Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction
Abstract
1. Introduction
1.1. BR Biosynthesis
1.2. Inactivation of BRs
1.3. BR Perception and Signal Transduction
2. Methods for Characterization of Small Molecules Interfering with BR Biosynthesis or Signaling
2.1. Determination of Potency
2.2. Target Identification
2.2.1. Quantification of Precursors
2.2.2. Feeding with Precursors
2.2.3. Mutant Rescue Experiments for BR Mimetics
2.2.4. In Vitro Enzyme Activity Assays
2.2.5. Determination of the Dissociation Constant
2.3. Specificity
3. Inhibitors of Brassinosteroid Biosynthesis
3.1. Brassinazole and BRZ2001
3.2. Brz220
3.3. The YCZ Series
3.4. Propiconazole
3.5. Triadimefon
3.6. Imazalil
3.7. Fenarimol and DDP4
3.8. Steroidal and Non-Steroidal Inhibitors of Steroid 5α-Reductase Activity
3.9. Spironolactone
3.10. Brassinopride
3.11. Voriconazole Impacts on BR Levels via Inhibition of Sterol Biosynthesis
4. Compounds Impacting on Brassinosteroid Signaling
4.1. Brassinolide-2,3-Acetonide: A Steroidal Inhibitor of BR Perception
4.2. Non-Steroidal Compounds Inhibiting or Activating BRI1
4.2.1. (E)-1,2-Bis[trans-(4aα,8aβ)-4-Oxo-6α,7α-dihydroxy-4a,5,6,7,8,8a-hexahydro-(3H)-naphthyl]-ethylene
4.2.2. Phenylfuran Derivatives
4.2.3. Benzene-1,4-Diyldimethanediyl Di-3-Ethyl-5-Methyl-1,2-Oxazole-4-Carboxylate
4.2.4. Turning an Antagonist to an Agonist: NSBR1
4.3. Compounds Targeting BIN2
4.3.1. Lithium
4.3.2. Bikinin and Methyliodobikinin
4.3.3. Brazide
4.4. Compounds with Unknown Targets
4.4.1. KM-01
4.4.2. F1874-108
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Mussig, C.; Shin, G.H.; Altmann, T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003, 133, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Garcia, M.P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-Garcia, S.; Russinova, E.; Cano-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Hacham, Y.; Holland, N.; Butterfield, C.; Ubeda-Tomas, S.; Bennett, M.J.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegon-Putze, I.; Bosch, N.; Ibanes, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146. [Google Scholar] [CrossRef]
- Kim, T.W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef]
- Gudesblat, G.E.; Schneider-Pizon, J.; Betti, C.; Mayerhofer, J.; Vanhoutte, I.; van Dongen, W.; Boeren, S.; Zhiponova, M.; de Vries, S.; Jonak, C.; et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 2012, 14, 548–554. [Google Scholar] [CrossRef]
- Khan, M.; Rozhon, W.; Bigeard, J.; Pflieger, D.; Husar, S.; Pitzschke, A.; Teige, M.; Jonak, C.; Hirt, H.; Poppenberger, B. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J. Biol. Chem. 2013, 288, 7519–7527. [Google Scholar] [CrossRef]
- Vogler, F.; Schmalzl, C.; Englhart, M.; Bircheneder, M.; Sprunck, S. Brassinosteroids promote Arabidopsis pollen germination and growth. Plant Reprod. 2014, 27, 153–167. [Google Scholar] [CrossRef]
- Iwasaki, T.; Hiroh Shibaoka, H. Brassinosteroids Act as Regulators of Tracheary-Element Differentiation in Isolated Zinnia Mesophyll Cells. Plant Cell Physiol. 1991, 32, 1007–1014. [Google Scholar] [CrossRef]
- Yamamoto, R.; Fujioka, S.; Demura, T.; Takatsuto, S.; Yoshida, S.; Fukuda, H. Brassinosteroid levels increase drastically prior to morphogenesis of tracheary elements. Plant Physiol. 2001, 125, 556–563. [Google Scholar] [CrossRef]
- Yamamoto, R.; Demura, T.; Fukuda, H. Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol. 1997, 38, 980–983. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Asami, T.; Yoshida, S. Brassinazole, an inhibitor of brassinosteroid biosynthesis, inhibits development of secondary xylem in cress plants (Lepidium sativum). Plant Cell Physiol. 2001, 42, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, M.; Nemeth, K.; Koncz-Kalman, Z.; Mathur, J.; Kauschmann, A.; Altmann, T.; Redei, G.P.; Nagy, F.; Schell, J.; Koncz, C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 1996, 85, 171–182. [Google Scholar] [CrossRef]
- Saito, M.; Kondo, Y.; Fukuda, H. BES1 and BZR1 Redundantly Promote Phloem and Xylem Differentiation. Plant Cell Physiol. 2018, 59, 590–600. [Google Scholar] [CrossRef]
- Lee, J.; Han, S.; Lee, H.Y.; Jeong, B.; Heo, T.Y.; Hyun, T.K.; Kim, K.; Je, B.I.; Lee, H.; Shim, D.; et al. Brassinosteroids facilitate xylem differentiation and wood formation in tomato. Planta 2019, 249, 1391–1403. [Google Scholar] [CrossRef]
- Chory, J.; Nagpal, P.; Peto, C.A. Phenotypic and Genetic Analysis of det2, a New Mutant That Affects Light-Regulated Seedling Development in Arabidopsis. Plant Cell 1991, 3, 445–459. [Google Scholar] [CrossRef]
- Fabregas, N.; Lozano-Elena, F.; Blasco-Escamez, D.; Tohge, T.; Martinez-Andujar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 4680. [Google Scholar] [CrossRef]
- Lima, J.V.; Lobato, A.K.S. Brassinosteroids improve photosystem II efficiency, gas exchange, antioxidant enzymes and growth of cowpea plants exposed to water deficit. Physiol. Mol. Biol. Plants 2017, 23, 59–72. [Google Scholar] [CrossRef]
- Eremina, M.; Unterholzner, S.J.; Rathnayake, A.I.; Castellanos, M.; Khan, M.; Kugler, K.G.; May, S.T.; Mayer, K.F.; Rozhon, W.; Poppenberger, B. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 2016, 113, E5982–E5991. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 Positively Regulates Freezing Tolerance via CBF-Dependent and CBF-Independent Pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Rozhon, W.; Poppenberger, B. Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 2016, 73, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance Through FERONIA Receptor-Like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Li, X.; Xia, X.J.; Zhou, Y.H.; Shi, K.; Chen, Z.; Yu, J.Q. H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot. 2014, 65, 4371–4383. [Google Scholar] [CrossRef]
- Ibanez, C.; Delker, C.; Martinez, C.; Burstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310. [Google Scholar] [CrossRef]
- Zhu, T.; Deng, X.; Zhou, X.; Zhu, L.; Zou, L.; Li, P.; Zhang, D.; Lin, H. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016, 6, 35392. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef]
- Singh, A.P.; Fridman, Y.; Holland, N.; Ackerman-Lavert, M.; Zananiri, R.; Jaillais, Y.; Henn, A.; Savaldi-Goldstein, S. Interdependent Nutrient Availability and Steroid Hormone Signals Facilitate Root Growth Plasticity. Dev. Cell 2018, 46, 59–72. [Google Scholar] [CrossRef]
- Nomura, T.; Kushiro, T.; Yokota, T.; Kamiya, Y.; Bishop, G.J.; Yamaguchi, S. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 2005, 280, 17873–17879. [Google Scholar] [CrossRef]
- Yokota, T.; Ohnishi, T.; Shibata, K.; Asahina, M.; Nomura, T.; Fujita, T.; Ishizaki, K.; Kohchi, T. Occurrence of brassinosteroids in non-flowering land plants, liverwort, moss, lycophyte and fern. Phytochemistry 2017, 136, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Fujioka, S.; Takatsuto, S.; Tsujimoto, M.; Choe, S. Castasterone is a likely end product of brassinosteroid biosynthetic pathway in rice. Biochem. Biophys. Res. Commun. 2008, 374, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Xin, P.; Yan, J.; Fan, J.; Chu, J.; Yan, C. An improved simplified high-sensitivity quantification method for determining brassinosteroids in different tissues of rice and Arabidopsis. Plant Physiol. 2013, 162, 2056–2066. [Google Scholar] [CrossRef] [PubMed]
- Marie-Andrée Hartmann, M.A. Plant sterols and the membrane environment. Trends Plant Sci. 1998, 3, 170–175. [Google Scholar] [CrossRef]
- Choe, S.; Dilkes, B.P.; Gregory, B.D.; Ross, A.S.; Yuan, H.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Tanaka, A.; Yoshida, S.; et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999, 119, 897–907. [Google Scholar] [CrossRef]
- Takahashi, T.; Gasch, A.; Nishizawa, N.; Chua, N.H. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 1995, 9, 97–107. [Google Scholar] [CrossRef]
- Choe, S.; Tanaka, A.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ross, A.S.; Tax, F.E.; Yoshida, S.; Feldmann, K.A. Lesions in the sterol delta reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000, 21, 431–443. [Google Scholar] [CrossRef]
- Choe, S.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Tissier, C.P.; Gregory, B.D.; Ross, A.S.; Tanaka, A.; Yoshida, S.; Tax, F.E.; et al. The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 1999, 11, 207–221. [Google Scholar]
- Carland, F.M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Nelson, T. The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 2002, 14, 2045–2058. [Google Scholar] [CrossRef]
- Clouse, S.D. Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 2002, 14, 1995–2000. [Google Scholar] [CrossRef]
- Diener, A.C.; Li, H.; Zhou, W.; Whoriskey, W.J.; Nes, W.D.; Fink, G.R. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 2000, 12, 853–870. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Schaller, H.; Goh, C.H.; Kwon, M.; Choe, S.; An, C.S.; Durst, F.; Feldmann, K.A.; Feyereisen, R. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 2005, 138, 2033–2047. [Google Scholar] [CrossRef] [PubMed]
- Schrick, K.; Mayer, U.; Horrichs, A.; Kuhnt, C.; Bellini, C.; Dangl, J.; Schmidt, J.; Jurgens, G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000, 14, 1471–1484. [Google Scholar] [PubMed]
