Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability
Abstract
:1. Introduction
2. Results and Discussion
2.1. Two-Pulse Electron Spin Echo Envelope Modulation (ESEEM) Measurements
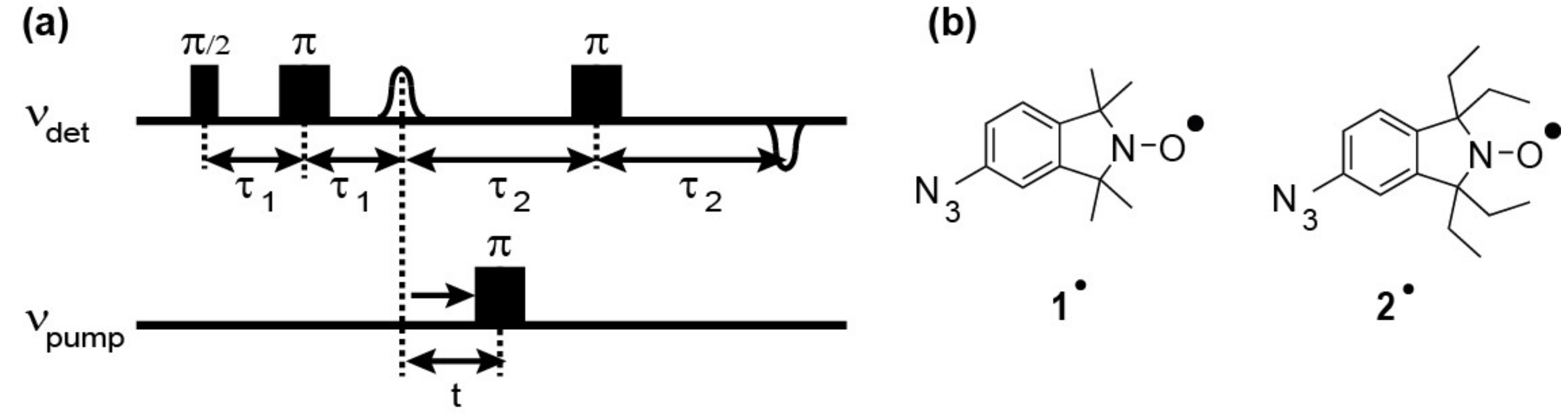
2.2. PELDOR Measurements
3. Materials and Methods
3.1. General Procedures RNA
3.1.1. RNA Sequence
3.1.2. Spin Labeling
3.1.3. EPR Sample Preparation
3.2. Chromatography
3.2.1. HPLC
3.2.2. LCMS
3.3. Spectroscopy
3.3.1. IR
3.3.2. CD
3.3.3. UV
3.4. EPR
3.4.1. cw EPR
3.4.2. Pulsed EPR
3.5. Stability Measurements
3.5.1. Sample Preparation for Measurements with Ascorbic Acid
3.5.2. HeLa Cell Lysis Preparation
3.5.3. Sample Preparation for Measurements in HeLa Lysate
3.5.4. EPR Stability Measurements
3.5.5. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Karijolich, J.; Yu, Y.-T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science: New York, NY, USA, 2014; ISBN 978-0-8153-4432-2. [Google Scholar]
- Lilley, D.M.J.; Eckstein, F. Ribozymes and RNA Catalysis; The Royal Society of Chemistry: Cambridge, UK, 2008; ISBN 978-0-85404-253-1. [Google Scholar]
- Breaker, R.R. Riboswitches and Translational Control. CSH Perspect. Biol. 2018, 10, 1–14. [Google Scholar] [CrossRef]
- Reyes, F.E.; Garst, A.D.; Batey, R.T. Strategies in RNA Crystallography. In Methods in Enzymology; Elsevier: San Diego, CA, USA, 2009; Chapter 6; pp. 119–139. ISBN 978-0-12-380922-3. [Google Scholar]
- Zheng, L.; Falschlunger, C.; Huang, K.; Mairhofer, E.; Yuan, S.; Wang, J.; Patel, D.J.; Micura, R.; Rem, A. Hatchet ribozyme structure and implications for cleavage mechanism. Proc. Natl. Acad. Sci. USA 2019, 116, 10783–10791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbert, H. CryoEM: A crystals to single particles round-trip. Curr. Opin. Struct. Biol. 2019, 58, 59–67. [Google Scholar] [CrossRef]
- Kaledhonkar, S.; Fu, Z.; Caban, K.; Li, W.; Chen, B.; Sun, M.; Gonzalez, R.L., Jr.; Frank, J. Late steps in bacterial translation initiation vizualized using time-resolved cryo-EM. Nature 2019, 570, 400–404. [Google Scholar] [CrossRef]
- Thompson, R.D.; Baisden, J.T.; Zhang, Q. NMR Characterization of RNA Small Molecule Interactions. Methods 2019, 167, 66–77. [Google Scholar] [CrossRef]
- Garcia-Lopez, A.; Tessaro, F.; Jonker, H.R.A.; Wacker, A.; Richter, C.; Comte, A.; Berntenis, N.; Schmucki, R.; Hatje, K.; Petermann, O.; et al. Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Goldfarb, D.; Stoll, S. EPR Spectroscopy: Fundamentals and Methods; John Wiley & Sons: Hoboken, NJ, USA, 2018; ISBN 978-1-119-16299-5. [Google Scholar]
- Hetzke, T.; Vogel, M.; Gophane, D.B.; Weigand, J.E.; Suess, B.; Sigurdsson, S.T.; Prisner, T.F. Influence of Mg2+ on the conformational flexibility of a tetracycline aptamer. RNA 2018, 25, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Lerner, E.; Corder, T.; Ingargiola, A.; Alhadid, Y.; Chung, S.Y.; Michalet, X.; Weiss, S. Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 2018, 359, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Andreo, A.Z.; Harm, U.; Klostermeier, D. Single-stranded regions modulate conformational dynamics and ATPase activity of elF4A to optimize 5′-UTR unwinding. Nucleic Acids Res. 2019, 47, 5260–5275. [Google Scholar] [CrossRef]
