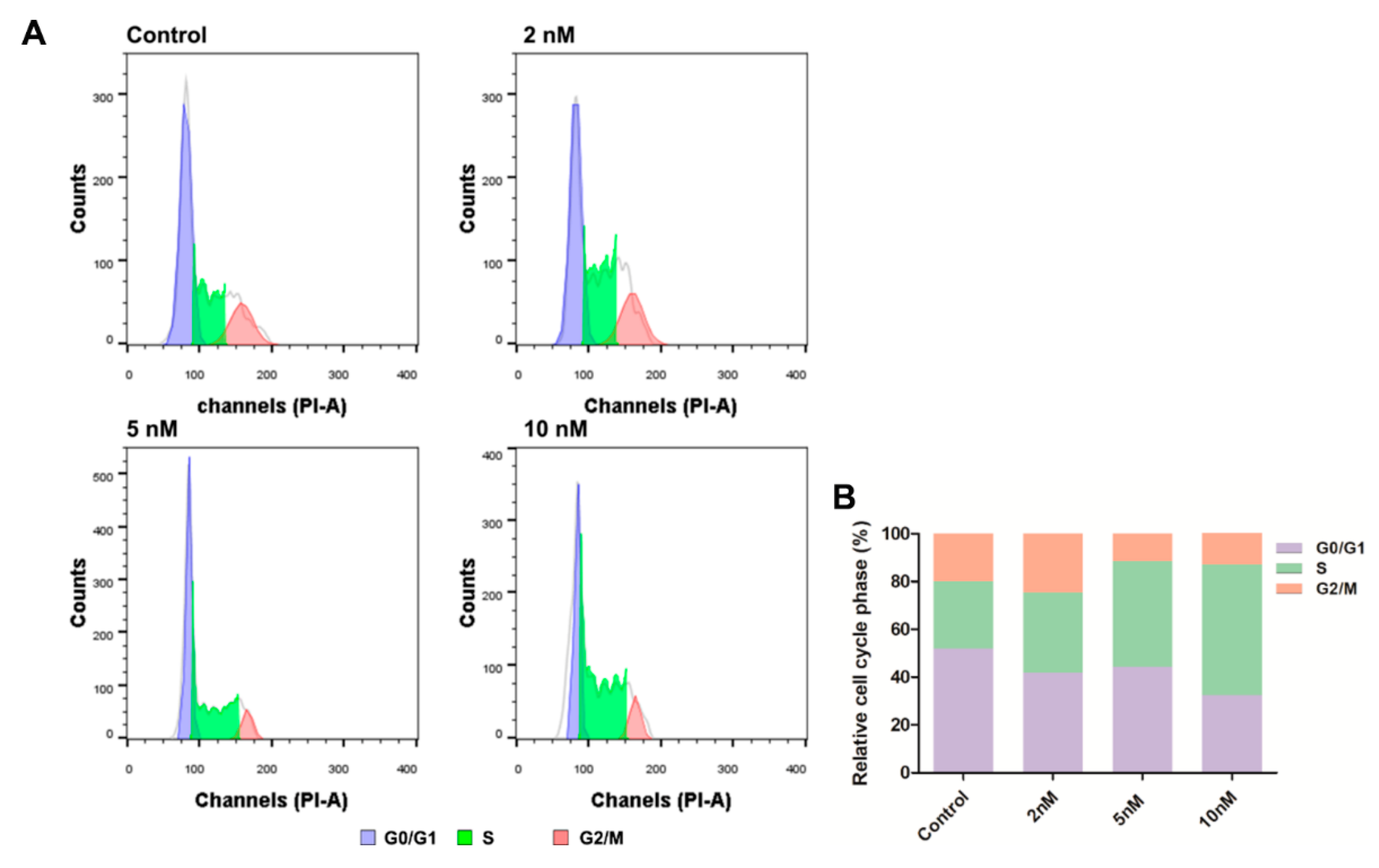
2.8. Cell Cycle Analysis
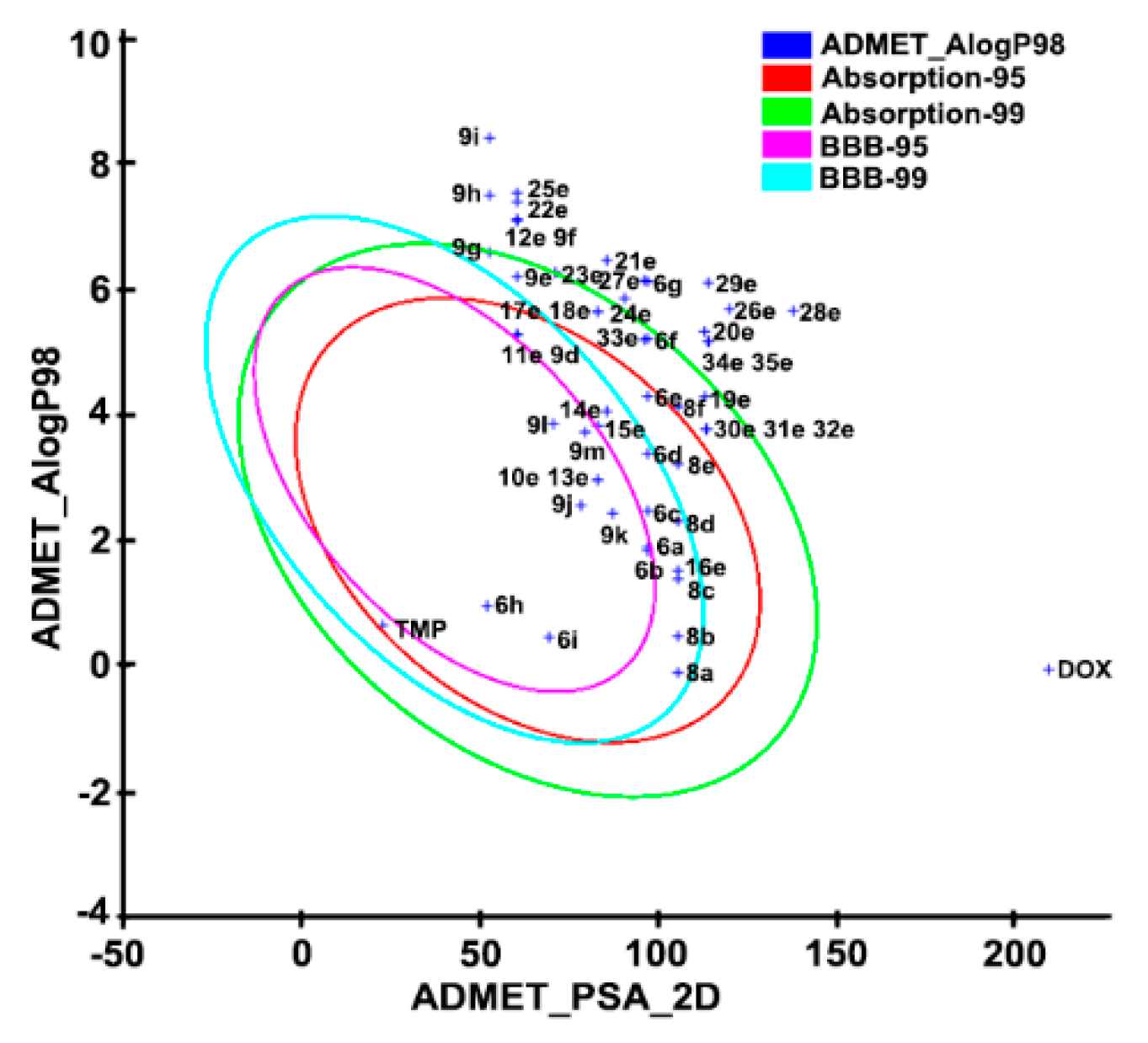
2.9. In Silico ADMET Prediction
3. Conclusions
4. Materials and Methods
4.1. Chemistry
4.2. General Synthetic Procedure for 6a–g
4.2.1. N1,N1,N2,N2-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Ethane-1,2-Diamine (6a)
4.2.2. N1,N1,N3,N3-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Propane-1,3-Diamine (6b)
4.2.3. N1,N1,N4,N4-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Butane-1,4-Diamine (6c)
4.2.4. N1,N1,N6,N6-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Hexane-1,6-Diamine (6d)
4.2.5. N1,N1,N8,N8-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Octane-1,8-Diamine (6e)
4.2.6. N1,N1,N10,N10-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Decane-1,10-Diamine (6f)
4.2.7. N1,N1,N12,N12-Tetrakis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Dodecane-1,12-Diamine (6g)
4.3. 1,4-Bis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Piperazine (6h)
4.4. 2-(Bis((3,5,6-Trimethylpyrazin-2-yl)Methyl)Amino)Ethanol (6i)
4.5. General Synthetic Procedure for 8a–f
4.5.1. N,N′-(Propane-1,3-Diyl)Bis(3,5,6-Trimethylpyrazine-2-Carboxamide) (8a)
4.5.2. N,N′-(Butane-1,4-Diyl)Bis(3,5,6-Trimethylpyrazine-2-Carboxamide) (8b)
4.5.3. N,N′-(Hexane-1,6-Diyl)Bis(3,5,6-Trimethylpyrazine-2-Carboxamide) (8c)
4.5.4. N,N′-(Octane-1,8-Diyl)Bis(3,5,6-Trimethylpyrazine-2-Carboxamide) (8d)
4.5.5. N,N′-(Decane-1,10-Diyl)Bis(3,5,6-Trimethylpyrazine-2-Carboxamide) (8e)
4.5.6. N,N′-(Dodecane-1,12-Diyl)Bis(3,5,6-Trimethylpyrazine-2-Carboxamide) (8f)
4.6. General Synthetic Procedure for 9e–35e
4.6.1. (2E,2′E)-N,N′-(Decane-1,10-Diyl)Bis(3-Phenylacrylamide) (9e)
4.6.2. N,N′-(Decane-1,10-Diyl)Dinicotinamide (10e)
4.6.3. N,N′-(Decane-1,10-Diyl)Dibenzamide (11e)
4.6.4. N,N′-(Decane-1,10-Diyl)Bis(2-Naphthamide) (12e)
4.6.5. N,N′-(Decane-1,10-Diyl)Diisonicotinamide (13e)
4.6.6. N,N′-(Decane-1,10-Diyl)Bis(Furan-2-Carboxamide) (14e)
4.6.7. N,N′-(Decane-1,10-Diyl)Dipicolinamide (15e)
4.6.8. N,N′-(Decane-1,10-Diyl)Bis(Pyrazine-2-Carboxamide) (16e)
4.6.9. N,N′-(Decane-1,10-Diyl)Bis(Quinoline-3-Carboxamide) (17e)
4.6.10. N,N′-(Decane-1,10-Diyl)Bis(Quinoline-6-Carboxamide) (18e)
4.6.11. N,N′-(Decane-1,10-Diyl)Bis(1H-Benzo[d]Imidazole-6-Carboxamide) (19e)
4.6.12. N,N′-(Decane-1,10-Diyl)Bis(2-Oxo-2H-Chromene-3-Carboxamide) (20e)
4.6.13. N,N′-(Decane-1,10-Diyl)Bis(Benzofuran-2-Carboxamide) (21e)
4.6.14. N,N′-(Decane-1,10-Diyl)Bis(Benzo[b]Thiophene-2-Carboxamide) (22e)
4.6.15. N,N′-(Decane-1,10-Diyl)Bis(1-Methyl-1H-Indole-3-Carboxamide) (23e)
4.6.16. N,N′-(Decane-1,10-Diyl)Bis(1H-Indole-6-Carboxamide) (24e)