- Jang, J.C.; Fujioka, S.; Tasaka, M.; Seto, H.; Takatsuto, S.; Ishii, A.; Aida, M.; Yoshida, S.; Sheen, J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000, 14, 1485–1497. [Google Scholar]
- Topping, J.F.; May, V.J.; Muskett, P.R.; Lindsey, K. Mutations in the HYDRA1 gene of Arabidopsis perturb cell shape and disrupt embryonic and seedling morphogenesis. Development 1997, 124, 4415–4424. [Google Scholar]
- Kim, T.W.; Hwang, J.Y.; Kim, Y.S.; Joo, S.H.; Chang, S.C.; Lee, J.S.; Takatsuto, S.; Kim, S.K. Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 2005, 17, 2397–2412. [Google Scholar] [CrossRef]
- Corey, E.J.; Matsuda, S.P.; Bartel, B. Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proc. Natl. Acad. Sci. USA 1993, 90, 11628–11632. [Google Scholar] [CrossRef]
- Lovato, M.A.; Hart, E.A.; Segura, M.J.; Giner, J.L.; Matsuda, S.P. Functional cloning of an Arabidopsis thaliana cDNA encoding cycloeucalenol cycloisomerase. J. Biol. Chem. 2000, 275, 13394–13397. [Google Scholar] [CrossRef]
- Ohnishi, T.; Szatmari, A.M.; Watanabe, B.; Fujita, S.; Bancos, S.; Koncz, C.; Lafos, M.; Shibata, K.; Yokota, T.; Sakata, K.; et al. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 2006, 18, 3275–3288. [Google Scholar] [CrossRef]
- Morikawa, T.; Mizutani, M.; Aoki, N.; Watanabe, B.; Saga, H.; Saito, S.; Oikawa, A.; Suzuki, H.; Sakurai, N.; Shibata, D.; et al. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell 2006, 18, 1008–1022. [Google Scholar] [CrossRef]
- Fujioka, S.; Li, J.; Choi, Y.H.; Seto, H.; Takatsuto, S.; Noguchi, T.; Watanabe, T.; Kuriyama, H.; Yokota, T.; Chory, J.; et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 1997, 9, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 1998, 10, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.T.; Tsukaya, H.; Uchimiya, H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998, 12, 2381–2391. [Google Scholar] [CrossRef] [PubMed]
- Rasbery, J.M.; Shan, H.; LeClair, R.J.; Norman, M.; Matsuda, S.P.; Bartel, B. Arabidopsis thaliana squalene epoxidase 1 is essential for root and seed development. J. Biol. Chem. 2007, 282, 17002–17013. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, R.; Wu, Y.; LoCascio, J.C.; Feldmann, K.A. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 1998, 10, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.; Fujioka, S.; Noguchi, T.; Takatsuto, S.; Yoshida, S.; Feldmann, K.A. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001, 26, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Fujioka, S.; Jeon, J.H.; Kim, H.B.; Takatsuto, S.; Yoshida, S.; An, C.S.; Choe, S. A double mutant for the CYP85A1 andCYP85A2 Genes of Arabidopsis exhibits a Brassinosteroid dwarf phenotype. J. Plant Biol. 2005, 48, 237–244. [Google Scholar] [CrossRef]
- Noguchi, T.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Yoshida, S.; Li, J.; Chory, J. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999, 120, 833–840. [Google Scholar] [CrossRef]
- Ohnishi, T.; Godza, B.; Watanabe, B.; Fujioka, S.; Hategan, L.; Ide, K.; Shibata, K.; Yokota, T.; Szekeres, M.; Mizutani, M. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 2012, 287, 31551–31560. [Google Scholar] [CrossRef]
- Shimada, Y.; Fujioka, S.; Miyauchi, N.; Kushiro, M.; Takatsuto, S.; Nomura, T.; Yokota, T.; Kamiya, Y.; Bishop, G.J.; Yoshida, S. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001, 126, 770–779. [Google Scholar] [CrossRef]
- Fujita, S.; Ohnishi, T.; Watanabe, B.; Yokota, T.; Takatsuto, S.; Fujioka, S.; Yoshida, S.; Sakata, K.; Mizutani, M. Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. 2006, 45, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, J. Regulation of brassinosteroid biosynthesis and inactivation. J. Integr. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Choe, S. The Regulation of Brassinosteroid Biosynthesis in Arabidopsis. Crit. Rev. Plant Sci. 2013, 32, 396–410. [Google Scholar] [CrossRef]
- Bishop, G.J.; Nomura, T.; Yokota, T.; Harrison, K.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Jones, J.D.; Kamiya, Y. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Fujioka, S.; Yokota, T.; Murofushi, N.; Sakurai, A. Identification of Brassinolide, Castasterone, Typhasterol, and Teasterone from the Pollen of Lilium elegans. Biosci. Biotechnol. Biochem. 1994, 58, 2075–2076. [Google Scholar] [CrossRef]
- Kang, J.G.; Yun, J.; Kim, D.H.; Chung, K.S.; Fujioka, S.; Kim, J.I.; Dae, H.W.; Yoshida, S.; Takatsuto, S.; Song, P.S.; et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 2001, 105, 625–636. [Google Scholar] [CrossRef]
- Youn, J.H.; Kim, T.W.; Joo, S.H.; Son, S.H.; Roh, J.; Kim, S.; Kim, T.W.; Kim, S.K. Function and molecular regulation of DWARF1 as a C-24 reductase in brassinosteroid biosynthesis in Arabidopsis. J. Exp. Bot. 2018, 69, 1873–1886. [Google Scholar] [CrossRef]
- Bancos, S.; Nomura, T.; Sato, T.; Molnar, G.; Bishop, G.J.; Koncz, C.; Yokota, T.; Nagy, F.; Szekeres, M. Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002, 130, 504–513. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- De Rybel, B.; Audenaert, D.; Vert, G.; Rozhon, W.; Mayerhofer, J.; Peelman, F.; Coutuer, S.; Denayer, T.; Jansen, L.; Nguyen, L.; et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 2009, 16, 594–604. [Google Scholar] [CrossRef]
- Coll-Garcia, D.; Mazuch, J.; Altmann, T.; Mussig, C. EXORDIUM regulates brassinosteroid-responsive genes. FEBS Lett. 2004, 563, 82–86. [Google Scholar] [CrossRef]
- Dejonghe, W.; Mishev, K.; Russinova, E. The brassinosteroid chemical toolbox. Curr. Opin. Plant Biol. 2014, 22, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Matsumoto, T.; Yamagami, A.; Ogawa, A.; Yamada, K.; Suzuki, R.; Sawada, T.; Fujioka, S.; Yoshizawa, Y.; Nakano, T. YCZ-18 is a new brassinosteroid biosynthesis inhibitor. PLoS ONE 2015, 10, e0120812. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Matsumoto, T.; Yamagami, A.; Hoshi, T.; Nakano, T.; Yoshizawa, Y. Fenarimol, a Pyrimidine-Type Fungicide, Inhibits Brassinosteroid Biosynthesis. Int. J. Mol. Sci. 2015, 16, 17273–17288. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Matsumoto, T.; Hoshi, T.; Yoshizawa, Y. In vitro and in vivo evidence for the inhibition of brassinosteroid synthesis by propiconazole through interference with side chain hydroxylation. Plant Signal Behav. 2016, 11, e1158372. [Google Scholar] [CrossRef][Green Version]
- Rosati, F.; Bardazzi, I.; De Blasi, P.; Simi, L.; Scarpi, D.; Guarna, A.; Serio, M.; Racchi, M.L.; Danza, G. 5alpha-Reductase activity in Lycopersicon esculentum: Cloning and functional characterization of LeDET2 and evidence of the presence of two isoenzymes. J. Steroid Biochem. Mol. Biol. 2005, 96, 287–299. [Google Scholar] [CrossRef]
- Li, J.; Biswas, M.G.; Chao, A.; Russell, D.W.; Chory, J. Conservation of function between mammalian and plant steroid 5alpha-reductases. Proc. Natl. Acad. Sci. USA 1997, 94, 3554–3559. [Google Scholar] [CrossRef]
- Rozhon, W.; Husar, S.; Kalaivanan, F.; Khan, M.; Idlhammer, M.; Shumilina, D.; Lange, T.; Hoffmann, T.; Schwab, W.; Fujioka, S.; et al. Genetic variation in plant CYP51s confers resistance against voriconazole, a novel inhibitor of brassinosteroid-dependent sterol biosynthesis. PLoS ONE 2013, 8, e53650. [Google Scholar] [CrossRef]
- Neff, M.M.; Nguyen, S.M.; Malancharuvil, E.J.; Fujioka, S.; Noguchi, T.; Seto, H.; Tsubuki, M.; Honda, T.; Takatsuto, S.; Yoshida, S.; et al. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 15316–15323. [Google Scholar] [CrossRef]
- Turk, E.M.; Fujioka, S.; Seto, H.; Shimada, Y.; Takatsuto, S.; Yoshida, S.; Wang, H.; Torres, Q.I.; Ward, J.M.; Murthy, G.; et al. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 2005, 42, 23–34. [Google Scholar] [CrossRef]
- Nakamura, M.; Satoh, T.; Tanaka, S.; Mochizuki, N.; Yokota, T.; Nagatani, A. Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo. J. Exp. Bot. 2005, 56, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nakazawa, M.; Shibata, K.; Yokota, T.; Ishikawa, A.; Suzuki, K.; Kawashima, M.; Ichikawa, T.; Shimada, H.; Matsui, M. shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J. 2005, 42, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Thornton, L.E.; Rupasinghe, S.G.; Peng, H.; Schuler, M.A.; Neff, M.M. Arabidopsis CYP72C1 is an atypical cytochrome P450 that inactivates brassinosteroids. Plant Mol. Biol. 2010, 74, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Turk, E.M.; Fujioka, S.; Seto, H.; Shimada, Y.; Takatsuto, S.; Yoshida, S.; Denzel, M.A.; Torres, Q.I.; Neff, M.M. CYP72B1 inactivates brassinosteroid hormones: An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 2003, 133, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kawabe, A.; Tokida-Segawa, A.; Shimizu, B.; Takatsuto, S.; Shimada, Y.; Fujioka, S.; Mizutani, M. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 2011, 67, 1–12. [Google Scholar] [CrossRef]
- Poppenberger, B.; Fujioka, S.; Soeno, K.; George, G.L.; Vaistij, F.E.; Hiranuma, S.; Seto, H.; Takatsuto, S.; Adam, G.; Yoshida, S.; et al. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 2005, 102, 15253–15258. [Google Scholar] [CrossRef]