- Chen, Y.; Pollack, L. SAXS Studies of RNA: Structures, dynamics, and interactions with partners. WIRES RNA 2016, 7, 512–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šponer, J.; Bussi, G.; Krepl, M.; Banáš, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurečka, P.; et al. RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunsicker-Wang, L.; Vogt, M.; DeRose, V.J. EPR Methods to Study Specific Metal-Ion Binding Sites in RNA. Meth. Enzymol. 2009, 468, 335–367. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, N.; Kraut, S.; Jäschke, A.; Schiemann, O. Characterizing multiple metal ion binding sites within a ribozyme by cadmium induced EPR silencing. HFSP J. 2007, 1, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Shelke, S.A.; Sigurdsson, S.T. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Structure and Bonding; Timmel, C., Harmer, C., Anderson, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 152, pp. 121–162. [Google Scholar] [CrossRef]
- Haugland, M.M.; Lovett, J.E.; Anderson, E.A. Advances in the synthesis of nitroxide radicals for use in biomolecule spin labelling. Chem. Soc. Rev. 2018, 47, 668–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevelev, G.Y.; Krumkacheva, O.A.; Lomzov, A.A.; Kuzhelev, A.A.; Rogozhinkova, O.Y.; Trukhin, D.V.; Troitykaya, T.I.; Tormyshev, V.M.; Fedin, M.V.; Pyshnyi, D.V.; et al. Physiological-Temperature Distance Measurement in Nucleic Acid using Triarylmethyl-Based Spin Labels and Pulsed Dipolar EPR Spectroscopy. J. Am. Chem. Soc. 2014, 134, 9874–9877. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y.; Borbat, P.; Zweier, J.L.; Freed, J.H.; Hubbell, W.L. Pulsed ESR Dipolar Spectroscopy for Distance Measurements in Immobilized Spin Labeled Proteins in Liquid Solution. J. Am. Chem. Soc. 2012, 134, 9950–9952. [Google Scholar] [CrossRef] [Green Version]
- Reginsson, G.W.; Kunjir, N.C.; Sigurdsson, S.T.; Schiemann, O. Trityl Radicals: Spin Labels for Nanometer-Distance Measurements. Chem. Eur. J. 2012, 18, 13580–13584. [Google Scholar] [CrossRef]
- Goldfarb, D. Gd3+ spin labeling for distance measurements by pulse EPR spectroscopy. Phys. Chem. Chem. Phys. 2014, 16, 9685–9699. [Google Scholar] [CrossRef]
- Wojciechowski, F.; Groß, A.; Holder, I.T.; Knörr, L.; Drescher, M.; Hartig, J.S. Pulsed EPR spectroscopy distance measurements of DNA internally labelled with Gd3+-DOTA. Chem. Commun. 2015, 51, 13850–13853. [Google Scholar] [CrossRef] [Green Version]
- Schiemann, O.; Weber, A.; Edwards, T.E.; Prisner, T.F.; Sigurdsson, S.T. Nanometer Distance Measurements on RNA Using PELDOR. J. Am. Chem. Soc. 2003, 125, 3435. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Jagtap, A.P.; Sigurdsson, S.T. Site-directed spin labeling of 2′-amino groups in RNA with isoindoline nitroxides that are resistant to reduction. Chem. Commun. 2015, 51, 13142–13145. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.Z.; Butcher, S.E.; Feigon, J.; Hubbell, W.L. Quantitative Analysis of the Isolated GAAA Tetraloop/Receptor Interaction in Solution: A Site-Directed Spin Labeling Study. Biochemistry 2001, 40, 6929–6936. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Popova, A.M.; Hidgen, K.; Qin, P.Z. A nucleotide-independent cyclic nitroxide label for monitoring segmental motions in nucleic acids. BMC Biophys. 2015, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Esquiaqui, J.M.; Sherman, E.M.; Ionescu, S.A.; Ye, J.-D.; Fanucci, G.E. characterizating the Dynamics of the Leader-Linker Interaction in the Glycine Riboswitch with Site-Directed Spin Labeling. Biochemistry 2014, 53, 3526–3528. [Google Scholar] [CrossRef]