4.6.17. (2E,2′E)-N,N′-(Decane-1,10-Diyl)Bis(3-(4-Chlorophenyl)Acrylamide) (25e)
4.6.18. (2E,2′E)-N,N′-(Decane-1,10-Diyl)Bis(3-(3-Hydroxy-4-Methoxyphenyl)Acrylamide) (26e)
4.6.19. (2E,2′E)-N,N′-(Decane-1,10-Diyl)Bis(3-(3,4-Dimethoxyphenyl)Acrylamide) (27e)
4.6.20. (2E,2′E)-N,N′-(Decane-1,10-Diyl)Bis(3-(4-Hydroxy-3,5-Dimethoxyphenyl)Acrylamide) (28e)
4.6.21. (2E,2′E)-N,N′-(Decane-1,10-Diyl)Bis(3-(3,4,5-Trimethoxyphenyl)Acrylamide) (29e)
4.6.22. N,N′-(Decane-1,10-Diyl)Bis(2-Aminobenzamide) (30e)
4.6.23. N,N′-(Decane-1,10-Diyl)Bis(4-Aminobenzamide) (31e)
4.6.24. N,N′-(Decane-1,10-Diyl)bis(3-Aminobenzamide) (32e)
4.6.25. N,N′-(Decane-1,10-Diyl)Bis(3,5-Dimethoxybenzamide) (33e)
4.6.26. N,N′-(Decane-1,10-Diyl)Bis(3,4,5-Trimethoxybenzamide) (34e)
4.6.27. N,N′-(Decane-1,10-Diyl)Bis(2,3,4-Trimethoxybenzamide) (35e)
4.7. General Synthetic Procedure for 9d, 9f
4.7.1. (2E,2′E)-N,N′-(Octane-1,8-Diyl)Bis(3-Phenylacrylamide) (9d)
4.7.2. (2E,2′E)-N,N′-(Dodecane-1,12-Diyl)Bis(3-Phenylacrylamide) (9f)
4.8. General Synthetic Procedure for 9g–9i
4.8.1. Octane-1,8-Diyl (2E,2′E)-Bis(3-Phenylacrylate) (9g)
4.8.2. Decane-1,10-Diyl (2E,2′E)-Bis(3-Phenylacrylate) (9h)
4.8.3. Dodecane-1,12-Diyl (2E,2′E)-Bis(3-Phenylacrylate) (9i)
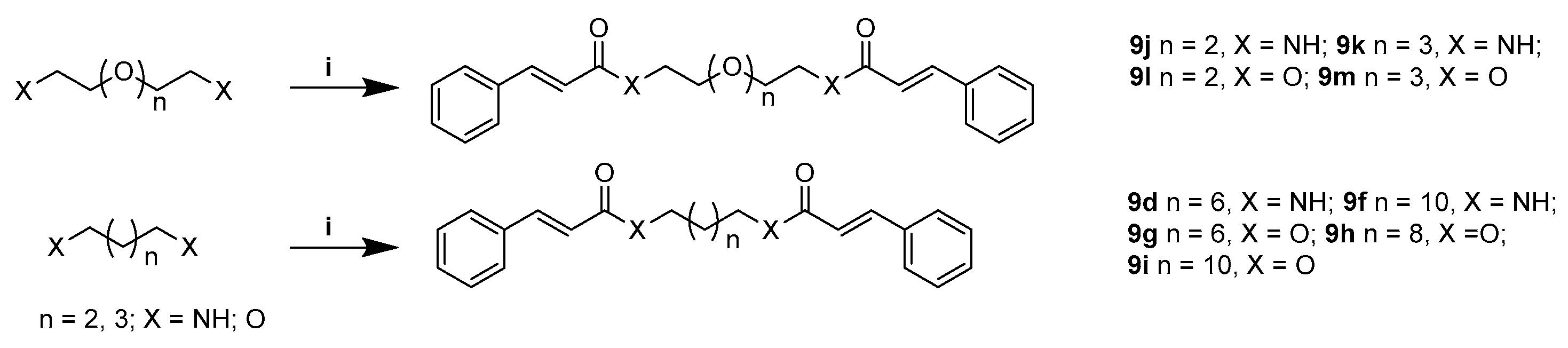
4.9. General Synthetic Procedure for 9j–9m
4.9.1. (2E,2′E)-N,N′-((Ethane-1,2-Diylbis(Oxy))Bis(Ethane-2,1-Diyl))Bis(3-Phenylacrylamide) (9j)
4.9.2. (2E,2′E)-N,N′-(((Oxybis(Ethane-2,1-Diyl))Bis(Oxy))Bis(Ethane-2,1-Diyl))Bis(3-Phenylacrylamide) (9k)
4.9.3. (Ethane-1,2-Diylbis(Oxy))Bis(Ethane-2,1-Diyl) (2E,2′E)-Bis(3-Phenylacrylate) (9l)
4.9.4. ((Oxybis(Ethane-2,1-Diyl))Bis(Oxy))Bis(Ethane-2,1-Diyl) (2E,2′E)-bis(3-Phenylacrylate) (9m)
4.10. Biological Assays
4.10.1. Cell Culture
4.10.2. Cell Viability Assay
4.10.3. Colony Formation Assay
4.10.4. Live/Dead Staining
4.10.5. Hoechst 33,342 Staining
4.10.6. Flow Cytometric Analysis of Apoptosis by Annexin V-FITC/PI Staining
4.10.7. Mitochondrial Membrane Potential (ΔΨm) Analysis
4.10.8. Cell Cycle Analysis
4.10.9. In Silico ADMET Prediction
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Et Biophys. Acta (Bba) - Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-Y.; Kao, T.-K.; Chen, W.-Y.; Ou, Y.-C.; Li, J.-R.; Liao, S.-L.; Raung, S.-L.; Chen, C.-J. Tetramethylpyrazine inhibits neutrophil activation following permanent cerebral ischemia in rats. Biochem. Biophys. Res. Commun. 2015, 463, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Miao, Q.; Miao, S.; Bi, L.; Zhang, S.; Yang, Q.; Zhou, X.; Zhang, M.; Xie, Y.; Zhang, J.; et al. Tetramethylpyrazine (TMP) exerts antitumor effects by inducing apoptosis and autophagy in hepatocellular carcinoma. Int. Immunopharmacol. 