- Husar, S.; Berthiller, F.; Fujioka, S.; Rozhon, W.; Khan, M.; Kalaivanan, F.; Elias, L.; Higgins, G.S.; Li, Y.; Schuhmacher, R.; et al. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Lacomme, C.; Roby, D. Molecular cloning of a sulfotransferase in Arabidopsis thaliana and regulation during development and in response to infection with pathogenic bacteria. Plant Mol. Biol. 1996, 30, 995–1008. [Google Scholar] [CrossRef]
- Rouleau, M.; Marsolais, F.; Richard, M.; Nicolle, L.; Voigt, B.; Adam, G.; Varin, L. Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J. Biol. Chem. 1999, 274, 20925–20930. [Google Scholar] [CrossRef]
- Marsolais, F.; Hernández Sebastià, C.; Rousseau, A.; Varin, L. Molecular and biochemical characterization of BNST4, an ethanol-inducible steroid sulfotransferase from Brassica napus, and regulation of BNST genes by chemical stress and during development. Plant Sci. 2004, 166, 1359–1370. [Google Scholar] [CrossRef]
- Marsolais, F.; Boyd, J.; Paredes, Y.; Schinas, A.M.; Garcia, M.; Elzein, S.; Varin, L. Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 2007, 225, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Cho, Y.H.; Kim, K.; Matsui, M.; Son, S.H.; Kim, S.K.; Fujioka, S.; Hwang, I. BAT1, a putative acyltransferase, modulates brassinosteroid levels in Arabidopsis. Plant J. 2013, 73, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Breuer, C.; Kawamura, A.; Jikumaru, Y.; Hanada, A.; Fujioka, S.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Kamiya, Y.; et al. Arabidopsis PIZZA has the capacity to acylate brassinosteroids. PLoS ONE 2012, 7, e46805. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wang, H.; Fujioka, S.; Zhou, T.; Tian, H.; Tian, W.; Wang, X. Homeostasis of brassinosteroids regulated by DRL1, a putative acyltransferase in Arabidopsis. Mol. Plant 2013, 6, 546–558. [Google Scholar] [CrossRef]
- Kolbe, A.; Schneider, B.; Porzel, A.; Schmidt, J.; Adam, G. Acyl-conjugated metabolites of brassinosteroids in cell suspension cultures of Ornithopus sativus. Phytochemistry 1995, 38, 633–636. [Google Scholar] [CrossRef]
- Soeno, K.; Asakawa, S.; Natsume, M.; Abe, H. Reversible conversion between teasterone and its ester conjugates in lily cell cultures. J. Pest. Sci. 2000, 25, 117–122. [Google Scholar] [CrossRef][Green Version]
- Roh, H.; Jeong, C.W.; Fujioka, S.; Kim, Y.K.; Lee, S.; Ahn, J.H.; Choi, Y.D.; Lee, J.S. Genetic evidence for the reduction of brassinosteroid levels by a BAHD acyltransferase-like protein in Arabidopsis. Plant Physiol. 2012, 159, 696–709. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Wang, R.; Li, W.; Rodermel, S.; Yu, F. Overexpression of a putative Arabidopsis BAHD acyltransferase causes dwarfism that can be rescued by brassinosteroid. J. Exp. Bot. 2012, 63, 5787–5801. [Google Scholar] [CrossRef]
- Yuan, T.; Fujioka, S.; Takatsuto, S.; Matsumoto, S.; Gou, X.; He, K.; Russell, S.D.; Li, J. BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J. 2007, 51, 220–233. [Google Scholar] [CrossRef]
- Jackson, R.G.; Kowalczyk, M.; Li, Y.; Higgins, G.; Ross, J.; Sandberg, G.; Bowles, D.J. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 2002, 32, 573–583. [Google Scholar] [CrossRef]
- Jackson, R.G.; Lim, E.K.; Li, Y.; Kowalczyk, M.; Sandberg, G.; Hoggett, J.; Ashford, D.A.; Bowles, D.J. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 2001, 276, 4350–4356. [Google Scholar] [CrossRef] [PubMed]
- George Thompson, A.M.; Iancu, C.V.; Neet, K.E.; Dean, J.V.; Choe, J.Y. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci. Rep. 2017, 7, 46629. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Doucet, C.J.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Ross, J.; Bowles, D.J. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 2002, 277, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 2008, 132, 417–425. [Google Scholar] [CrossRef]
- Dong, T.; Xu, Z.Y.; Park, Y.; Kim, D.H.; Lee, Y.; Hwang, I. Abscisic acid uridine diphosphate glucosyltransferases play a crucial role in abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2014, 165, 277–289. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, J.P.; Li, D.K.; Luo, Q.; Yan, Q.; Liu, Z.B.; Ye, L.M.; Wang, J.M.; Li, X.F.; Yang, Y. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–471. [Google Scholar] [CrossRef]
- She, J.; Han, Z.; Kim, T.W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Wang, J.; Yang, M.; Wang, Z.Y.; et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar] [CrossRef]
- She, J.; Han, Z.; Zhou, B.; Chai, J. Structural basis for differential recognition of brassinolide by its receptors. Protein Cell 2013, 4, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Goshe, M.B.; Soderblom, E.J.; Phinney, B.S.; Kuchar, J.A.; Li, J.; Asami, T.; Yoshida, S.; Huber, S.C.; Clouse, S.D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 2005, 17, 1685–1703. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Wang, X.; Kota, U.; Goshe, M.B.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kota, U.; He, K.; Blackburn, K.; Li, J.; Goshe, M.B.; Huber, S.C.; Clouse, S.D. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 2008, 15, 220–235. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef]
- Wang, H.; Yang, C.; Zhang, C.; Wang, N.; Lu, D.; Wang, J.; Zhang, S.; Wang, Z.X.; Ma, H.; Wang, X. Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 2011, 21, 825–834. [Google Scholar] [CrossRef]
- Russinova, E.; Borst, J.W.; Kwaaitaal, M.; Cano-Delgado, A.; Yin, Y.; Chory, J.; de Vries, S.C. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 2004, 16, 3216–3229. [Google Scholar] [CrossRef]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2006, 18, 626–638. [Google Scholar] [CrossRef]
- Geldner, N.; Hyman, D.L.; Wang, X.; Schumacher, K.; Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007, 21, 1598–1602. [Google Scholar] [CrossRef]
- Cano-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-Garcia, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef]
- Zhou, A.; Wang, H.; Walker, J.C.; Li, J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004, 40, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Guan, S.; Burlingame, A.L.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Youn, J.H.; Park, C.H.; Kim, T.W.; Guan, S.; Xu, S.; Burlingame, A.L.; Kim, Y.P.; Kim, S.K.; Wang, Z.Y.; et al. Oligomerization between BSU1 Family Members Potentiates Brassinosteroid Signaling in Arabidopsis. Mol. Plant 2016, 9, 178–181. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H.; Vafeados, D.; Chory, J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001, 127, 14–22. [Google Scholar] [CrossRef]
- Choe, S.; Schmitz, R.J.; Fujioka, S.; Takatsuto, S.; Lee, M.O.; Yoshida, S.; Feldmann, K.A.; Tax, F.E. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol. 2002, 130, 1506–1515. [Google Scholar] [CrossRef]
- Perez-Perez, J.M.; Ponce, M.R.; Micol, J.L. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 2002, 242, 161–173. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef]
- Rozhon, W.; Mayerhofer, J.; Petutschnig, E.; Fujioka, S.; Jonak, C. ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 2010, 62, 215–223. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Chory, J. Downstream nuclear events in brassinosteroid signalling. Nature 2006, 441, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Piao, H.L.; Pih, K.T.; Lim, J.H.; Kang, S.G.; Jin, J.B.; Kim, S.H.; Hwang, I. An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 1999, 119, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Piao, H.L.; Lim, J.H.; Kim, S.J.; Cheong, G.W.; Hwang, I. Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 2001, 27, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Wrzaczek, M.; Rozhon, W.; Jonak, C. A Proteasome-regulated glycogen synthase kinase-3 modulates disease response in plants. J. Biol. Chem. 2007, 282, 5249–5255. [Google Scholar] [CrossRef]
- Kempa, S.; Rozhon, W.; Samaj, J.; Erban, A.; Baluska, F.; Becker, T.; Haselmayer, J.; Schleiff, E.; Kopka, J.; Hirt, H.; et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. Plant J. 2007, 49, 1076–1090. [Google Scholar] [CrossRef]
- Dal Santo, S.; Stampfl, H.; Krasensky, J.; Kempa, S.; Gibon, Y.; Petutschnig, E.; Rozhon, W.; Heuck, A.; Clausen, T.; Jonak, C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 2012, 24, 3380–3392. [Google Scholar] [CrossRef]
- Zhao, J.; Peng, P.; Schmitz, R.J.; Decker, A.D.; Tax, F.E.; Li, J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002, 130, 1221–1229. [Google Scholar] [CrossRef]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef]
- Ryu, H.; Kim, K.; Cho, H.; Park, J.; Choe, S.; Hwang, I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 2007, 19, 2749–2762. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Rozhon, W.; Poppenberger, B. Analysis of In Vitro DNA Interactions of Brassinosteroid-Controlled Transcription Factors Using Electrophoretic Mobility Shift Assay. Methods Mol. Biol. 2017, 1564, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Sun, Y.; Wang, Z.Y. Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol. Biol. 2012, 876, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Liu, S.; Guo, H.; Li, L.; Schnable, P.; Yin, Y. Identification of Brassinosteroid Target Genes by Chromatin Immunoprecipitation Followed by High-Throughput Sequencing (ChIP-seq) and RNA-Sequencing. Methods Mol. Biol. 2017, 1564, 63–79. [Google Scholar] [CrossRef]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef]