- Sicoli, G.; Wachowius, F.; Bennati, M.; Höbartner, C. Probing Secondary Structures of Spin-Labeled RNA by Pulsed EPR Spectroscopy. Angew. Chem. Int. Ed. 2010, 49, 6443–6447. [Google Scholar] [CrossRef] [Green Version]
- Schiemann, K.; Piton, N.; Plackmeyer, J.; Bode, B.E.; Prisner, T.F.; Engels, J.W. Spin labeling of oligonucleotides with the nitroxide TPA and use of PELDOR, a pulse EPR method, to measure intramolecular distances. Nat. Protoc. 2007, 2, 904–923. [Google Scholar] [CrossRef]
- Höbartner, C.; Sicoli, G.; Wachowius, F.; Gophane, D.B.; Sigurdsson, S.T. Synthesis and Characterization of RNA Containing a Rigid and Nonperturbing Cytidine-Derived Spin Label. J. Org. Chem. 2012, 77, 7749–7754. [Google Scholar] [CrossRef]
- Kerzhner, M.; Abdullin, D.; Więcek, J.; Matsuoka, H.; Hagelueken, G.; Schiemann, O.; Famulok, M. Post-synthetic Spin Labeling of RNA through Click Chemistry for PELDOR Measurements. Chem. Eur. J. 2016, 22, 12113–12121. [Google Scholar] [CrossRef]
- Weinreich, T.; Jaumann, E.A.; Scheffer, U.; Prisner, T.F.; Göbel, M.W. A Cytidine Phosporamidite with Protected nitroxide Spin Label: Synthesis of a Full-Length TAR RNA and Investigation by In-Line Probing and EPR Spectroscopy. Chem. Eur. J. 2018, 24, 6202–6207. [Google Scholar] [CrossRef]
- Juliusson, H.Y.; Segler, A.-L.J.; Sigurdsson, S.T. Benzoyl-Protected Hydroxylamines for Improved Chemical Synthesis of Oligonucleotides Containing Nitroxides Spin Labels. Eur. J. Org. Chem. 2019, 23, 3799–3805. [Google Scholar] [CrossRef]
- Schmidt, T.; Wälti, M.A.; Baber, J.L.; Hustedt, E.J.; Clore, G.M. Long Distance Measurements up to 160 Å in the GroEL Tetradecamer Using Q-Band DEER EPR Spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 15905–15909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristić, I.; Endeward, B.; Margraf, D.; Marko, A.; Prisner, T.F. EPR Spectroscopy Applications in Chemistry and Biology; Klostermeier, D., Hamman, D., Eds.; Springer: Berlin, Heidelberg, Germany, 2012; Volume 321, pp. 159–198. ISBN 978-3-642-28346-8. [Google Scholar]
- Schiemann, O. Mapping Global Folds in Oligonulceotides by Pulsed Electron-electron Double Resonance. In Methods in Enzymology; Elsevier: San Diego, CA, USA, 2009; ISBN 978-0-12-380922-3. [Google Scholar]
- Marx, L.; Chiarelli, R.; Guiberteau, T.; Rassat, A. A comparative study of the reduction by ascorbate of 1,1,3,3-tetraethylisoindolin-2-yloxyl and of 1,1,3,3-tetramethylisoindolin-2-yloxyl. J. Chem. Soc. Perkin Trans. 2000, 1, 1181–1182. [Google Scholar] [CrossRef]
- Paletta, J.T.; Pink, M.; Foley, B.; Rajca, S.; Rajca, A. Synthesis and Reduction Kinetics of Sterically Shielded Pyrrolidine Nitroxides. Org. Lett. 2012, 16, 5298–5300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Paletta, J.T.; Berg, K.; Reinhart, E.; Rajca, S.; Rajca, A. Synthesis of Unnatural Amino Acids Functionalized with sterically Shielded Pyrroline Nitroxides. Org. Lett. 2014, 16, 5298–5300. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, A.P.; Krstic, I.; Kunjir, N.C.; Hänsel, R.; Prisner, T.F.; Sigurdsson, S.T. Sterically shielded spin labels for in-cell EPR spectroscopy: Analysis of stability in reducing environment. Free Radic. Res. 2015, 49, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, G.; Bonucci, A.; Casano, G.; Gerbaud, G.; Abel, S.; Thomé, V.; Kodjabachian, L.; Magalon, A.; Guigliarelli, B.; Belle, V.; et al. A Bioresistant Nitroxide Spin Label for In-Cell EPR Spectroscopy: In Vitro and In Oocytes Protein Structural Dynamics Studies. Angew. Chem. Int. Ed. 2018, 57, 1366–1370. [Google Scholar] [CrossRef]