2015, 26, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Yan, X.; Chen, W.; Gao, J.; Qian, L.; Qiu, S. Antihepatocellular Carcinoma Potential of Tetramethylpyrazine Induces Cell Cycle Modulation and Mitochondrial-Dependent Apoptosis: Regulation of p53 Signaling Pathway in HepG2 Cells In Vitro. Integr. Cancer Ther. 2016, 15, 226–236. [Google Scholar] [CrossRef]
- Ai, Y.; Zhu, B.; Ren, C.; Kang, F.; Li, J.; Huang, Z.; Lai, Y.; Peng, S.; Ding, K.; Tian, J.; et al. Discovery of New Monocarbonyl Ligustrazine–Curcumin Hybrids for Intervention of Drug-Sensitive and Drug-Resistant Lung Cancer. J. Med. Chem. 2016, 59, 1747–1760. [Google Scholar] [CrossRef]
- Xu, B.; Yan, W.-Q.; Xu, X.; Wu, G.-R.; Zhang, C.-Z.; Han, Y.-T.; Chu, F.-H.; Zhao, R.; Wang, P.-L.; Lei, H.-M. Combination of amino acid/dipeptide with ligustrazine-betulinic acid as antitumor agents. Eur. J. Med. Chem. 2017, 130, 26–38. [Google Scholar] [CrossRef]
- Wang, P.-L.; Cheng, Y.-T.; Xu, K.; An, Y.-W.; Wang, W.; Li, Q.-S.; Han, Q.-J.; Li, Q.; Zhang, H.-G.; Lei, H.-M. Synthesis and Antitumor Evaluation of One Novel Tetramethylpyrazine-Rhein Derivative. Asian J. Chem. 2013, 25, 4885–4888. [Google Scholar] [CrossRef]
- Chow, L.M.C.; Chan, T.H. Novel Classes of Dimer Antitumour Drug Candidates. Curr. Pharm. Des. 2009, 15, 659–674. [Google Scholar] [CrossRef]
- Joshi, A.; Vance, D.; Rai, P.; Thiyagarajan, A.; Kane, R.S. The Design of Polyvalent Therapeutics. Chem. –A Eur. J. 2008, 14, 7738–7747. [Google Scholar] [CrossRef]
- Beekman, A.C.; Barentsen, A.R.W.; Woerdenbag, H.J.; Van Uden, W.; Pras, N.; Konings, A.W.T.; El-Feraly, F.S.; Galal, A.M.; Wikström, H.V. Stereochemistry-Dependent Cytotoxicity of Some Artemisinin Derivatives. J. Nat. Prod. 1997, 60, 325–330. [Google Scholar] [CrossRef]
- Hu, Y.; Li, C.; Kulkarni, B.A.; Strobel, G.; Lobkovsky, E.; Torczynski, R.M.; Porco, J.A. Exploring Chemical Diversity of Epoxyquinoid Natural Products: Synthesis and Biological Activity of (−)-Jesterone and Related Molecules. Org. Lett. 2001, 3, 1649–1652. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, D.T.; Goldbohm, R.A.; van Poppel, G.; Verhagen, H.; van den Brandt, P.A. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol. Biomark. Amp; Prev. 1996, 5, 733–748. [Google Scholar]
- Chaires, J.B.; Leng, F.; Przewloka, T.; Fokt, I.; Ling, Y.H.; Perezsoler, R.; Priebe, W. Structure-based design of a new bisintercalating anthracycline antibiotic. J. Med. Chem. 1997, 40, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Zha, G.-F.; Qin, H.-L.; Youssif, B.G.M.; Amjad, M.W.; Raja, M.A.G.; Abdelazeem, A.H.; Bukhari, S.N.A. Discovery of potential anticancer multi-targeted ligustrazine based cyclohexanone and oxime analogs overcoming the cancer multidrug resistance. Eur. J. Med. Chem. 2017, 135, 34–48. [Google Scholar] [CrossRef]