- Ye, H.; Li, L.; Guo, H.; Yin, Y. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 20142–20147. [Google Scholar] [CrossRef]
- Poppenberger, B.; Rozhon, W.; Khan, M.; Husar, S.; Adam, G.; Luschnig, C.; Fujioka, S.; Sieberer, T. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 2011, 30, 1149–1161. [Google Scholar] [CrossRef]
- Khan, M.; Rozhon, W.; Unterholzner, S.J.; Chen, T.; Eremina, M.; Wurzinger, B.; Bachmair, A.; Teige, M.; Sieberer, T.; Isono, E.; et al. Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 2014, 5, 4687. [Google Scholar] [CrossRef]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef]
- Martinez, C.; Espinosa-Ruiz, A.; de Lucas, M.; Bernardo-Garcia, S.; Franco-Zorrilla, J.M.; Prat, S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Fan, M.; Bai, M.Y.; Kim, J.G.; Wang, T.; Oh, E.; Chen, L.; Park, C.H.; Son, S.H.; Kim, S.K.; Mudgett, M.B.; et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.J.; Hangarter, R.P.; Estelle, M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998, 116, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.M.; Ostin, A.; Sandberg, G.; Romano, C.P.; Estelle, M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7197–7202. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three Auxin Response Factors Promote Hypocotyl Elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, F.; Wang, S.; Su, Y.; Ji, X.; Jiang, P.; Chen, R.; Hou, S.; Ding, Y. MLK1 and MLK2 Coordinate RGA and CCA1 Activity to Regulate Hypocotyl Elongation in Arabidopsis thaliana. Plant Cell 2018, 30, 67–82. [Google Scholar] [CrossRef]
- Cole, B.; Kay, S.A.; Chory, J. Automated analysis of hypocotyl growth dynamics during shade avoidance in Arabidopsis. Plant J. 2011, 65, 991–1000. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Zhou, X.Y.; Song, L.; Xue, H.W. Brassinosteroids regulate the differential growth of Arabidopsis hypocotyls through auxin signaling components IAA19 and ARF7. Mol. Plant 2013, 6, 887–904. [Google Scholar] [CrossRef]
- Chen, I.J.; Lo, W.S.; Chuang, J.Y.; Cheuh, C.M.; Fan, Y.S.; Lin, L.C.; Wu, S.J.; Wang, L.C. A chemical genetics approach reveals a role of brassinolide and cellulose synthase in hypocotyl elongation of etiolated Arabidopsis seedlings. Plant Sci. 2013, 209, 46–57. [Google Scholar] [CrossRef]
- Deslauriers, S.D.; Larsen, P.B. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol. Plant 2010, 3, 626–640. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Song, K.; Tsoy, Y.; Kim, J.Y.; Kwon, Y.J.; Kang, M.; Edberg Hansen, M.A. Robust dose-response curve estimation applied to high content screening data analysis. Source Code Biol. Med. 2014, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Prinz, H. Hill coefficients, dose-response curves and allosteric mechanisms. J. Chem. Biol. 2010, 3, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Yamada, K.; Yoshizawa, Y.; Oh, K. Comparison of Effect of Brassinosteroid and Gibberellin Biosynthesis Inhibitors on Growth of Rice Seedlings. Rice Sci. 2016, 23, 51–55. [Google Scholar] [CrossRef]
- Wada, K.; Marumo, S.; Abe, H.; Morishita, T.; Nakamura, K.; Uchiyama, M.; Mori, M. A Rice Lamina Inclination Test - A Micro-quantitative Bioassay for Brassinosteroids. Agric. Biol. Chem. 1984, 48, 719–726. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Jang, S. Rice Lamina Joint Inclination Assay. Bio-Protocol 2017, 7, e2409. [Google Scholar] [CrossRef]
- Liu, S.; Yu, C.; Tian, H.; Hu, T.; He, Y.; Li, Z.; Tan, W.; Zhang, L.; Duan, L. A Novel Bikinin Analogue for Arabidopsis and Rice with Superior Plant Growth-Promoting Activity. J. Plant Growth Regul. 2018, 37, 166–173. [Google Scholar] [CrossRef]
- Sekimata, K.; Han, S.Y.; Yoneyama, K.; Takeuchi, Y.; Yoshida, S.; Asami, T. A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J. Agric. Food Chem. 2002, 50, 3486–3490. [Google Scholar] [CrossRef]
- Sekimata, K.; Ohnishi, T.; Mizutani, M.; Todoroki, Y.; Han, S.Y.; Uzawa, J.; Fujioka, S.; Yoneyama, K.; Takeuchi, Y.; Takatsuto, S.; et al. Brz220 interacts with DWF4, a cytochrome P450 monooxygenase in brassinosteroid biosynthesis, and exerts biological activity. Biosci. Biotechnol. Biochem. 2008, 72, 7–12. [Google Scholar] [CrossRef]
- Yamada, K.; Yajima, O.; Yoshizawa, Y.; Oh, K. Synthesis and biological evaluation of novel azole derivatives as selective potent inhibitors of brassinosteroid biosynthesis. Bioorg. Med. Chem. 2013, 21, 2451–2461. [Google Scholar] [CrossRef]
- Oh, K.; Yamada, K.; Asami, T.; Yoshizawa, Y. Synthesis of novel brassinosteroid biosynthesis inhibitors based on the ketoconazole scaffold. Bioorg. Med. Chem. Lett. 2012, 22, 1625–1628. [Google Scholar] [CrossRef]
- Fujioka, S.; Takatsuto, S.; Yoshida, S. An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 2002, 130, 930–939. [Google Scholar] [CrossRef] [PubMed]
- He, J.X.; Fujioka, S.; Li, T.C.; Kang, S.G.; Seto, H.; Takatsuto, S.; Yoshida, S.; Jang, J.C. Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 2003, 131, 1258–1269. [Google Scholar] [CrossRef] [PubMed]
- Swaczynová, J.; Novák, O.; Hauserová, E.; Fuksová, K.; Šíša, M.; Ladislav Kohout, L.; Strnad, M. New Techniques for the Estimation of Naturally Occurring Brassinosteroids. Plant Growth Regul. 2007, 21, 1–14. [Google Scholar] [CrossRef]
- Asami, T.; Mizutani, M.; Fujioka, S.; Goda, H.; Min, Y.K.; Shimada, Y.; Nakano, T.; Takatsuto, S.; Matsuyama, T.; Nagata, N.; et al. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J. Biol. Chem. 2001, 276, 25687–25691. [Google Scholar] [CrossRef] [PubMed]
- Asami, T.; Mizutani, M.; Shimada, Y.; Goda, H.; Kitahata, N.; Sekimata, K.; Han, S.Y.; Fujioka, S.; Takatsuto, S.; Sakata, K.; et al. Triadimefon, a fungicidal triazole-type P450 inhibitor, induces brassinosteroid deficiency-like phenotypes in plants and binds to DWF4 protein in the brassinosteroid biosynthesis pathway. Biochem. J. 2003, 369, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Asami, T.; Min, Y.K.; Sekimata, K.; Shimada, Y.; Wang, J.M.; Fujioka, S.; Yoshida, S. Mode of Action of Brassinazole: A Specific Inhibitor of Brassinosteroid Biosynthesis. ACS Symp. Ser. 2000, 774, 269–280. [Google Scholar] [CrossRef]
- Rozhon, W.; Wang, W.; Berthiller, F.; Mayerhofer, J.; Chen, T.; Petutschnig, E.; Sieberer, T.; Poppenberger, B.; Jonak, C. Bikinin-like inhibitors targeting GSK3/Shaggy-like kinases: Characterisation of novel compounds and elucidation of their catabolism in planta. BMC Plant Biol. 2014, 14, 172. [Google Scholar] [CrossRef]
- Dixon, M. The determination of enzyme inhibitor constants. Biochem. J. 1953, 55, 170–171. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 1974, 137, 143–144. [Google Scholar] [CrossRef]
- Baici, A. The specific velocity plot. A graphical method for determining inhibition parameters for both linear and hyperbolic enzyme inhibitors. Eur. J. Biochem. 1981, 119, 9–14. [Google Scholar] [CrossRef]
- Yoshino, M.; Murakami, K. A graphical method for determining inhibition constants. J. Enzym. Inhib. Med. Chem. 2009, 24, 1288–1290. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, M. A graphical method for determining inhibition parameters for partial and complete inhibitors. Biochem. J. 1987, 248, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Di Trani, J.M.; De Cesco, S.; O’Leary, R.; Plescia, J.; do Nascimento, C.J.; Moitessier, N.; Mittermaier, A.K. Rapid measurement of inhibitor binding kinetics by isothermal titration calorimetry. Nat. Commun. 2018, 9, 893. [Google Scholar] [CrossRef] [PubMed]
- Damian, L. Isothermal titration calorimetry for studying protein-ligand interactions. Methods Mol. Biol. 2013, 1008, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, T.H.; Padrick, S.B.; Gardner, K.H.; Brautigam, C.A. On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 2016, 496, 79–93. [Google Scholar] [CrossRef]
- Pattnaik, P. Surface plasmon resonance: Applications in understanding receptor-ligand interaction. Appl. Biochem. Biotechnol. 2005, 126, 79–92. [Google Scholar] [CrossRef]
- Neumann, T.; Junker, H.D.; Schmidt, K.; Sekul, R. SPR-based fragment screening: Advantages and applications. Curr. Top. Med. Chem. 2007, 7, 1630–1642. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Poulos, T.L. Current Approaches for Investigating and Predicting Cytochrome P450 3A4-Ligand Interactions. Adv. Exp. Med. Biol. 2015, 851, 83–105. [Google Scholar] [CrossRef]
- Tamaskovic, R.; Simon, M.; Stefan, N.; Schwill, M.; Pluckthun, A. Designed ankyrin repeat proteins (DARPins) from research to therapy. Methods Enzym. 2012, 503, 101–134. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Rozhon, W.; Papacek, M.; Ciomas, J.; Lange, T.; Kugler, K.G.; Mayer, K.F.; Sieberer, T.; Poppenberger, B. Brassinosteroids Are Master Regulators of Gibberellin Biosynthesis in Arabidopsis. Plant Cell 2015, 27, 2261–2272. [Google Scholar] [CrossRef]