- Bleicken, S.; Assafa, T.E.; Zhang, H.; Elsner, C.; Ritsch, I.; Pink, M.; Rajca, S.; Jeschke, G.; Rajca, A.; Bordignon, E. gem-Diethyl Pyrroline Nitroxide Spin Labels: Synthesis, EPR Characterization, Rotamer Libraries and Biocompatibility. Chemistryopen 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Braun, T.S.; Widder, P.; Osswald, U.; Groß, L.; Williams, L.; Schmidt, M.; Helmle, I.; Summerer, D.; Drescher, M. Isoindoline-Base Nitroxides as Bioresistant Spin Labels for Protein Labeling via Cysteines and Alkyne Bearing Noncanonical Amino Acids. ChemBioChem 2019, in press. [Google Scholar] [CrossRef]
- Haugland, M.M.; El-Sagheer, A.H.; Porter, R.J.; Peña, J.; Brown, T.; Anderson, E.A.; Lovett, J.E. 2′-Alkynylnucleotides: A Sequence- and Spin Label-Flexible Strategy for EPR Spectroscopy in DNA. J. Am. Chem. Soc. 2016, 138, 9069–9072. [Google Scholar] [CrossRef] [Green Version]
- Kerzhner, M.; Matsuoka, H.; Wuebben, C.; Famulok, M.; Schiemann, O. High-Yield Spin Labeling of Long RNAs for Electron Paramagnetic Resonance Spectroscopy. Biochemistry 2018, 57, 2923–2931. [Google Scholar] [CrossRef] [PubMed]
- Sproules, S. Molecules as electron spin qubits. In Electron Paramagnetic Resononance; Chechik, V., Murphy, D.M., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2017; Volume 25, ISBN 978-1-78262-857-6. [Google Scholar]
- Eaton, S.S.; Eaton, G.R. Distance Measurements in Biological Systems by EPR; Berliner, L.J., Eaton, G.R., Eaton, S.S., Eds.; Kluwer: New York, NY, USA, 2000; ISBN 0-306-47109-4. [Google Scholar]
- Lindgren, M.; Eaton, G.R.; Eaton, S.S.; Jonsson, B.-H.; Hammarström, P.; Svensson, M.; Carlsson, U. Electron spin echo decay as a probe of aminoxyl environment in spin-labeled mutants of human carbonic anhydrase II. J. Chem. Soc. Perkin Trans. 1997, 2, 2549–2554. [Google Scholar] [CrossRef]
- Huber, M.; Lindgren, M.; Hammarström, P.; Mårtensson, L.-G.; Carlsson, U.; Eaton, G.R.; Eaton, S.S. Phase memory relaxation times of spin labels in human carbonic anhydrase II: Pulsed EPR to determine spin label location. Biophys. Chem. 2001, 94, 245–256. [Google Scholar] [CrossRef]
- Ward, R.; Bowman, A.; Sozudogru, E.; El-Mkami, H.; Owen-Hughes, T.; Norman, D.G. EPR Distance measurements in deuterated proteins. J. Magn. Reson. 2010, 207, 164–167. [Google Scholar] [CrossRef] [PubMed]
- El-Mkami, H.; Ward, R.; Bowman, A.; Owen-Hughes, T.; Norman, D.G. The spatial effect of protein deuteration on nitroxide spin-label relaxation: Implication for EPR distance measurement. J. Magn. Reson. 2014, 248, 36–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zecevic, A.A.; Eaton, G.R.; Eaton, S.S.; Lindgren, M. Dephasing of electron spin echoes for nitroxyl radicals in glassy solvents by non-methyl and methyl protons. Mol. Phys. 1998, 95, 1255–1263. [Google Scholar] [CrossRef]
- Jeschke, G.; Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007, 9, 1895–1910. [Google Scholar] [CrossRef]