- Wang, P.; She, G.; Yang, Y.; Li, Q.; Zhang, H.; Liu, J.; Cao, Y.; Xu, X.; Lei, H. Synthesis and Biological Evaluation of New Ligustrazine Derivatives as Anti-Tumor Agents. Molecules 2012, 17, 4972–4985. [Google Scholar] [CrossRef]
- Xu, B.; Chu, F.; Zhang, Y.; Wang, X.; Li, Q.; Liu, W.; Xu, X.; Xing, Y.; Chen, J.; Wang, P.; et al. A Series of New Ligustrazine-Triterpenes Derivatives as Anti-Tumor Agents: Design, Synthesis, and Biological Evaluation. Int. J. Mol. Sci. 2015, 16, 21035–21055. [Google Scholar] [CrossRef]
- Taylor, A.P.; Robinson, R.P.; Fobian, Y.M.; Blakemore, D.C.; Jones, L.H.; Fadeyi, O. Modern advances in heterocyclic chemistry in drug discovery. Org. Biomol. Chem. 2016, 14, 6611–6637. [Google Scholar] [CrossRef]
- Klein, B.; Berkowitz, J. Pyrazines. I. Pyrazine-N-oxides. Preparation and Spectral Characteristics1. J. Am. Chem. Soc. 1959, 81, 5160–5166. [Google Scholar] [CrossRef]
- Cheng, X.-C.; Liu, X.-Y.; Xu, W.-F.; Guo, X.-L.; Zhang, N.; Song, Y.-N. Ligustrazine derivatives. Part 3: Design, synthesis and evaluation of novel acylpiperazinyl derivatives as potential cerebrocardiac vascular agents. Bioorganic Med. Chem. 2009, 17, 3018–3024. [Google Scholar] [CrossRef]
- Wu, G.-R.; Xu, B.; Yang, Y.-Q.; Zhang, X.-Y.; Fang, K.; Ma, T.; Wang, H.; Xue, N.-N.; Chen, M.; Guo, W.-B.; et al. Synthesis and biological evaluation of podophyllotoxin derivatives as selective antitumor agents. Eur. J. Med. Chem. 2018, 155, 183–196. [Google Scholar] [CrossRef]
- van Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cell Sensitivity Assays: The MTT Assay. In Cancer Cell Culture: Methods and Protocols; Cree, I.A., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 237–245. [Google Scholar]
- Yi, B.; Liu, D.; He, M.; Li, Q.; Liu, T.; Shao, J. Role of the ROS/AMPK signaling pathway in tetramethylpyrazine-induced apoptosis in gastric cancer cells. Oncol. Lett. 2013, 6, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zeng, L.; Pan, L.; Yuan, S.; Wu, M.; Kong, X. Tetramethylpyrazine regulates breast cancer cell viability, migration, invasion and apoptosis by affecting the activity of Akt and caspase-3. Oncol. Lett. 2018, 15, 4557–4563. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Xu, X.; Li, G.; Gu, S.; Xu, K.; Gong, Y.; Xu, B.; Wang, M.; Zhang, H.; Zhang, Y.; et al. Amino acid derivatives of ligustrazine-oleanolic acid as new cytotoxic agents. Molecules 2014, 19, 18215–18231. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Zhang, L.; Yu, H.-X.; Lu, R.-R.; Bao, J.-D.; Tan, C.; Sun, Z. The mechanism underlying proliferation-inhibitory and apoptosis-inducing effects of curcumin on papillary thyroid cancer cells. Food Chem. 2012, 132, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Meena, A.; Yadav, D.K.; Srivastava, A.; Khan, F.; Chanda, D.; Chattopadhyay, S.K. In Silico Exploration of Anti-Inflammatory Activity of Natural Coumarinolignoids. Chem. Biol. Drug Des. 2011, 78, 567–579. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminformatics 2018, 10, 29. [Google Scholar] [CrossRef]
- Egan, W.J.; Lauri, G. Prediction of intestinal permeability. Adv. Drug Deliv. Rev. 2002, 54, 273–289. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
Sample Availability: Samples of all compounds are available from the authors. |
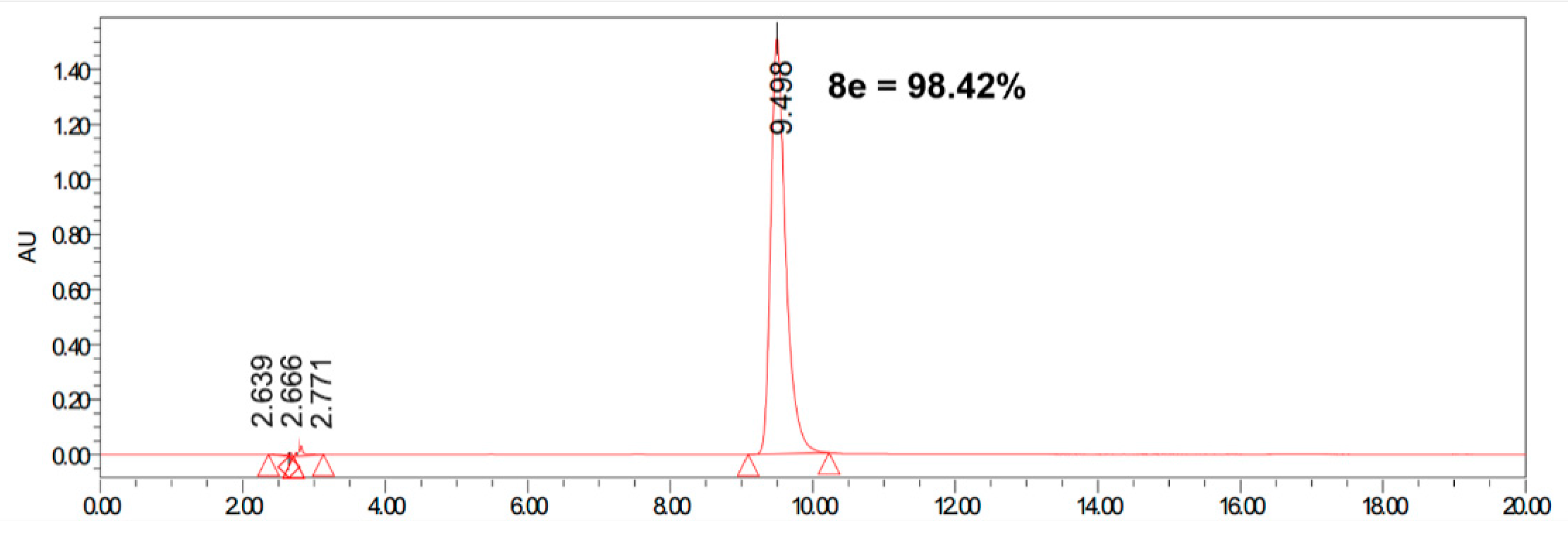
| Comp. | Antiproliferative Activity IC50 ± SEM (µM) | ||||||
|---|---|---|---|---|---|---|---|
| HeLa[a] | Hep G2[b] | MCF-7[c] | FaDu[d] | A549[e] | MCF 10A[f] | SI[g] | |
| 6a | >100 | >100 | >100 | >100 | >100 | 90.29 ± 3.41 | NCh |
| 6b | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 6c | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 6d | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 6e | 62.67 ± 4.19 | 50.77 ± 1.35 | 66.36 ± 0.05 | 49.50 ± 1.61 | >100 | 85.44 ± 2.28 | 1.73 |
| 6f | 10.16 ± 0.55 | 10.72 ± 0.22 | 20.83 ± 0.14 | 10.94 ± 0.12 | 20.05 ± 0.41 | 12.97 ± 0.55 | 1.19 |
| 6g | 7.89 ± 0.86 | 9.62 ± 0.11 | 9.83 ± 0.17 | 6.80 ± 0.05 | 9.23 ± 0.16 | 6.57 ± 0.30 | 0.97 |
| 6h | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 6i | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 8a | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 8b | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| 8c | >100 | >100 | >100 | 80.41 ± 1.36 | >100 | 46.59 ± 0.65 | NCh |
| 8d | 3.31 ± 0.14 | >20 | 0.480 ± 0.003 | 0.11 ± 0.03 | 6.35 ± 0.05 | 2.69 ± 0.46 | 24.45 |
| 8e | 1.42 ± 0.71 | >20 | 0.037 ± 0.001 | 0.00136 ± 0.00035 | 1.05 ± 0.05 | 0.047 ± 0.008 | 34.56 |
| 8f | 4.96 ± 1.62 | >20 | 0.158 ± 0.009 | 0.174 ± 0.010 | 2.46 ± 0.05 | 0.051 ± 0.021 | 0.29 |
| 9e | >100 | >100 | >100 | 0.054 ± 0.002 | 11.27 ± 0.81 | >100 | >1851.85 |
| 10e | >100 | >100 | >100 | 0.25 ± 0.02 | 2.94 ± 0.25 | 38.60 ± 2.48 | 154.40 |
| 11e | >100 | >100 | >100 | 0.50 ± 0.06 | 64.30 ± 3.46 | 80.54 ± 7.60 | 161.08 |
| 12e | >100 | >100 | >100 | 1.33 ± 0.07 | 8.77 ± 0.47 | >100 | >75.19 |
| 13e | >100 | >100 | >100 | 48.31 ± 3.17 | >100 | >100 | >2.07 |
| 14e | >100 | 97.75 ± 1.55 | >100 | 6.33 ± 0.65 | 44.37 ± 0.29 | 19.82 ± 1.41 | 3.13 |