- Tong, H.; Chu, C. Reply: Brassinosteroid Regulates Gibberellin Synthesis to Promote Cell Elongation in Rice: Critical Comments on Ross and Quittenden’s Letter. Plant Cell 2016, 28, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Rozhon, W.; Poppenberger, B. Reply: Interaction between Brassinosteroids and Gibberellins: Synthesis or Signaling? In Arabidopsis, Both! Plant Cell 2016, 28, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Debeaujon, I.; Koornneef, M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000, 122, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; van der Veen, J.H. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Appl. Genet. 1980, 58, 257–263. [Google Scholar] [CrossRef] [PubMed]
- van Der Schaar, W.; Alonso-Blanco, C.; Leon-Kloosterziel, K.M.; Jansen, R.C.; van Ooijen, J.W.; Koornneef, M. QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 1997, 79, 190–200. [Google Scholar] [CrossRef][Green Version]
- Papi, M.; Sabatini, S.; Bouchez, D.; Camilleri, C.; Costantino, P.; Vittorioso, P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000, 14, 28–33. [Google Scholar] [CrossRef]
- Steber, C.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef]
- Yoshida, Y.; Noshiro, M.; Aoyama, Y.; Kawamoto, T.; Horiuchi, T.; Gotoh, O. Structural and evolutionary studies on sterol 14-demethylase P450 (CYP51), the most conserved P450 monooxygenase: II. Evolutionary analysis of protein and gene structures. J. Biochem. 1997, 122, 1122–1128. [Google Scholar] [CrossRef]
- Saito, S.; Okamoto, M.; Shinoda, S.; Kushiro, T.; Koshiba, T.; Kamiya, Y.; Hirai, N.; Todoroki, Y.; Sakata, K.; Nambara, E.; et al. A plant growth retardant, uniconazole, is a potent inhibitor of ABA catabolism in Arabidopsis. Biosci. Biotechnol. Biochem. 2006, 70, 1731–1739. [Google Scholar] [CrossRef]
- Asami, T.; Yoshida, S. Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 1999, 4, 348–353. [Google Scholar] [CrossRef]
- Izumi, K.; Yamaguchi, I.; Wada, A.; Oshio, H.; Takahashi, N. Effects of a New Plant Growth Retardant (E)-1-(4-Chlorophenyl)-4,4-dimethyl-2-(1,2,4-triazol-1-yl)-1-penten-3-ol (S-3307) on the Growth and Gibberellin Content of Rice Plants. Plant Cell Physiol. 1984, 25, 611–617. [Google Scholar] [CrossRef]
- Izumi, K.; Kamiya, Y.; Sakurai, A.; Oshio, H.; Takahashi, N. Studies of Sites of Action of a New Plant Growth Retardant (E)-1-(4-Chlorophenyl)-4,4-dimethyl-2-(1,2,4-triazol-1-yl)-1-penten-3-ol (S-3307) and Comparative Effects of Its Stereoisomers in a Cell-Free System from Cucurbita maxima. Plant Cell Physiol. 1985, 26, 821–827. [Google Scholar] [CrossRef]
- Miyazaki, S.; Katsumata, T.; Natsume, M.; Kawaide, H. The CYP701B1 of Physcomitrella patens is an ent-kaurene oxidase that resists inhibition by uniconazole-P. FEBS Lett. 2011, 585, 1879–1883. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Poole, A.; Peacock, W.J.; Dennis, E.S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999, 119, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Nakamura, Y.; Takahashi, N.; Nonaka, M.; Sekimoto, H.; Oshio, H.; Takatsuto, S. Inconsistency between browth and endogenous levels of gibberellins, brassinosteroids, and sterols in Pisum sativum treated with uniconazole antipodes. In Gibberellins; Takahashi, N., Phinney, B.O., Macmillan, J., Eds.; Springer: New York, NY, USA, 1991. [Google Scholar] [CrossRef]
- Kitahata, N.; Saito, S.; Miyazawa, Y.; Umezawa, T.; Shimada, Y.; Min, Y.K.; Mizutani, M.; Hirai, N.; Shinozaki, K.; Yoshida, S.; et al. Chemical regulation of abscisic acid catabolism in plants by cytochrome P450 inhibitors. Bioorg. Med. Chem. 2005, 13, 4491–4498. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Millar, A.A.; Jacobsen, J.V.; Ross, J.J.; Helliwell, C.A.; Poole, A.T.; Scofield, G.; Reid, J.B.; Gubler, F. Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 2006, 45, 942–954. [Google Scholar] [CrossRef]
- Yang, S.H.; Zeevaart, J.A. Expression of ABA 8′-hydroxylases in relation to leaf water relations and seed development in bean. Plant J. 2006, 47, 675–686. [Google Scholar] [CrossRef]
- Eggels, S.; Avramova, V.; Schon, C.C.; Poppenberger, B.; Rozhon, W. Assay for abscisic acid 8′-hydroxylase activity of cloned plant cytochrome P450 oxidases in Saccharomyces cerevisiae. Anal. Biochem. 2018, 553, 24–27. [Google Scholar] [CrossRef]
- Izumi, K.; Nakagawa, S.; Kobayashi, M.; Oshio, M.; Sakurai, A.; Takahashi, N. Levels of IAA, Cytokinins, ABA and Ethylene in Rice Plants as Affected by a Gibberellin Biosynthesis Inhibitor, Uniconazole-P. Plant Cell Physiol. 1988, 29, 97–104. [Google Scholar] [CrossRef]
- Best, N.B.; Johal, G.; Dilkes, B.P. Phytohormone inhibitor treatments phenocopy brassinosteroid–gibberellin dwarf mutant interactions in maize. Plant Direct 2017, 1, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.K.; Asami, T.; Fujioka, S.; Murofushi, N.; Yamaguchi, I.; Yoshida, S. New lead compounds for brassinosteroid biosynthesis inhibitors. Bioorg. Med. Chem. Lett. 1999, 9, 425–430. [Google Scholar] [CrossRef]
- Asami, T.; Min, Y.K.; Nagata, N.; Yamagishi, K.; Takatsuto, S.; Fujioka, S.; Murofushi, N.; Yamaguchi, I.; Yoshida, S. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000, 123, 93–100. [Google Scholar] [CrossRef]
- Fujiyama, K.; Hino, T.; Kanadani, M.; Watanabe, B.; Jae Lee, H.; Mizutani, M.; Nagano, S. Structural insights into a key step of brassinosteroid biosynthesis and its inhibition. Nat. Plants 2019, 5, 589–594. [Google Scholar] [CrossRef]
- Werbrouck, S.P.; Saibo, N.J.; Dhuyvetter, H.; De Schepper, S.; Van Der Straeten, D.; Debergh, P.C. Physiological and morphological evidence of brassinosteroid-biosynthesis inhibition by the fungicide imazalil. Physiol. Plant. 2003, 119, 69–77. [Google Scholar] [CrossRef]
- Liu, J.; Nie, M.; Zhou, Q.; Gao, S.; Jiang, W.; Chung, L.W.; Tang, W.; Ding, K. Enantioselective palladium-catalyzed diboration of 1,1-disubstituted allenes. Chem. Sci. 2017, 8, 5161–5165. [Google Scholar] [CrossRef]
- Matsumoto, T.; Yamada, K.; Iwasaki, I.; Yoshizawa, Y.; Oh, K. New Compounds Induce Brassinosteroid Deficient-like Phenotypes in Rice. Plants 2013, 2, 521–529. [Google Scholar] [CrossRef]
- Sekimata, K.; Kimura, T.; Kaneko, I.; Nakano, T.; Yoneyama, K.; Takeuchi, Y.; Yoshida, S.; Asami, T. A specific brassinosteroid biosynthesis inhibitor, Brz2001: Evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta 2001, 213, 716–721. [Google Scholar] [CrossRef]
- Takakusagi, Y.; Manita, D.; Kusayanagi, T.; Izaguirre-Carbonell, J.; Takakusagi, K.; Kuramochi, K.; Iwabata, K.; Kanai, Y.; Sakaguchi, K.; Sugawara, F. Mapping a disordered portion of the Brz2001-binding site on a plant monooxygenase, DWARF4, using a quartz-crystal microbalance biosensor-based T7 phage display. Assay Drug Dev. Technol. 2013, 11, 206–215. [Google Scholar] [CrossRef]
- Xia, X.J.; Wang, Y.J.; Zhou, Y.H.; Tao, Y.; Mao, W.H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.Q. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Terakado, J.; Fujihara, S.; Goto, S.; Kuratani, R.; Suzuki, A.; Yoshida, S.; Yoneyama, T. Systemic Effect of a Brassinosteroid on Root Nodule Formation in Soybean as Revealed by the Application of Brassinolide and Brassinazole. Soil Sci. Plant Nutr. 2005, 51, 389–395. [Google Scholar] [CrossRef]
- Sekimata, K.; Uzawa, J.; Han, S.Y.; Yoneyama, K.; Takeuchi, Y.; Yoshida, S.; Asami, T. Brz220 a novel brassinosteroid biosynthesis inhibitor: Stereochemical structure–activity relationship. Tetrahedron Asymmetry 2002, 13, 1875–1878. [Google Scholar] [CrossRef]
- Scheinfeld, N. Ketoconazole: A review of a workhorse antifungal molecule with a focus on new foam and gel formulations. Drugs Today (Barc) 2008, 44, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Foy, D.S.; Trepanier, L.A. Antifungal treatment of small animal veterinary patients. Vet. Clin. N. Am. Small Anim. Pract. 2010, 40, 1171–1188. [Google Scholar] [CrossRef]
- Vanden Bossche, H.; Engelen, M.; Rochette, F. Antifungal agents of use in animal health--chemical, biochemical and pharmacological aspects. J. Vet. Pharm. 2003, 26, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Faergemann, J.; Borgers, M.; Degreef, H. A new ketoconazole topical gel formulation in seborrhoeic dermatitis: An updated review of the mechanism. Expert Opin. Pharm. 2007, 8, 1365–1371. [Google Scholar] [CrossRef]
- Green, C.A.; Farr, P.M.; Shuster, S. Treatment of seborrhoeic dermatitis with ketoconazole: II. Response of seborrhoeic dermatitis of the face, scalp and trunk to topical ketoconazole. Br. J. Dermatol. 1987, 116, 217–221. [Google Scholar] [CrossRef]
- Torres, M.A.; Mohamed, J.; Cavazos-Adame, H.; Martinez, L.A. Topical ketoconazole for fungal keratitis. Am. J. Ophthalmol. 1985, 100, 293–298. [Google Scholar] [CrossRef]
- Gupta, A.K.; Lyons, D.C. The Rise and Fall of Oral Ketoconazole. J. Cutan. Med. Surg. 2015, 19, 352–357. [Google Scholar] [CrossRef]