- Kulik, L.V.; Dzuba, S.A.; Grigoryev, I.A.; Tsvetkov, Y.D. Electron dipole-dipole interaction in ESEEM of nitroxide biradicals. Chem. Phys. Lett. 2001, 343, 315–324. [Google Scholar] [CrossRef]
- Abdullin, D.; Hagelueken, G.; Hunter, R.I.; Smith, G.M.; Schiemann, O. Geometric model-based fitting algorithm for orientation-selective PELDOR data. Mol. Phys. 2014, 113, 544–560. [Google Scholar] [CrossRef]
- Goddard-Boger, E.D.; Stick, R.V. An Efficient, Inexpensive, and Shelf-Stable Diazotransfer Reagent: Imidazole-1-sulfonyl Azide Hydrochloride. Org. Lett. 2007, 9, 3797–3800. [Google Scholar] [CrossRef]
- Stoll, S.; Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Gafurov, M.; Lyubenova, S.; Denysenkov, V.; Ouari, O.; Karoui, H.; Le Moigne, F.; Tordo, P.; Prisner, T. EPR Charaterization of a Rigid Bis-TEMPO-Bis-Ketal for Dynamic Nuclear Polarization. App. Magn. Reson. 2009, 37, 505–514. [Google Scholar] [CrossRef]
- Windle, J.J. Hyperfine Coupling Constantes for Nitroxide Spin Probes in Water and Carbon Tetrachloride. J. Magn. Reson. 1981, 45, 432–439. [Google Scholar] [CrossRef]
- Hagelueken, G.; Ward, R.; Naismith, J.H.; Schiemann, O. MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL. Appl. Magn. Reson. 2012, 42, 377–391. [Google Scholar] [CrossRef] [Green Version]
- Hagelueken, G.; Abdullin, D.; Ward, R.; Schiemann, O. mtsslSuite: In silico spin labelling, trilateration and distance-constrained rigid body docking. Mol. Phys. 2013, 111, 2757–2766. [Google Scholar] [CrossRef] [Green Version]
- Hagelueken, G.; Abdullin, D.; Schiemann, O. mtsslSuite: Probing Biomolecular Conformation by Spin-Labeling Studies. In Methods in Enzymology; Elsevier: Wlaltham, MA, USA, 2015; ISBN 978-0-12-802834-6. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |





| Parameter | A2 | B2 | ||
|---|---|---|---|---|
| D2O | D2O + 17% H2O | D2O | D2O + 17% H2O | |
| 0.56 | 0.57 | 0.47 | 0.42 | |
| /µs | 20.7 | 13.4 | 21.1 | 13.7 |
| 1.9 | 1.8 | 2.0 | 2.2 | |
| /µs | 1.0 | 1.0 | 1.0 | 1.0 |
| 0.8 | 0.8 | 0.8 | 0.8 | |
| Parameter | A2 | B2 | ||||
|---|---|---|---|---|---|---|
| r | Δr | (nm) | 4.74 (0.06) * | 0.22 (0.07) | 4.80 (0.06) | 0.22 (0.08) |
| ξ | Δξ | (°) | 37/143 (10) | 7 (10) | 44/136 (10) | 4 (15) |
| β | Δβ | (°) | 56/124 (5) | 25 (15) | 34/146 (5) | 10 (15) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wuebben, C.; Blume, S.; Abdullin, D.; Brajtenbach, D.; Haege, F.; Kath-Schorr, S.; Schiemann, O. Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability. Molecules 2019, 24, 4482. https://doi.org/10.3390/molecules24244482
Wuebben C, Blume S, Abdullin D, Brajtenbach D, Haege F, Kath-Schorr S, Schiemann O. Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability. Molecules. 2019; 24(24):4482. https://doi.org/10.3390/molecules24244482
Chicago/Turabian StyleWuebben, Christine, Simon Blume, Dinar Abdullin, Dominik Brajtenbach, Florian Haege, Stephanie Kath-Schorr, and Olav Schiemann. 2019. "Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability" Molecules 24, no. 24: 4482. https://doi.org/10.3390/molecules24244482
APA StyleWuebben, C., Blume, S., Abdullin, D., Brajtenbach, D., Haege, F., Kath-Schorr, S., & Schiemann, O. (2019). Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability. Molecules, 24(24), 4482. https://doi.org/10.3390/molecules24244482