| 15e | 37.74 ± 4.32 | 26.34 ± 5.36 | 21.48 ± 1.47 | 8.85 ± 0.45 | 23.08 ± 0.12 | 54.65 ± 5.92 | 6.18 |
| 16e | >100 | >100 | >100 | 88.11 ± 1.37 | >100 | >100 | NCh |
| 17e | >20 | >20 | >20 | 0.236 ± 0.005 | >20 | >20 | >84.75 |
| 18e | >20 | >20 | >20 | 0.697 ± 0.021 | >20 | >20 | >28.69 |
| 19e | >20 | >20 | >20 | 8.74 ± 0.44 | >20 | >20 | >2.29 |
| 20e | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 21e | >20 | >20 | >20 | 0.665 ± 0.028 | >20 | >20 | >30.08 |
| 22e | >20 | >20 | >20 | 0.020 ± 0.002 | >20 | >20 | >1000 |
| 23e | >20 | >20 | >20 | 0.638 ± 0.089 | >20 | >20 | >31.35 |
| 24e | >20 | >20 | >20 | 5.296 ± 0.366 | >20 | >20 | >3.78 |
| 25e | >20 | >20 | >20 | 0.056 ± 0.016 | >20 | >20 | >357.14 |
| 26e | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 27e | >20 | >20 | >20 | 4.621 ± 0.539 | >20 | >20 | >4.33 |
| 28e | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 29e | >20 | >20 | >20 | 5.067 ± 0.461 | >20 | >20 | >3.95 |
| 30e | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 31e | >20 | >20 | >20 | 5.594 ± 0.628 | >20 | >20 | >3.58 |
| 32e | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 33e | >20 | >20 | >20 | 10.393 ± 0.949 | >20 | >20 | >1.92 |
| 34e | >20 | >20 | >20 | 5.853 ± 0.408 | >20 | >20 | >3.42 |
| 35e | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 9d | >20 | >20 | >20 | 1.054 ± 0.091 | >20 | >20 | >18.98 |
| 9f | >20 | >20 | >20 | 0.027 ± 0.002 | >20 | >20 | >740.74 |
| 9g | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 9h | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 9i | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 9j | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 9k | >20 | >20 | >20 | 4.447 ± 0.208 | >20 | >20 | >4.50 |
| 9l | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| 9m | >20 | >20 | >20 | >20 | >20 | >20 | NCh |
| TMP | >100 | >100 | >100 | >100 | >100 | >100 | NCh |
| DOX | 11.39 ± 0.48 | 28.04 ± 1.09 | 6 ± 0.07 | 1.27 ± 0.13 | >100 | 0.55 ± 0.03 | 0.43 |
| Comp. | HBDa | HBAb | RBNc | logPd | logSe | PSAf | HIAg | PPBh | BBBi | CYP3A4j | T1/2 (h)k | HTl | LD50(mg/kg)m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6a | 0 | 10 | 11 | 1.84 | −2.23 | 96.79 | 0.81 | 0.68 | 0.94 | 0 | 2.02 | 0 | 459.04 |
| 6b | 0 | 10 | 12 | 1.90 | −1.93 | 96.79 | 0.80 | 0.67 | 0.91 | 0 | 2.13 | 0 | 429.48 |
| 6c | 0 | 10 | 13 | 2.48 | −2.05 | 96.79 | 0.80 | 0.68 | 0.91 | 0 | 2.14 | 0 | 462.17 |
| 6d | 0 | 10 | 15 | 3.39 | −2.02 | 96.79 | 0.80 | 0.65 | 0.91 | 0 | 2.12 | 0 | 447.58 |
| 6e | 0 | 10 | 17 | 4.31 | −1.90 | 96.79 | 0.80 | 0.65 | 0.91 | 0 | 2.14 | 0 | 425.74 |