- Venkateswarlu, K.; Denning, D.W.; Kelly, S.L. Inhibition and interaction of cytochrome P450 of Candida krusei with azole antifungal drugs. J. Med. Vet. Mycol. 1997, 35, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Lepesheva, G.I.; Hargrove, T.Y.; Kleshchenko, Y.; Nes, W.D.; Villalta, F.; Waterman, M.R. CYP51: A major drug target in the cytochrome P450 superfamily. Lipids 2008, 43, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, T.; Matsumoto, T.; Yamada, K.; Yoshizawa, Y.; Oh, K. Synthesis of New Brassinosteroid Biosynthesis Inhibitor with Coumarin Moiety as a Fluorescent Probe. Int. J. Chem. Eng. Appl. 2015, 5, 379–383. [Google Scholar] [CrossRef][Green Version]
- Yamada, K.; Yoshizawa, Y.; Oh, K. Synthesis of 2RS,4RS-1-[2-phenyl-4-[2-(2-trifluromethoxy-phenoxy)-ethyl]-1,3-dioxolan-2-yl-methyl]-1H-1,2,4-triazole derivatives as potent inhibitors of brassinosteroid biosynthesis. Molecules 2012, 17, 4460–4473. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Yamada, K.; Yoshizawa, Y. Asymmetric synthesis and effect of absolute stereochemistry of YCZ-2013, a brassinosteroid biosynthesis inhibitor. Bioorg. Med. Chem. Lett. 2013, 23, 6915–6919. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Nacev, B.A.; Bhat, S.; Liu, J.O. Impact of Absolute Stereochemistry on the Antiangiogenic and Antifungal Activities of Itraconazole. ACS Med. Chem. Lett. 2010, 1, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, N.V.; Jenkins, J.E.E.; Collins, B.; Groves, S.J. Powdery mildew (Erysiphe graminis) on winter wheat: Control with fungicides and the effects on yield. Crop Prot. 1994, 13, 93–98. [Google Scholar] [CrossRef]
- Švec, M.; Miklovičová, M.; Sýkora, M.; Krippel, E. Fungicide sensitivity of populations of wheat powdery mildew (Erysiphe graminis f.sp. Tritici) in Central Europe in 1993. Pestic. Sci. 1995, 43, 47–52. [Google Scholar] [CrossRef]
- Gooding, M.J.; Smith, S.P.; Davies, W.P.; Kettlewell, P.S. Effects of late-season applications of propiconazole and tridemorph on disease, senescence, grain development and the breadmaking quality of winter wheat. Crop Prot. 1994, 13, 362–370. [Google Scholar] [CrossRef]
- Olvång, H. Sensitivity of Drechslera teres and Septoria nodorum to sterol-biosynthesis inhibitors. Neth. J. Plant Pathol. 1988, 94, 57–68. [Google Scholar] [CrossRef]
- Loughman, R.; Thomas, G.J. Fungicide and cultivar control of Septoria diseases of wheat. Crop Prot. 1992, 11, 349–354. [Google Scholar] [CrossRef]
- Wicks, T.J. Fungicidal control of leaf spot (Septoria apiicola) of celery. Aust. J. Exp. Agric. 1989, 29, 261–266. [Google Scholar] [CrossRef]
- Clarkson, J.P.; Kennedy, R.; Phleps, K.; Davies, J.; Bowtell, J. Quantifying the effect of reduced doses of propiconazole (Tilt) and initial disease incidence on leek rust development. Plant Pathol. 1997, 46, 952–963. [Google Scholar] [CrossRef]
- Dickens, J.S. Studies on the chemical control of chrysanthemum white rust caused by Puccinia horiana. Plant Pathol. 1990, 39, 434–442. [Google Scholar] [CrossRef]
- Jørgensen, L.N.; Nielsen, B.J. Control of yellow rust (Puccinia striiformis) on winter wheat by ergosterol inhibitors at full and reduced dosages. Crop Prot. 1994, 13, 323–330. [Google Scholar] [CrossRef]
- Boyacioglu, D.; Hettiarachchy, N.S.; Stack, R.W. Effect of three systemic fungicides on deoxynivalenol (vomitoxin) production by Fusarium graminearum in wheat. Can. J. Plant Sci. 1992, 72, 93–101. [Google Scholar] [CrossRef]
- Nel, B.; Steinberg, C.; Labuschagne, N.; Viljoen, A. Evaluation of fungicides and sterilants for potential application in the management of Fusarium wilt of banana. Crop Prot. 2007, 26, 697–705. [Google Scholar] [CrossRef]
- Paul, P.A.; Lipps, P.E.; Hershman, D.E.; McMullen, M.P.; Draper, M.A.; Madden, L.V. Efficacy of Triazole-Based Fungicides for Fusarium Head Blight and Deoxynivalenol Control in Wheat: A Multivariate Meta-Analysis. Phytopathology 2008, 98, 999–1011. [Google Scholar] [CrossRef]
- Yoshida, Y.; Aoyama, Y. Sterol 14alpha-demethylase and its inhibition: Structural considerations on interaction of azole antifungal agents with lanosterol 14alpha-demethylase (P-450(14DM)) of yeast. Biochem. Soc. Trans. 1991, 19, 778–782. [Google Scholar] [CrossRef]
- Wiggins, T.E.; Baldwin, B.C. Binding of azole fungicides related to diclobutrazol to cytochrome P-450. Pestic. Sci. 1984, 15, 206–209. [Google Scholar] [CrossRef]
- Warrilow, A.G.; Parker, J.E.; Kelly, D.E.; Kelly, S.L. Azole affinity of sterol 14alpha-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob. Agents Chemother. 2013, 57, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.M.; Goldstone, J.V.; Lamb, D.C.; Kubota, A.; Lemaire, B.; Stegeman, J.J. Identification, modeling and ligand affinity of early deuterostome CYP51s, and functional characterization of recombinant zebrafish sterol 14alpha-demethylase. Biochim. Biophys. Acta 2014, 1840, 1825–1836. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hartwig, T.; Corvalan, C.; Best, N.B.; Budka, J.S.; Zhu, J.Y.; Choe, S.; Schulz, B. Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 2012, 7, e36625. [Google Scholar] [CrossRef] [PubMed]
- Leath, S.; Bowen, K.L. Effects of powdery mildew, triadimenol seed treatment, and triadimefon foliar sprays on yield of winter wheat in North Carolina. Phytopathology 1989, 79, 152–155. [Google Scholar] [CrossRef]
- Frank, J.A.; Ayers, J.E. Effect of triadimenol seed treatment on powdery mildew epidemics on winter wheat. Phytopathology 1986, 76, 254–257. [Google Scholar] [CrossRef]
- Lipps, P.E.; Madden, L.V. Effect of fungicide application timing on control of powdery mildew and grain yield of winter wheat. Plant Dis. 1989, 73, 991–994. [Google Scholar] [CrossRef]
- Milus, E.A. Effect of foliar fungicides on disease control, yield and test weight of soft red winter wheat. Crop Prot. 1994, 13, 291–295. [Google Scholar] [CrossRef]
- Mueller, D.S.; Jeffers, S.N.; Buck, J.W. Effect of Timing of Fungicide Applications on Development of Rusts on Daylily, Geranium, and Sunflower. Plant Dis. 2004, 88, 657–661. [Google Scholar] [CrossRef]
- Fletcher, R.A.; Arnold, V. Stimulation of cytokinins and chlorophyll synthesis in cucumber cotyledons by triadimefon. Physiol. Plant. 1986, 66, 197–201. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Gopi, R.; Panneerselvam, R. Growth and photosynthetic pigments responses of two varieties of Catharanthus roseus to triadimefon treatment. C. R. Biol. 2008, 331, 272–277. [Google Scholar] [CrossRef]
- Buchenauer, H.; Röhner, E. Effect of triadimefon and triadimenol on growth of various plant species as well as on gibberellin content and sterol metabolism in shoots of barley seedlings. Pestic. Biochem. Physiol. 1981, 15, 58–70. [Google Scholar] [CrossRef]
- Eckert, J.W.; Sievert, J.R.; Ratnayake, M. Reduction of imazalil effectiveness against citrus green mold in california packinghouses by resistant biotypes of Penicillium digitatum. Plant Dis. 1994, 78, 971–974. [Google Scholar] [CrossRef]
- Bus, V.G.; Bongers, A.J.; Risse, L.A. Occurrence of Penicillium digitatum and P. italicum resistant to benomyl, thiabendazole and imazalil on citrus fruit from different geographic origins. Plant Dis. 1991, 75, 1098–1100. [Google Scholar] [CrossRef]
- Cabras, P.; Schirra, M.; Pirisi, F.M.; Garau, V.L.; Angioni, A. Factors affecting imazalil and thiabendazole uptake and persistence in citrus fruits following dip treatments. J. Agric. Food Chem. 1999, 47, 3352–3354. [Google Scholar] [CrossRef]
- Boubaker, H.; Saadi, B.; Boudyach, E.H.; Benaoumar, A.A. Sensitivity of Penicillium digitatum and P. italicum to Imazalil and Thiabendazole in Morocco. Plant Pathol. J. 2009, 8, 152–158. [Google Scholar] [CrossRef][Green Version]
- Siegel, M.R.; Ragsdale, N.N. Antifungal mode of action of imazalil. Pestic. Biochem. Physiol. 1978, 9, 48–56. [Google Scholar] [CrossRef]
- Kerkenaar, A.; Van Rossum, J.M.; Versluis, G.G.; Marsman, J.W. Effect of fenpropimorph and imazalil on sterol biosynthesis in Penicillium italicum. Pestic. Sci. 1984, 15, 177–187. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, L.; Zhang, J.; Han, R.; Li, S.; Yuan, Y.; Wan, J.; Xiao, W.; Liu, D. Optimised expression and spectral analysis of the target enzyme CYP51 from Penicillium digitatum with possible new DMI fungicides. Pest. Manag. Sci. 2010, 66, 1344–1350. [Google Scholar] [CrossRef]
- Fletcher, J.T.; Smewin, B.J.; Cook, R.T. Tomato powdery mildew. Plant Pathol. J. 1988, 37, 594–598. [Google Scholar] [CrossRef]
- Schwabe, W.F.S.; Jones, A.L.; Jonker, J.P. Greenhouse Evaluation of the Curative and Protective Action of Sterol-Inhibiting Fungicides against Apple Scab. Phytopathology 1984, 74, 249–252. [Google Scholar] [CrossRef]
- Carisse, O.; Pelletier, J.R. Sensitivity distribution of Venturia inaequalis to fenarimol in Québec apple orchards. Phytoprotection 1994, 75, 35–43. [Google Scholar] [CrossRef]
- Köller, W.; Wilcox, W.F. Interactive Effects of Dodine and the DMI Fungicide Fenarimol in the Control of Apple Scab. Plant Dis. 2000, 84, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.C.; Szkolnik, M.; Meyer, F.W. Suppression of cedar apple rust pycnia on apple leaves following postinfection applications of fenarimol and triforine. Phytopathology 1978, 68, 1805–1809. [Google Scholar] [CrossRef]
- Burden, R.S.; Cooke, D.T.; Carter, G.A. Inhibitors of sterol biosynthesis and growth in plants and fungi. Phytochemistry 1989, 28, 1791–1804. [Google Scholar] [CrossRef]
- Coolbaugh, R.C.; Swanson, D.I.; West, C.A. Comparative Effects of Ancymidol and Its Analogs on Growth of Peas and Ent-Kaurene Oxidation in Cell-Free Extracts of Immature Marah macrocarpus Endosperm. Plant Physiol. 1982, 69, 707–711. [Google Scholar] [CrossRef]
- Taylor, H.M.; Jones, C.D.; Davenport, J.D.; Hirsch, K.S.; Kress, T.J.; Weaver, D. Aromatase inhibition by 5-substituted pyrimidines and dihydropyrimidines. J. Med. Chem. 1987, 30, 1359–1365. [Google Scholar] [CrossRef]
- Wang, J.M.; Asami, T.; Yoshida, S.; Murofushi, N. Biological evaluation of 5-substituted pyrimidine derivatives as inhibitors of brassinosteroid biosynthesis. Biosci. Biotechnol. Biochem. 2001, 65, 817–822. [Google Scholar] [CrossRef]
- Jenkins, E.P.; Hsieh, C.L.; Milatovich, A.; Normington, K.; Berman, D.M.; Francke, U.; Russell, D.W. Characterization and chromosomal mapping of a human steroid 5 alpha-reductase gene and pseudogene and mapping of the mouse homologue. Genomics 1991, 11, 1102–1112. [Google Scholar] [CrossRef]
- Thigpen, A.E.; Davis, D.L.; Milatovich, A.; Mendonca, B.B.; Imperato-McGinley, J.; Griffin, J.E.; Francke, U.; Wilson, J.D.; Russell, D.W. Molecular genetics of steroid 5 alpha-reductase 2 deficiency. J. Clin. Investig. 1992, 90, 799–809. [Google Scholar] [CrossRef]
- Uemura, M.; Tamura, K.; Chung, S.; Honma, S.; Okuyama, A.; Nakamura, Y.; Nakagawa, H. Novel 5 alpha-steroid reductase (SRD5A3, type-3) is overexpressed in hormone-refractory prostate cancer. Cancer Sci. 2008, 99, 81–86. [Google Scholar] [CrossRef]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Xiao, Y.; Li, X.; Lu, X.; Deng, W.; Li, D.; Hou, L.; Hu, M.; Li, Y.; Pei, Y. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 2007, 51, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Prada, I.; Kortekamp, A. Effect of sterol biosynthesis inhibitors and azole-type inducers on growth and development of Plasmopara viticola on grapevine. Vitis 2015, 54, 91–96. [Google Scholar]
- Asami, T.; Oh, K.; Jikumaru, Y.; Shimada, Y.; Kaneko, I.; Nakano, T.; Takatsuto, S.; Fujioka, S.; Yoshida, S. A mammalian steroid action inhibitor spironolactone retards plant growth by inhibition of brassinosteroid action and induces light-induced gene expression in the dark. J. Steroid Biochem. Mol. Biol. 2004, 91, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Pitt, B.; Zannad, F.; Remme, W.J.; Cody, R.; Castaigne, A.; Perez, A.; Palensky, J.; Wittes, J. The effect of spironolactone on morbidity and mortality in patients with severe heart failure. Randomized Aldactone Evaluation Study Investigators. N. Engl. J. Med. 1999, 341, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Adlin, E.V.; Marks, A.D.; Channick, B.J. Spironolactone and hydrochlorothiazide in essential hypertension. Blood pressure response and plasma renin activity. Arch. Intern. Med. 1972, 130, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Chapman, N.; Dobson, J.; Wilson, S.; Dahlof, B.; Sever, P.S.; Wedel, H.; Poulter, N.R.; Anglo-Scandinavian Cardiac Outcomes Trial, I. Effect of spironolactone on blood pressure in subjects with resistant hypertension. Hypertension 2007, 49, 839–845. [Google Scholar] [CrossRef]
- Engbaek, M.; Hjerrild, M.; Hallas, J.; Jacobsen, I.A. The effect of low-dose spironolactone on resistant hypertension. J. Am. Soc. Hypertens. 2010, 4, 290–294. [Google Scholar] [CrossRef]
- Colussi, G.; Rombola, G.; De Ferrari, M.E.; Macaluso, M.; Minetti, L. Correction of hypokalemia with antialdosterone therapy in Gitelman’s syndrome. Am. J. Nephrol. 1994, 14, 127–135. [Google Scholar] [CrossRef]
- Yongsiri, S.; Thammakumpee, J.; Prongnamchai, S.; Tengpraettanakorn, P.; Chueansuwan, R.; Tangjaturonrasme, S.; Dinchuthai, P. Randomized, double-blind, placebo-controlled trial of spironolactone for hypokalemia in continuous ambulatory peritoneal dialysis patients. Apher. Dial. 2015, 19, 81–86. [Google Scholar] [CrossRef]
- Messina, M.; Manieri, C.; Biffignandi, P.; Massucchetti, C.; Novi, R.F.; Molinatti, G.M. Antiandrogenic properties of spironolactone. Clinical trial in the management of female hirsutism. J. Endocrinol. Investig. 1983, 6, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Fejes-Toth, G.; Pearce, D.; Naray-Fejes-Toth, A. Subcellular localization of mineralocorticoid receptors in living cells: Effects of receptor agonists and antagonists. Proc. Natl. Acad. Sci. USA 1998, 95, 2973–2978. [Google Scholar] [CrossRef] [PubMed]
- Sam, K.M.; Auger, S.; Luu-The, V.; Poirier, D. Steroidal spiro-gamma-lactones that inhibit 17 beta-hydroxysteroid dehydrogenase activity in human placental microsomes. J. Med. Chem. 1995, 38, 4518–4528. [Google Scholar] [CrossRef]
- Tremblay, M.R.; Luu-The, V.; Leblanc, G.; Noel, P.; Breton, E.; Labrie, F.; Poirier, D. Spironolactone-related inhibitors of type II 17beta-hydroxysteroid dehydrogenase: Chemical synthesis, receptor binding affinities, and proliferative/antiproliferative activities. Bioorg. Med. Chem. 1999, 7, 1013–1023. [Google Scholar] [CrossRef]
- Poirier, D.; Bydal, P.; Tremblay, M.R.; Sam, K.M.; Luu-The, V. Inhibitors of type II 17beta-hydroxysteroid dehydrogenase. Mol. Cell. Endocrinol. 2001, 171, 119–128. [Google Scholar] [CrossRef]
- Mathur, J.; Molnar, G.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Yokota, T.; Adam, G.; Voigt, B.; Nagy, F.; Maas, C.; et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998, 14, 593–602. [Google Scholar] [CrossRef]
- Gendron, J.M.; Haque, A.; Gendron, N.; Chang, T.; Asami, T.; Wang, Z.Y. Chemical genetic dissection of brassinosteroid-ethylene interaction. Mol. Plant 2008, 1, 368–379. [Google Scholar] [CrossRef]
- De Grauwe, L.; Vandenbussche, F.; Tietz, O.; Palme, K.; Van Der Straeten, D. Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 2005, 46, 827–836. [Google Scholar] [CrossRef]
- Herbrecht, R.; Denning, D.W.; Patterson, T.F.; Bennett, J.E.; Greene, R.E.; Oestmann, J.W.; Kern, W.V.; Marr, K.A.; Ribaud, P.; Lortholary, O.; et al. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N. Engl. J. Med. 2002, 347, 408–415. [Google Scholar] [CrossRef]
- Fukuoka, T.; Johnston, D.A.; Winslow, C.A.; de Groot, M.J.; Burt, C.; Hitchcock, C.A.; Filler, S.G. Genetic basis for differential activities of fluconazole and voriconazole against Candida krusei. Antimicrob. Agents Chemother. 2003, 47, 1213–1219. [Google Scholar] [CrossRef]
- Parker, J.E.; Warrilow, A.G.; Price, C.L.; Mullins, J.G.; Kelly, D.E.; Kelly, S.L. Resistance to antifungals that target CYP51. J. Chem. Biol. 2014, 7, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Heintz, R.; Benveniste, P. Plant sterol metabolism. Enzymatic cleavage of the 9beta, 19beta-cyclopropane ring of cyclopropyl sterols in bramble tissue cultures. J. Biol. Chem. 1974, 249, 4267–4274. [Google Scholar] [PubMed]
- Hubert Schaller, H.; Maillot-Vernier, P.; Benveniste, P.; Belliard, G. Sterol composition of tobacco calli selected for resistance to fenpropimorph. Phytochemistry 1991, 30, 2547–2554. [Google Scholar] [CrossRef]
- Back, T.G.; Pharis, R.P. Structure-Activity Studies of Brassinosteroids and the Search for Novel Analogues and Mimetics with Improved Bioactivity. J. Plant Growth Regul. 2003, 22, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Duran, M.I.; Gonzalez, C.; Acosta, A.; Olea, A.F.; Diaz, K.; Espinoza, L. Synthesis of Five Known Brassinosteroid Analogs from Hyodeoxycholic Acid and Their Activities as Plant-Growth Regulators. Int. J. Mol. Sci. 2017, 18, 516. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, D.; Sun, X.; Ding, T.; Lei, B.; Zhang, C. Structure-activity relationship of brassinosteroids and their agricultural practical usages. Steroids 2017, 124, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Irani, N.G.; Di Rubbo, S.; Mylle, E.; Van den Begin, J.; Schneider-Pizon, J.; Hnilikova, J.; Sisa, M.; Buyst, D.; Vilarrasa-Blasi, J.; Szatmari, A.M.; et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 2012, 8, 583–589. [Google Scholar] [CrossRef]
- Irani, N.G.; Di Rubbo, S.; Russinova, E. In vivo imaging of brassinosteroid endocytosis in Arabidopsis. Methods Mol. Biol. 2014, 1209, 107–117. [Google Scholar] [CrossRef]
- Muto, T.; Todoroki, Y. Brassinolide-2,3-acetonide: A brassinolide-induced rice lamina joint inclination antagonist. Bioorg. Med. Chem. 2013, 21, 4413–4419. [Google Scholar] [CrossRef][Green Version]
- Andersen, D.L.; Back, T.G.; Janzen, L.; Michalak, K.; Pharis, R.P.; Sung, G.C. Design, synthesis, and bioactivity of the first nonsteroidal mimetics of brassinolide. J. Org. Chem. 2001, 66, 7129–7141. [Google Scholar] [CrossRef]
- Sugiura, A.; Horoiwa, S.; Aoki, T.; Takimoto, S.; Yamagami, A.; Nakano, T.; Nakagawa, Y.; Miyagawa, H. Discovery of a nonsteroidal brassinolide-like compound, NSBR1. J. Pest. Sci. 2017, 42, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Takimoto, S.; Sugiura, A.; Minami, S.; Tasaka, T.; Nakagawa, Y.; Miyagawa, H. In silico exploration for agonists/antagonists of brassinolide. Bioorg. Med. Chem. Lett. 2016, 26, 1709–1714. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Heng, N.; Dang, X.; Liu, J.; Yao, X.; Zhang, C. Structure based in silico identification of potentially non-steroidal brassinosteroids mimics. Mol. Biosyst. 2017, 13, 1364–1369. [Google Scholar] [CrossRef] [PubMed]