| 6f | 0 | 10 | 19 | 5.22 | −1.67 | 96.79 | 0.80 | 0.66 | 0.91 | 0 | 2.18 | 0 | 460.97 |
| 6g | 0 | 10 | 21 | 6.13 | −1.35 | 96.79 | 0.80 | 0.67 | 0.91 | 0 | 2.23 | 0 | 508.77 |
| 6h | 0 | 6 | 4 | 0.97 | −2.49 | 51.75 | 0.79 | 0.67 | 0.98 | 0 | 2.00 | 0 | 664.69 |
| 6i | 1 | 6 | 6 | 0.47 | −1.52 | 69.21 | 0.78 | 0.70 | 0.75 | 0 | 1.77 | 0 | 1011.08 |
| 8a | 2 | 6 | 6 | −0.09 | −1.53 | 105.27 | 0.69 | 0.78 | 0.98 | 0 | 1.81 | 1 | 1027.40 |
| 8b | 2 | 6 | 7 | 0.49 | −1.87 | 105.27 | 0.72 | 0.79 | 0.98 | 0 | 1.88 | 1 | 988.28 |
| 8c | 2 | 6 | 9 | 1.40 | −2.30 | 105.27 | 0.72 | 0.80 | 0.98 | 0 | 1.89 | 1 | 1033.87 |
| 8d | 2 | 6 | 11 | 2.31 | −2.67 | 105.27 | 0.72 | 0.83 | 0.98 | 0 | 1.92 | 1 | 1099.10 |
| 8e | 2 | 6 | 13 | 3.22 | −2.97 | 105.27 | 0.72 | 0.85 | 0.98 | 0 | 2.03 | 1 | 1278.93 |
| 8f | 2 | 6 | 15 | 4.14 | −3.21 | 105.27 | 0.72 | 0.85 | 0.98 | 0 | 2.01 | 1 | 1318.54 |
| 9e | 2 | 2 | 15 | 6.21 | −4.45 | 60.22 | 0.69 | 0.89 | 0.99 | 0 | 1.93 | 1 | 961.82 |
| 10e | 2 | 4 | 13 | 2.97 | −2.60 | 82.74 | 0.70 | 0.91 | 0.99 | 0 | 1.85 | 1 | 1075.58 |
| 11e | 2 | 2 | 13 | 5.27 | −4.27 | 60.22 | 0.68 | 0.91 | 1.00 | 0 | 1.96 | 1 | 1653.45 |
| 12e | 2 | 2 | 13 | 7.09 | −6.11 | 60.22 | 0.69 | 0.92 | 0.99 | 0 | 2.12 | 1 | 1818.99 |
| 13e | 2 | 4 | 13 | 2.97 | −2.60 | 82.74 | 0.72 | 0.91 | 0.99 | 0 | 1.89 | 0 | 1070.63 |
| 14e | 2 | 2 | 13 | 4.06 | −3.66 | 85.33 | 0.41 | 0.90 | 1.00 | 0 | 1.78 | 0 | 1145.12 |
| 15e | 2 | 4 | 13 | 3.83 | −3.35 | 82.74 | 0.70 | 0.91 | 0.99 | 0 | 1.89 | 1 | 1128.86 |
| 16e | 2 | 6 | 13 | 1.53 | −1.68 | 105.27 | 0.68 | 0.85 | 0.99 | 0 | 1.82 | 1 | 1461.77 |
| 17e | 2 | 4 | 13 | 5.65 | −5.10 | 82.74 | 0.70 | 0.95 | 0.98 | 0 | 2.22 | 1 | 1217.90 |
| 18e | 2 | 4 | 13 | 5.65 | −5.10 | 82.74 | 0.72 | 0.96 | 0.95 | 0 | 1.99 | 1 | 1254.91 |
| 19e | 4 | 4 | 13 | 4.31 | −4.38 | 112.85 | 0.69 | 0.90 | 0.94 | 0 | 1.91 | 1 | 1167.64 |
| 20e | 2 | 6 | 13 | 5.33 | −4.57 | 112.68 | 0.43 | 0.93 | 0.98 | 0 | 2.29 | 0 | 973.08 |
| 21e | 2 | 2 | 13 | 6.46 | −6.02 | 85.33 | 0.47 | 0.94 | 0.99 | 0 | 2.00 | 1 | 966.72 |
| 22e | 2 | 2 | 13 | 7.40 | −6.99 | 60.22 | 0.69 | 0.91 | 0.99 | 0 | 2.07 | 1 | 904.88 |
| 23e | 2 | 2 | 13 | 6.27 | −5.55 | 70.92 | 0.75 | 0.95 | 0.99 | 0 | 2.27 | 1 | 995.92 |
| 24e | 4 | 2 | 13 | 5.86 | −5.15 | 90.33 | 0.72 | 0.95 | 0.96 | 0 | 2.01 | 1 | 1072.61 |
| 25e | 2 | 2 | 15 | 7.54 | −5.78 | 60.22 | 0.67 | 0.89 | 0.99 | 0 | 1.98 | 1 | 1322.11 |
| 26e | 4 | 6 | 17 | 5.69 | −3.28 | 119.71 | 0.45 | 0.87 | 0.85 | 0 | 1.93 | 1 | 1249.90 |
| 27e | 2 | 6 | 19 | 6.14 | −3.64 | 95.94 | 0.54 | 0.82 | 0.98 | 0 | 1.82 | 1 | 1217.58 |
| 28e | 4 | 8 | 19 | 5.66 | −3.13 | 137.57 | 0.46 | 0.77 | 0.94 | 0 | 2.02 | 1 | 1163.97 |
| 29e | 2 | 8 | 21 | 6.11 | −3.08 | 113.80 | 0.54 | 0.73 | 0.96 | 0 | 1.95 | 0 | 927.46 |
| 30e | 4 | 4 | 13 | 3.78 | −2.74 | 113.30 | 0.59 | 0.90 | 0.99 | 0 | 1.96 | 1 | 849.92 |
| 31e | 4 | 4 | 13 | 3.78 | −2.69 | 113.30 | 0.64 | 0.90 | 0.97 | 0 | 1.93 | 0 | 951.43 |
| 32e | 4 | 4 | 13 | 3.78 | −2.72 | 113.30 | 0.64 | 0.90 | 0.98 | 0 | 1.94 | 0 | 853.84 |
| 33e | 2 | 6 | 17 | 5.21 | −3.97 | 95.94 | 0.53 | 0.81 | 0.99 | 0 | 1.76 | 0 | 1184.47 |
| 34e | 2 | 8 | 19 | 5.18 | −3.62 | 113.80 | 0.54 | 0.76 | 0.98 | 0 | 1.63 | 0 | 1073.30 |
| 35e | 2 | 8 | 19 | 5.18 | −3.67 | 113.80 | 0.53 | 0.76 | 0.97 | 0 | 1.72 | 0 | 1061.01 |
| 9d | 2 | 2 | 13 | 5.30 | −4.11 | 60.22 | 0.69 | 0.91 | 0.99 | 0 | 1.92 | 1 | 1523.97 |
| 9f | 2 | 2 | 17 | 7.12 | −4.73 | 60.22 | 0.69 | 0.87 | 0.99 | 0 | 1.95 | 0 | 1003.19 |
| 9g | 0 | 4 | 15 | 6.59 | −5.34 | 52.46 | 0.55 | 0.88 | 0.98 | 0 | 1.96 | 1 | 6181.36 |
| 9h | 0 | 4 | 17 | 7.50 | −5.69 | 52.46 | 0.55 | 0.86 | 0.98 | 0 | 1.96 | 1 | 4624.51 |
| 9i | 0 | 4 | 19 | 8.42 | −5.96 | 52.46 | 0.55 | 0.85 | 0.98 | 0 | 1.96 | 0 | 3127.76 |
| 9j | 2 | 4 | 13 | 2.57 | −2.06 | 78.08 | 0.37 | 0.86 | 0.99 | 0 | 1.97 | 1 | 1574.67 |
| 9k | 2 | 5 | 16 | 2.44 | −1.41 | 87.01 | 0.37 | 0.85 | 0.99 | 0 | 1.90 | 0 | 1693.04 |
| 9l | 0 | 6 | 15 | 3.86 | −2.96 | 70.32 | 0.50 | 0.88 | 0.98 | 0 | 1.89 | 1 | 4511.05 |
| 9m | 0 | 7 | 18 | 3.73 | −2.14 | 79.25 | 0.50 | 0.86 | 0.98 | 0 | 1.83 | 0 | 4493.16 |
| DOX | 6 | 12 | 5 | −0.04 | −4.80 | 209.31 | 0.02 | 0.78 | 0.02 | 0 | 2.71 | 1 | 324.50 |
| TMP | 0 | 2 | 0 | 0.66 | −1.51 | 22.52 | 0.93 | 0.49 | 0.99 | 0 | 1.77 | 0 | 1194.46 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Hong, G.; Li, G.; Wang, W.; Liu, T. Novel Homo-Bivalent and Polyvalent Compounds Based on Ligustrazine and Heterocyclic Ring as Anticancer Agents. Molecules 2019, 24, 4505. https://doi.org/10.3390/molecules24244505
Wang J, Hong G, Li G, Wang W, Liu T. Novel Homo-Bivalent and Polyvalent Compounds Based on Ligustrazine and Heterocyclic Ring as Anticancer Agents. Molecules. 2019; 24(24):4505. https://doi.org/10.3390/molecules24244505
Chicago/Turabian StyleWang, Jiawen, Ge Hong, Guoliang Li, Wenzhi Wang, and Tianjun Liu. 2019. "Novel Homo-Bivalent and Polyvalent Compounds Based on Ligustrazine and Heterocyclic Ring as Anticancer Agents" Molecules 24, no. 24: 4505. https://doi.org/10.3390/molecules24244505
APA StyleWang, J., Hong, G., Li, G., Wang, W., & Liu, T. (2019). Novel Homo-Bivalent and Polyvalent Compounds Based on Ligustrazine and Heterocyclic Ring as Anticancer Agents. Molecules, 24(24), 4505. https://doi.org/10.3390/molecules24244505