- Van Lookeren Campagne, M.M.; Wang, M.; Spek, W.; Peters, D.; Schaap, P. Lithium respecifies cyclic AMP-induced cell-type specific gene expression in Dictyostelium. Dev. Genet. 1988, 9, 589–596. [Google Scholar] [CrossRef]
- Kao, K.R.; Masui, Y.; Elinson, R.P. Lithium-induced respecification of pattern in Xenopus laevis embryos. Nature 1986, 322, 371–373. [Google Scholar] [CrossRef]
- Bosch, F.; Gomez-Foix, A.M.; Arino, J.; Guinovart, J.J. Effects of lithium ions on glycogen synthase and phosphorylase in rat hepatocytes. J. Biol. Chem. 1986, 261, 16927–16931. [Google Scholar]
- Klein, P.S.; Melton, D.A. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 1996, 93, 8455–8459. [Google Scholar] [CrossRef]
- Stambolic, V.; Ruel, L.; Woodgett, J.R. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr. Biol. 1996, 6, 1664–1668. [Google Scholar] [CrossRef]
- Ryves, W.J.; Harwood, A.J. Lithium inhibits glycogen synthase kinase-3 by competition for magnesium. Biochem. Biophys. Res. Commun. 2001, 280, 720–725. [Google Scholar] [CrossRef]
- Siegfried, E.; Chou, T.B.; Perrimon, N. wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell 1992, 71, 1167–1179. [Google Scholar] [CrossRef]
- Pierce, S.B.; Kimelman, D. Regulation of Spemann organizer formation by the intracellular kinase Xgsk-3. Development 1995, 121, 755–765. [Google Scholar] [PubMed]
- He, X.; Saint-Jeannet, J.P.; Woodgett, J.R.; Varmus, H.E.; Dawid, I.B. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature 1995, 374, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Frame, S.; Cohen, P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001, 359, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Woodgett, J.R.; Cohen, P. Multisite phosphorylation of glycogen synthase. Molecular basis for the substrate specificity of glycogen synthase kinase-3 and casein kinase-II (glycogen synthase kinase-5). Biochim. Biophys. Acta 1984, 788, 339–347. [Google Scholar] [CrossRef]
- Yan, Z.; Zhao, J.; Peng, P.; Chihara, R.K.; Li, J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009, 150, 710–721. [Google Scholar] [CrossRef]
- Zhang, S.; Cai, Z.; Wang, X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 4543–4548. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Huang, L.C.; Wei, K.; Yu, J.W.; Zhang, C.Q.; Liu, Q.Q. Light involved regulation of BZR1 stability and phosphorylation status to coordinate plant growth in Arabidopsis. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef]
- Huang, H.Y.; Jiang, W.B.; Hu, Y.W.; Wu, P.; Zhu, J.Y.; Liang, W.Q.; Wang, Z.Y.; Lin, W.H. BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 2013, 6, 456–469. [Google Scholar] [CrossRef]
- Shahzad, B.; Tanveer, M.; Hassan, W.; Shah, A.N.; Anjum, S.A.; Cheema, S.A.; Ali, I. Lithium toxicity in plants: Reasons, mechanisms and remediation possibilities - A review. Plant Physiol. Biochem. 2016, 107, 104–115. [Google Scholar] [CrossRef]
- Bingham, F.T.; Bradford, G.R.; Page, A.L. Toxicity of lithium to plants. Calif. Agric. 1964, 18, 6–7. [Google Scholar]
- Shahzad, B.; Mughal, M.N.; Tanveer, M.; Gupta, D.; Abbas, G. Is lithium biologically an important or toxic element to living organisms? An overview. Environ. Sci. Pollut. Res. Int. 2017, 24, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium transport in plant cells. Biochim. Biophys. Acta 2000, 1465, 140–151. [Google Scholar] [CrossRef]
- Gillaspy, G.E.; Keddie, J.S.; Oda, K.; Gruissem, W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell 1995, 7, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Bueso, E.; Alejandro, S.; Carbonell, P.; Perez-Amador, M.A.; Fayos, J.; Belles, J.M.; Rodriguez, P.L.; Serrano, R. The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J. 2007, 52, 1052–1065. [Google Scholar] [CrossRef]
- Lehfeldt, C.; Shirley, A.M.; Meyer, K.; Ruegger, M.O.; Cusumano, J.C.; Viitanen, P.V.; Strack, D.; Chapple, C. Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 2000, 12, 1295–1306. [Google Scholar] [CrossRef]
- Jung, P.J.M.; Leipner, J.; Lachia, M.D.; De Masmaeker, A.; McLachlan, M.M.W. Plant Growth Regulating Compounds. U.S. Patent 9,345,244 B342, 24 May 2016. [Google Scholar]
- Liu, S.; He, Y.; Tian, H.; Yu, C.; Tan, W.; Li, Z.; Duan, L. Application of Brassinosteroid Mimetics Improves Growth and Tolerance of Maize to Nicosulfuron Toxicity. J. Plant Growth Regul. 2019, 38, 701–712. [Google Scholar] [CrossRef]
- Kim, S.K.; Asano, T.; Marumo, S. Biological Activity of Brassinosteroid Inhibitor KM-01 Produced by a Fungus Drechslera avenae. Biosci. Biotechnol. Biochem. 1995, 58, 1394–1397. [Google Scholar] [CrossRef]
- Kim, S.K.; Hatori, M.; Ojika, M.; Sakagami, Y.; Marumo, S. KM-01, a brassinolide inhibitor, its production, isolation and structure from two fungi Drechslera avenae and Pycnoporus coccineus. Bioorg. Med. Chem. 1998, 6, 1975–1982. [Google Scholar] [CrossRef]
- Larsen, P.B. Brassinosteroid Mimetics. International Patent WO 206/183519, 17 November 2016. [Google Scholar]
- Song, L.I.; Zhou, X.Y.; Li, L.I.; Xue, L.J.; Yang, X.I.; Xue, H.W. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant 2009, 2, 755–772. [Google Scholar] [CrossRef]
- von Sivers, L.; Jaspar, H.; Johst, B.; Roese, M.; Bitterlich, M.; Franken, P.; Kuhn, C. Brassinosteroids Affect the Symbiosis Between the AM Fungus Rhizoglomus irregularis and Solanaceous Host Plants. Front. Plant Sci. 2019, 10, 571. [Google Scholar] [CrossRef]
- Wang, X.; Teng, Y.; Zhang, N.; Christie, P.; Li, Z.; Luo, Y.; Wang, J. Rhizobial symbiosis alleviates polychlorinated biphenyls-induced systematic oxidative stress via brassinosteroids signaling in alfalfa. Sci. Total Environ. 2017, 592, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, T.; Yasuda, D.; Mori, T.; Yoshimitsu, Y.; Nakamura, Y.; Yoshida, S.; Asami, T.; Okamoto, S.; Matsuo, T. Characterization of brassinosteroid-regulated proteins in a nuclear-enriched fraction of Arabidopsis suspension-cultured cells. Plant Physiol. Biochem. 2011, 49, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, T.; Yoshimitsu, Y.; Nakamura, Y.; Okamoto, S.; Matsuo, T. Does brassinosteroid function require chromatin remodeling? Plant Signal. Behav. 2011, 6, 1824–1827. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nie, S.; Huang, S.; Wang, S.; Mao, Y.; Liu, J.; Ma, R.; Wang, X. Enhanced brassinosteroid signaling intensity via SlBRI1 overexpression negatively regulates drought resistance in a manner opposite of that via exogenous BR application in tomato. Plant Physiol. Biochem. 2019, 138, 36–47. [Google Scholar] [CrossRef]
- Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Asami, T.; Yu, J.Q. A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 2008, 59, 2299–2308. [Google Scholar] [CrossRef]
- Zhu, T.; Tan, W.R.; Deng, X.G.; Zheng, T.; Zhang, D.W.; Lin, H.H. Effects of brassinosteroids on quality attributes and ethylene synthesis in postharvest tomato fruit. Postharvest Biol. Technol. 2015, 100, 196–204. [Google Scholar] [CrossRef]
- Chai, Y.; Zhang, Q.; Tian, L.; Li, C.L.; Xing, Y.; Qin, L.; Shen, Y.Y. Brassinosteroid is involved in strawberry fruit ripening. Plant Growth Regul. 2013, 69, 63–69. [Google Scholar] [CrossRef]
- Symons, G.M.; Davies, C.; Shavrukov, Y.; Dry, I.B.; Reid, J.B.; Thomas, M.R. Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol. 2006, 140, 150–158. [Google Scholar] [CrossRef]
- Pereira-Netto, A.B.; Roessner, U.; Fujioka, S.; Bacic, A.; Asami, T.; Yoshida, S.; Clouse, S.D. Shooting control by brassinosteroids: Metabolomic analysis and effect of brassinazole on Malus prunifolia, the Marubakaido apple rootstock. Tree Physiol. 2009, 29, 607–620. [Google Scholar] [CrossRef]
- Deng, X.G.; Zhu, T.; Peng, X.J.; Xi, D.H.; Guo, H.; Yin, Y.; Zhang, D.W.; Lin, H.H. Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana. Sci. Rep. 2016, 6, 20579. [Google Scholar] [CrossRef]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed]
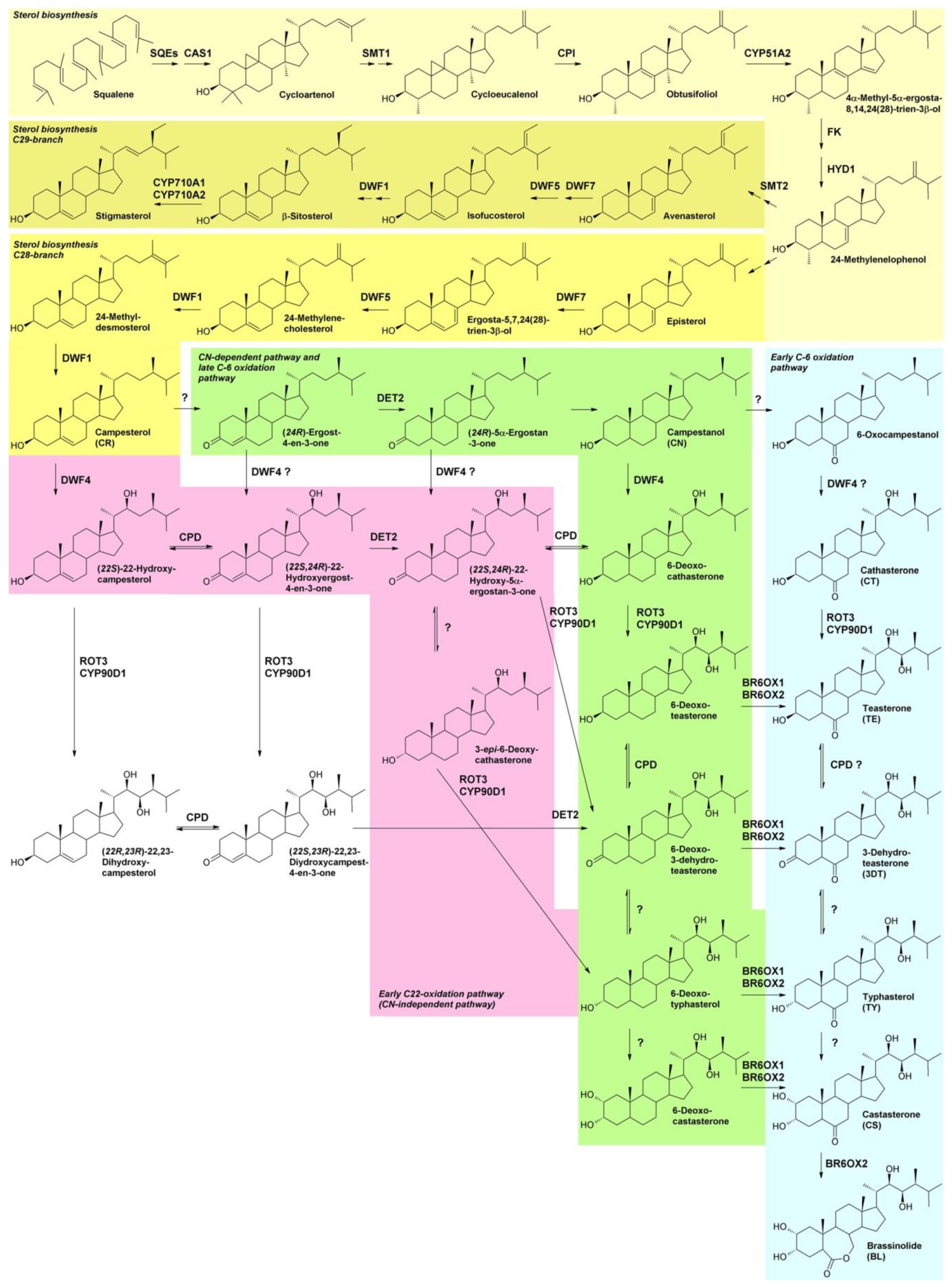
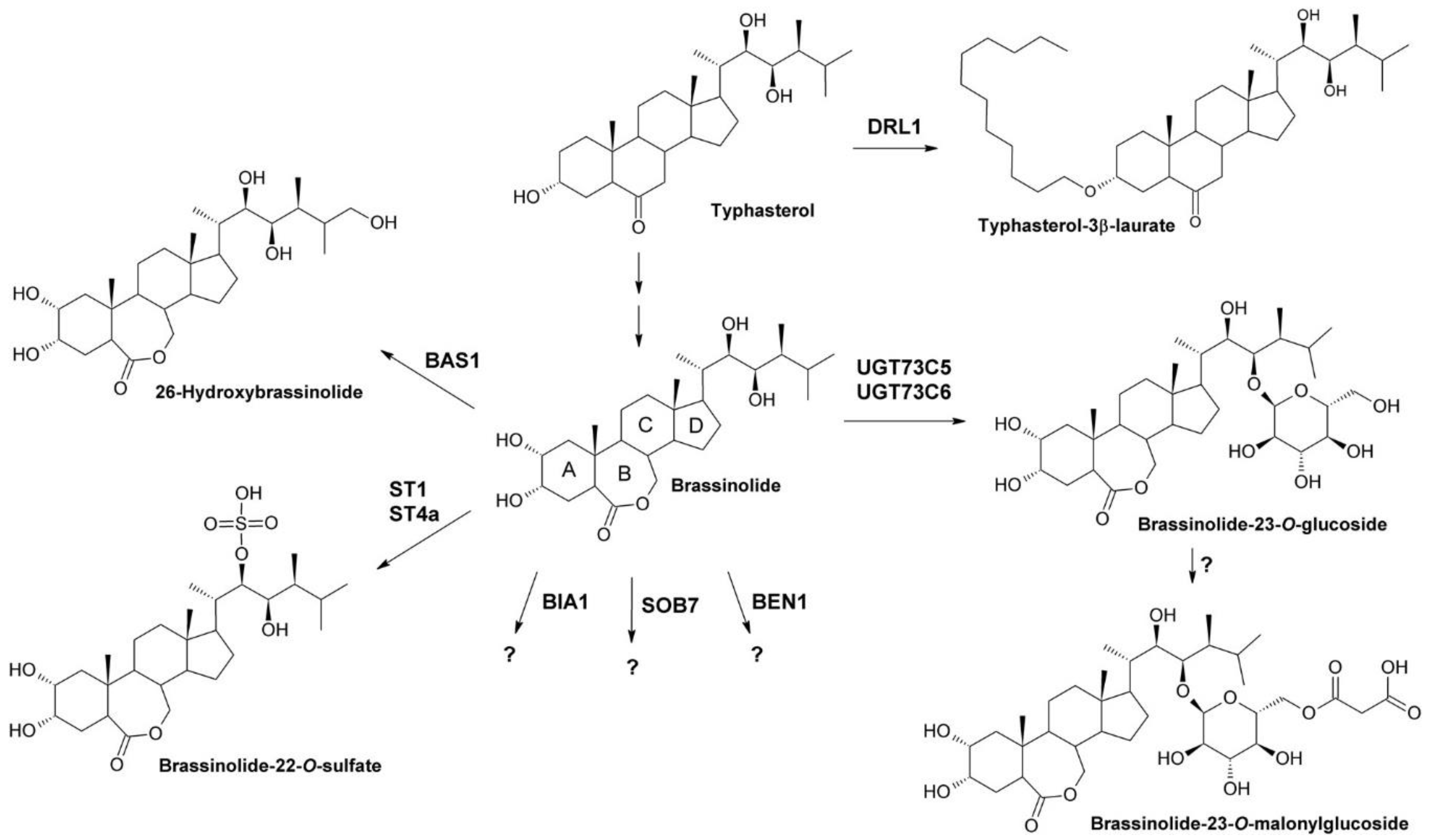
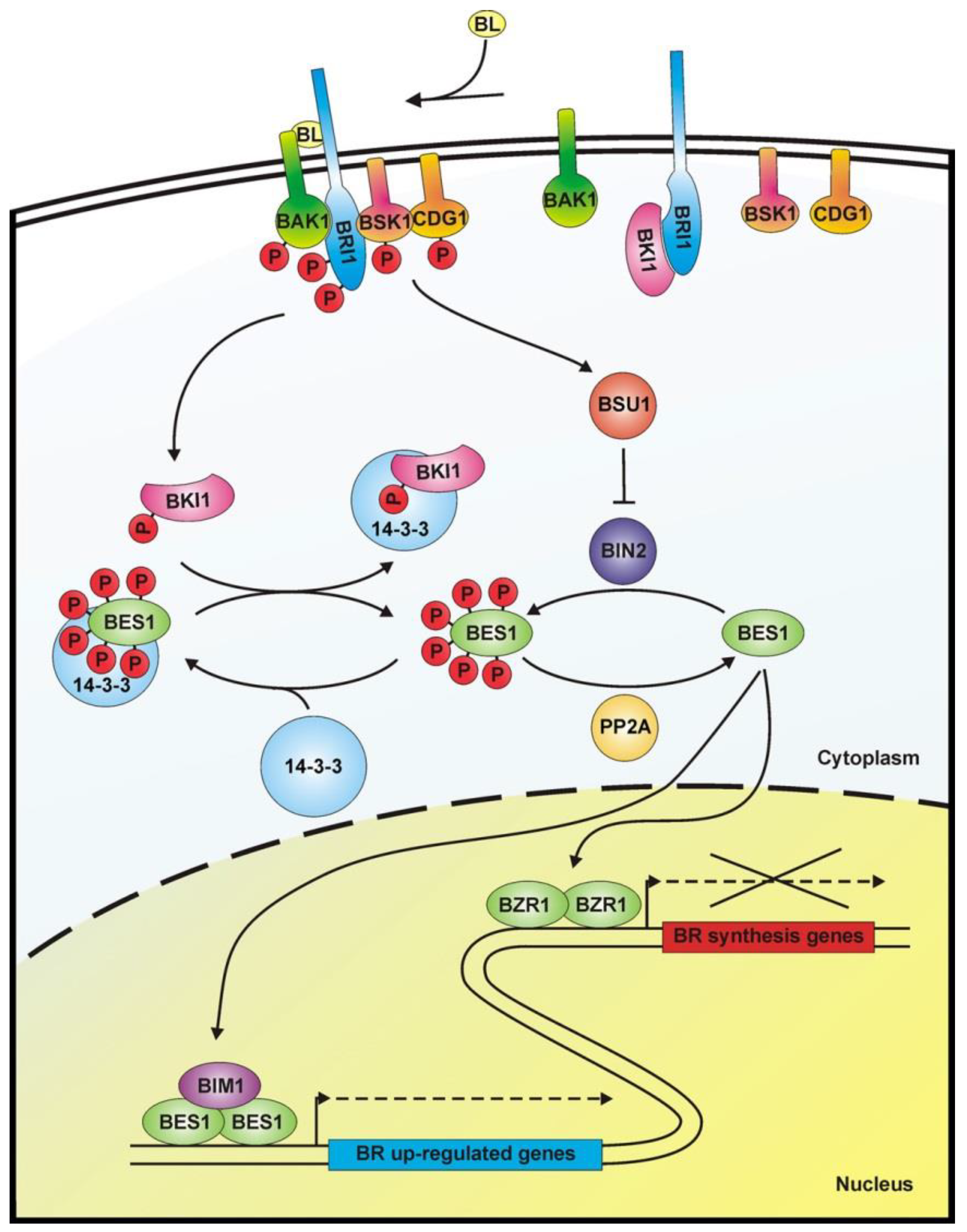
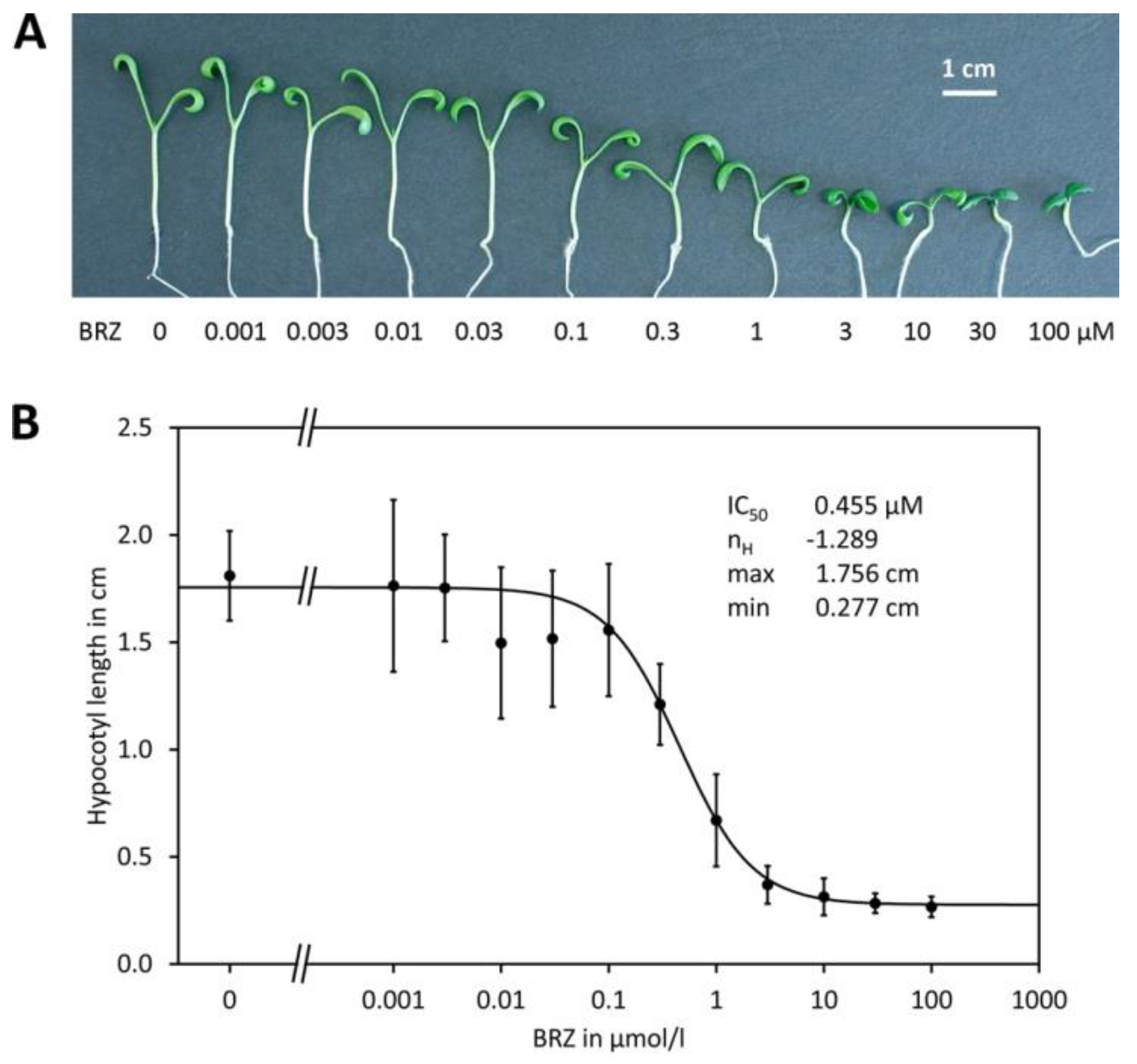
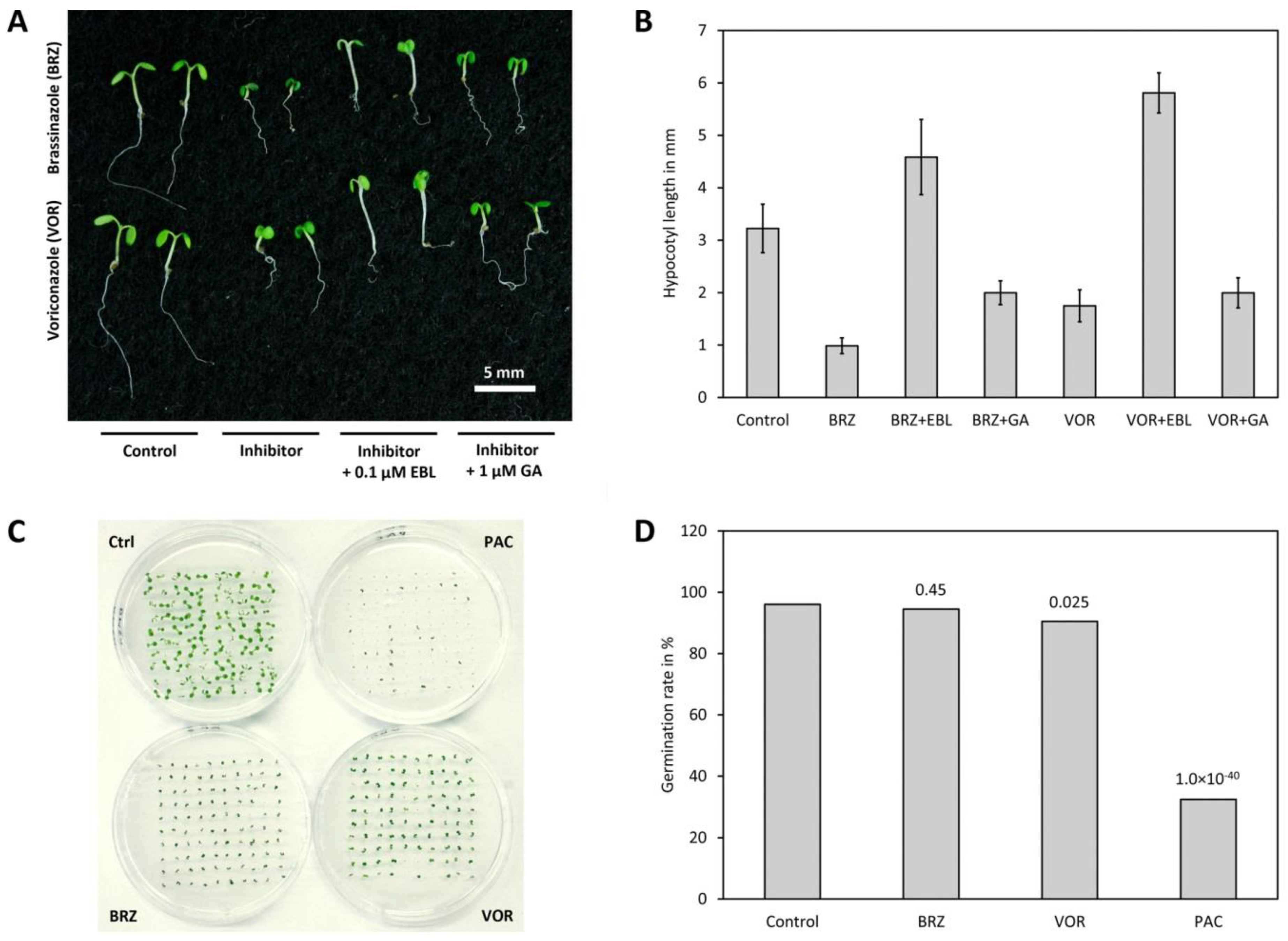
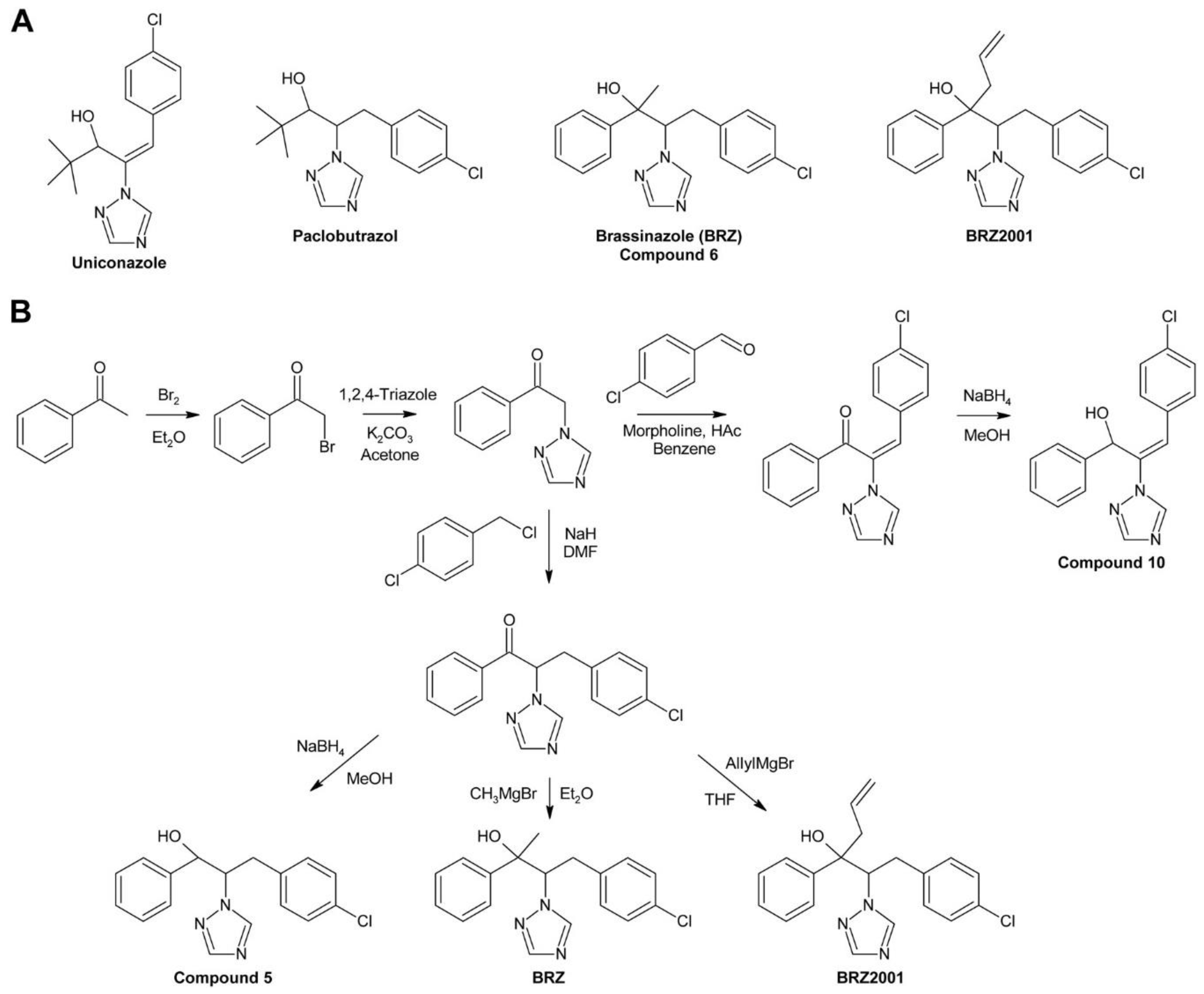
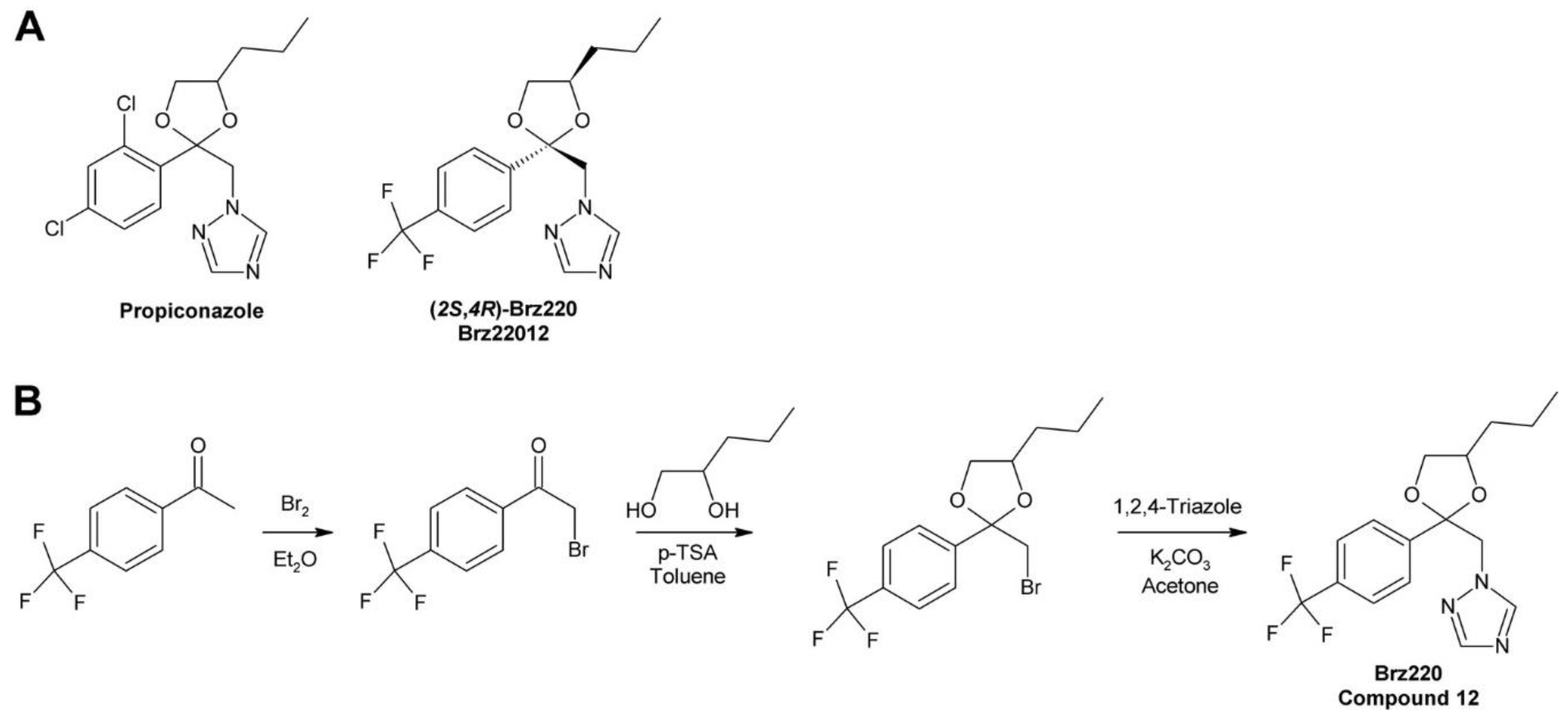
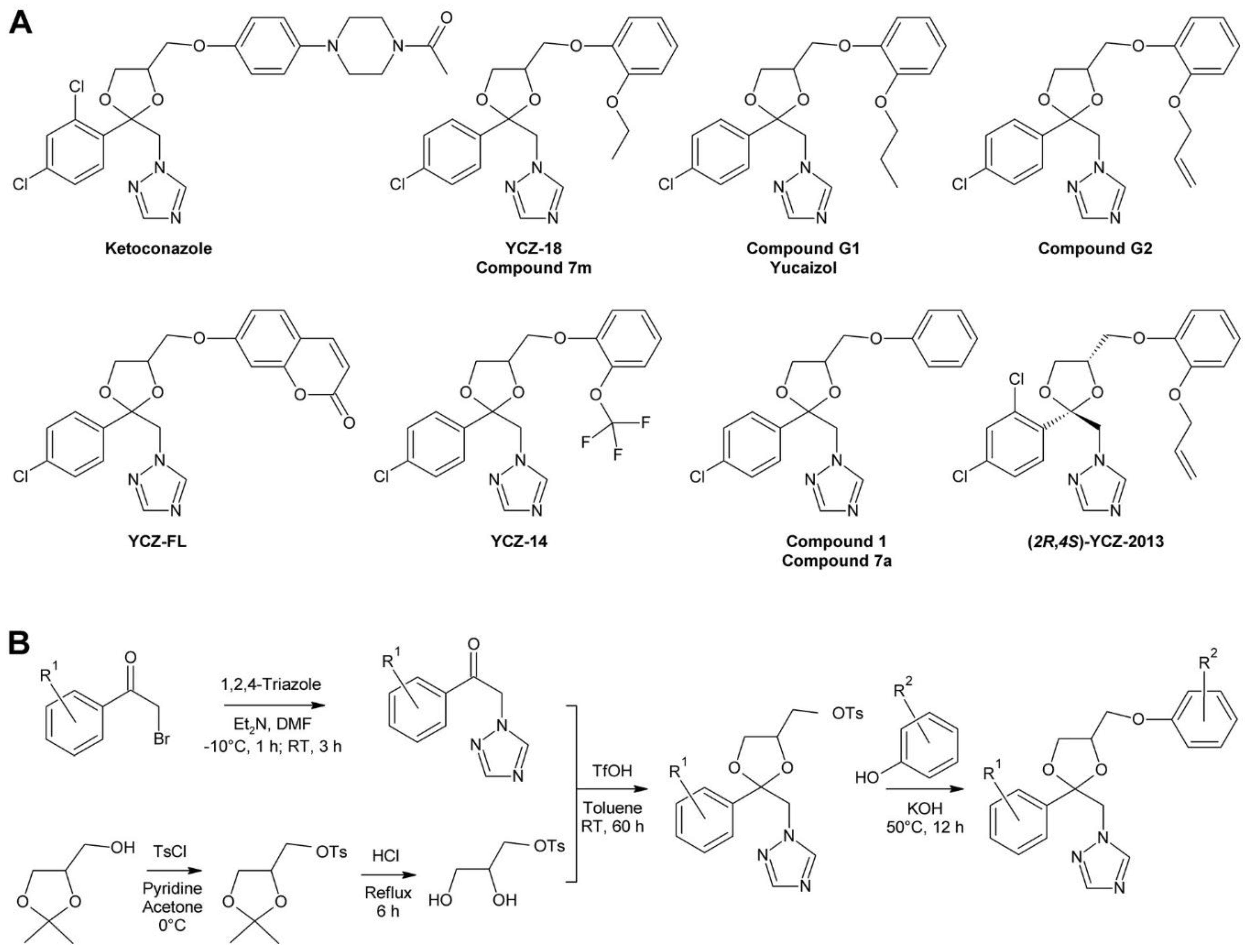
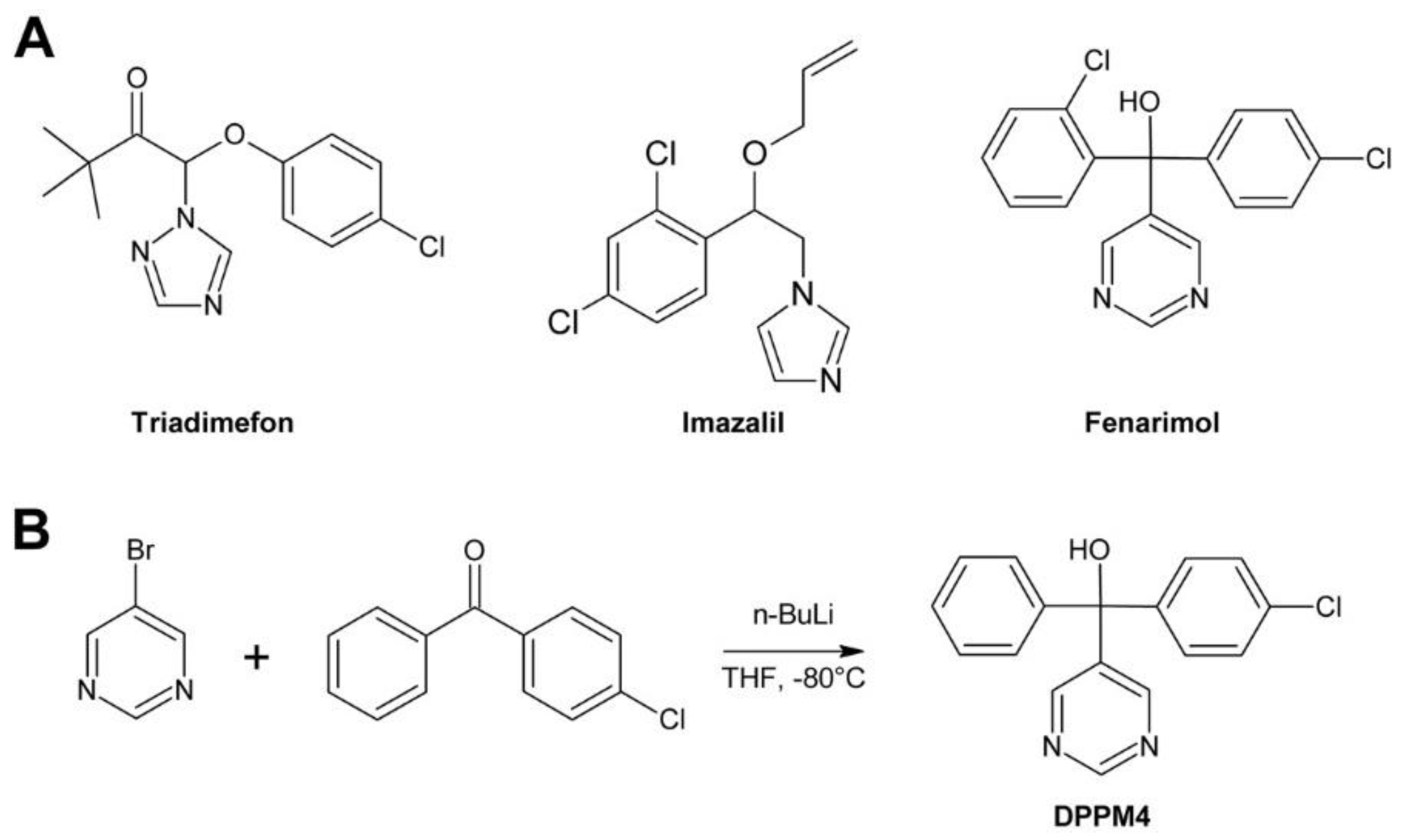
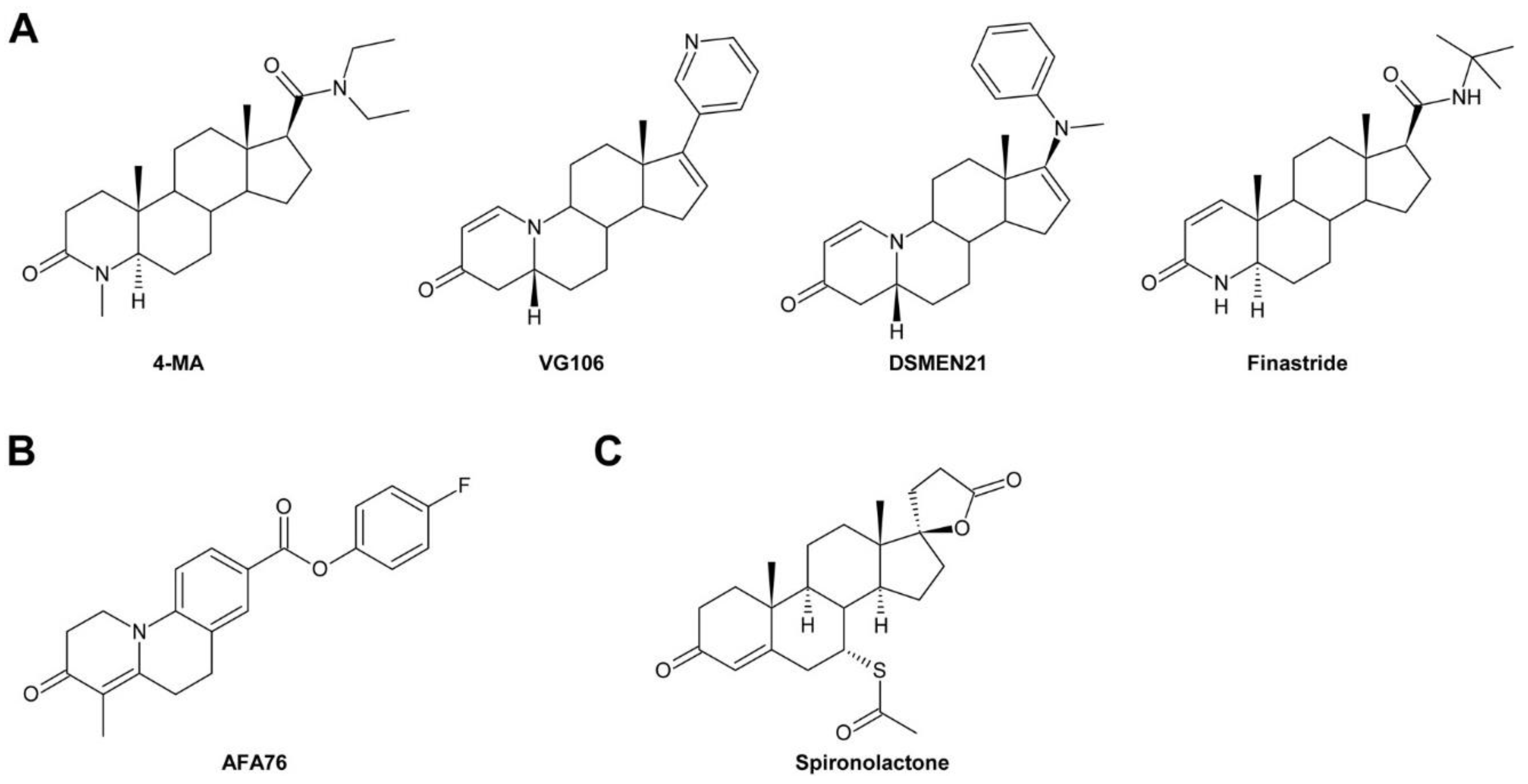
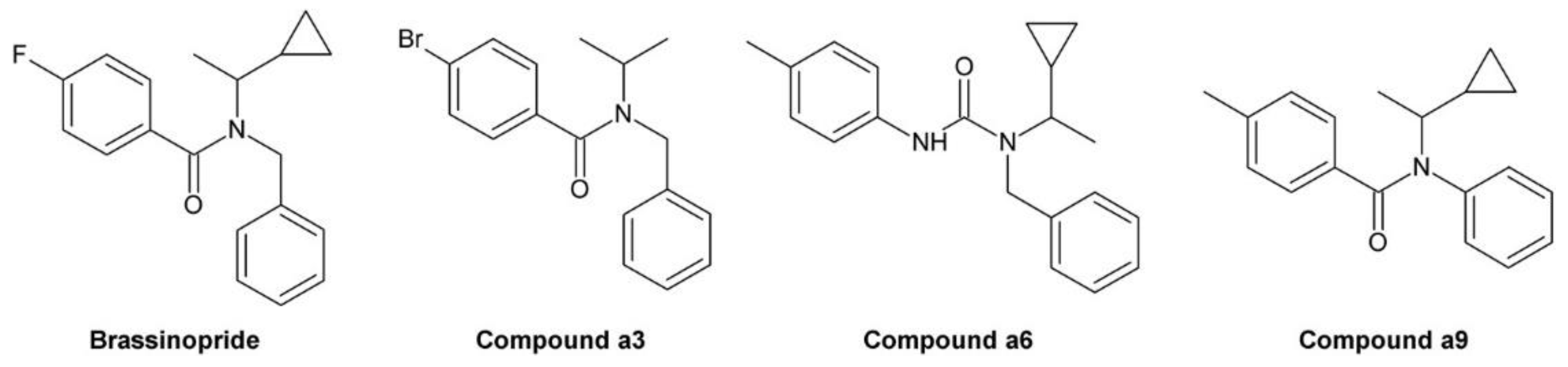
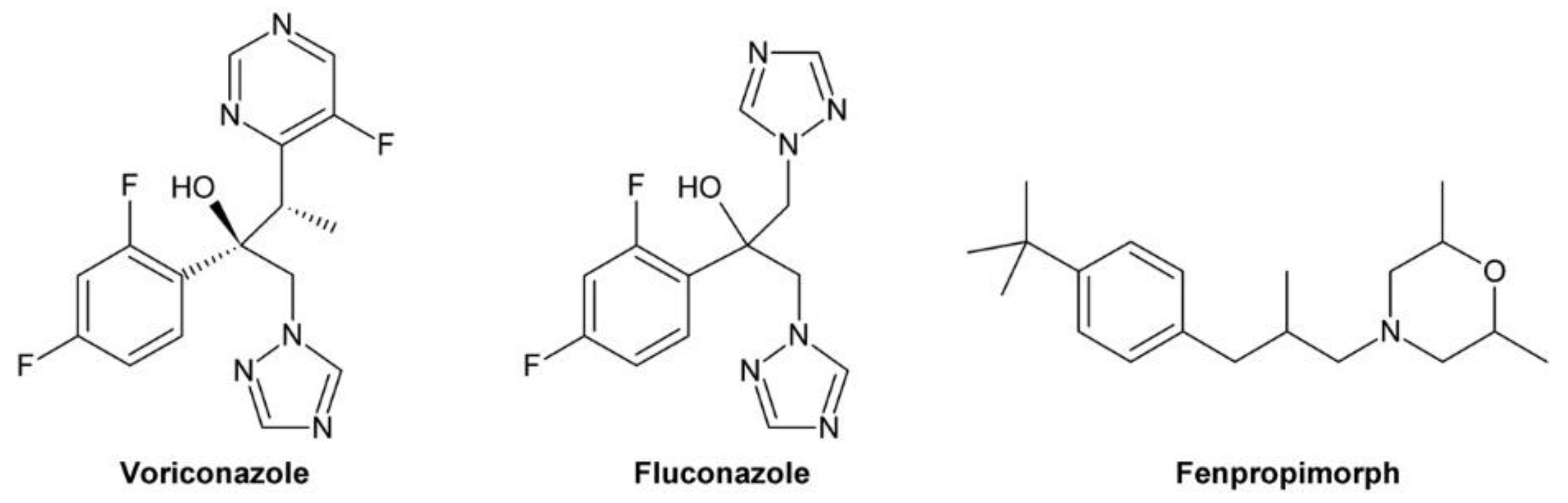
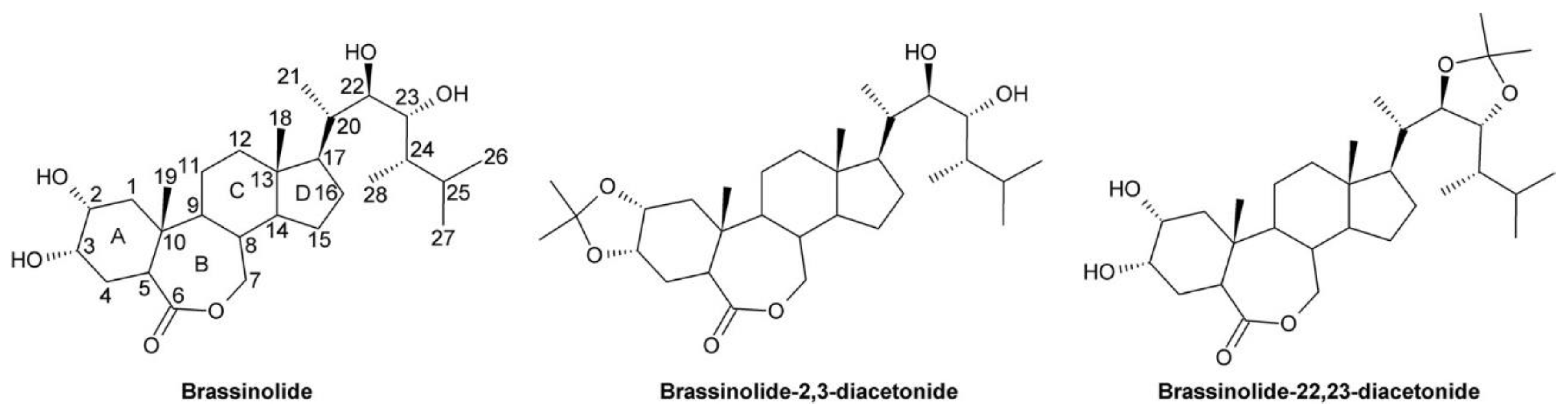
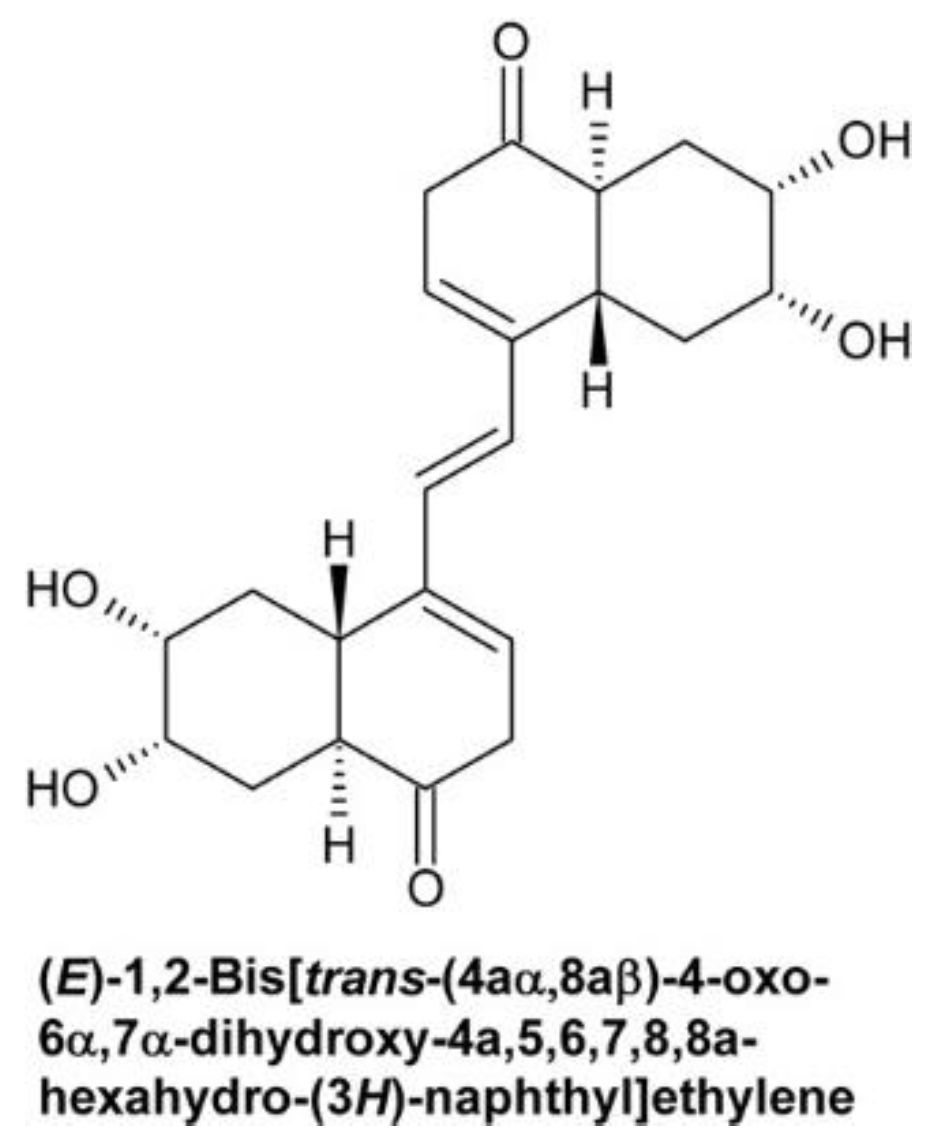
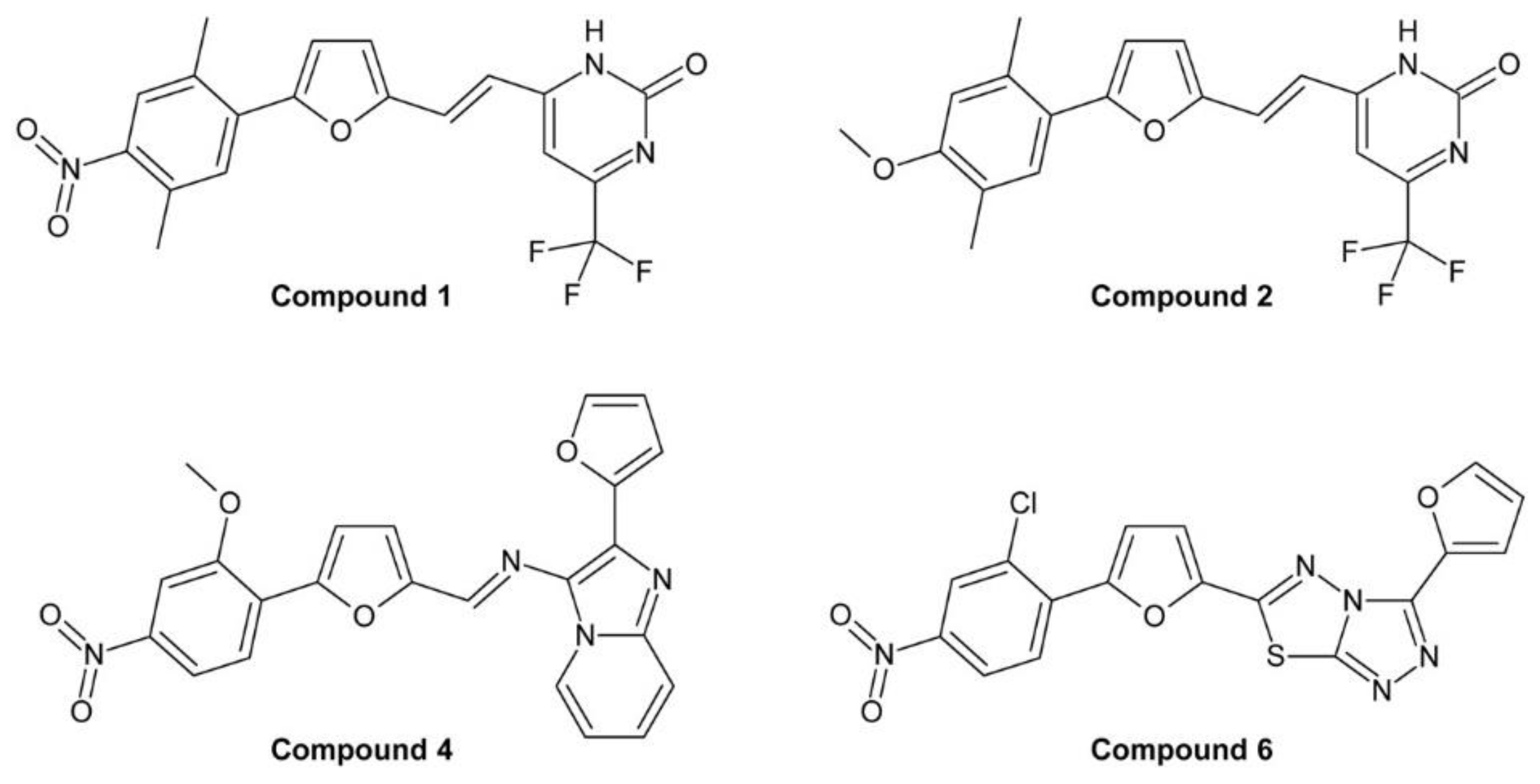
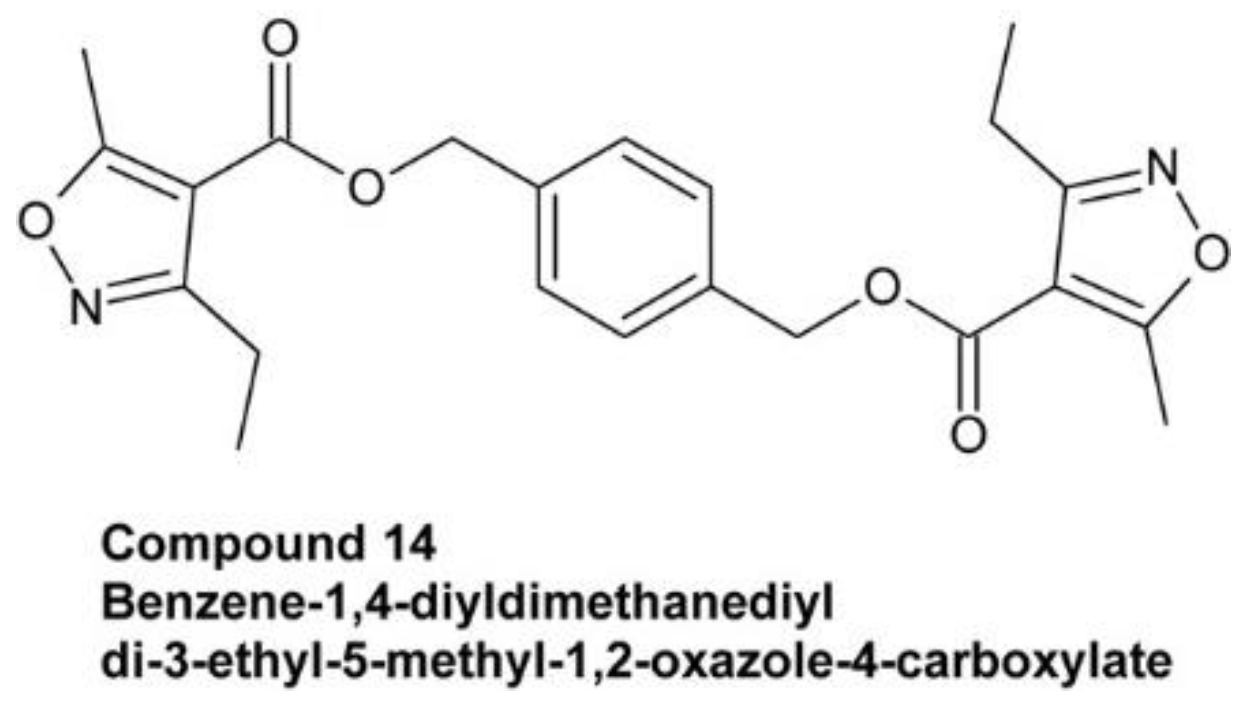

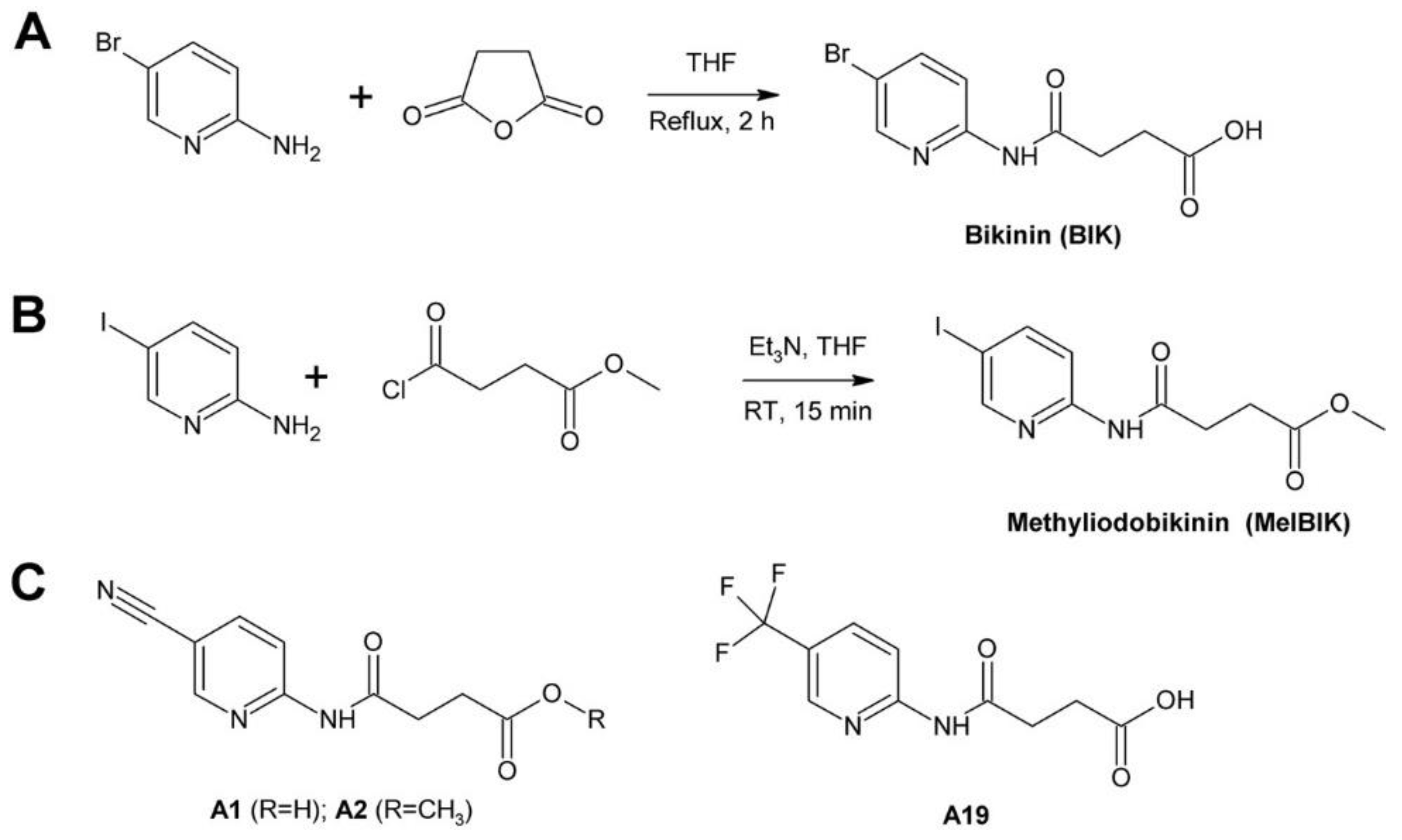
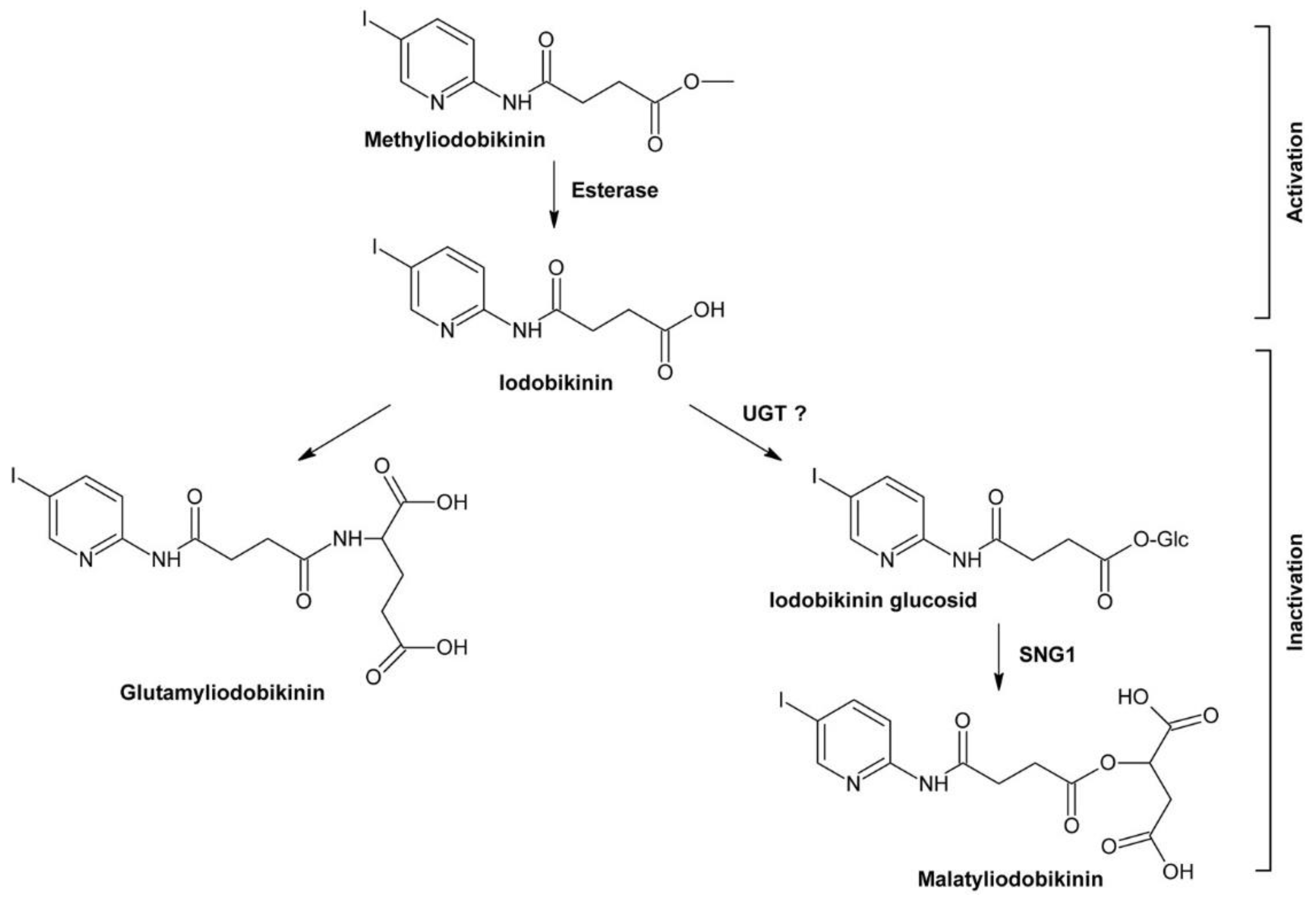
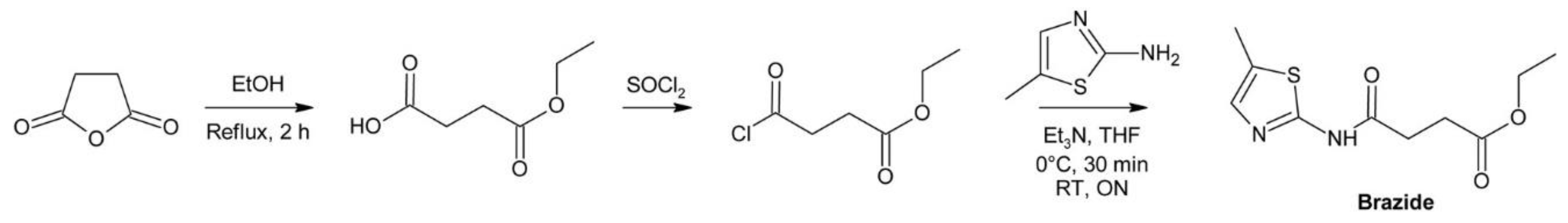
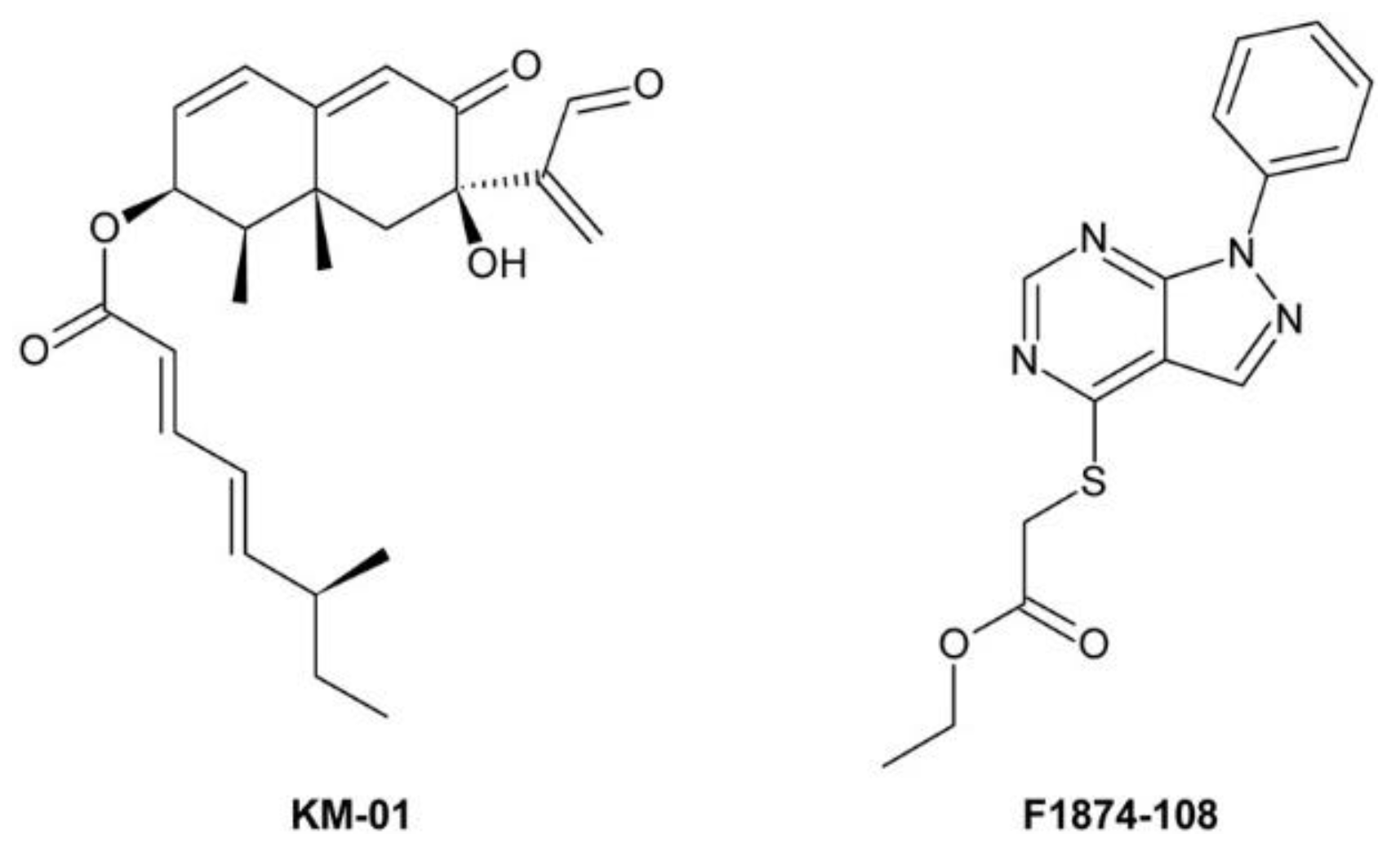
| Abbreviation; Alternative Name | Name | A. thaliana Locus | References |
|---|---|---|---|
| BR6OX1; CYP85A1 | BRASSINOSTEROID-6-OXIDASE 1 | AT5G38970 | [30] |
| BR6OX2; CYP85A2 | BRASSINOSTEROID-6-OXIDASE 2 | AT3G30180 | [30,46] |
| CPD; CYP90A1 | CONSTITUTIVE PHOTOMORPHOGENIC DWARF | AT5G05690 | [15] |
| CAS1 | CYCLOARTENOL SYNTHASE 1 | AT2G07050 | [47] |
| CPI1 | CYCLOPROPYL ISOMERASE | AT5G50375 | [48] |
| CYP51A2; CYP51G1 | CYTOCHROME P450 51A2 | AT1G11680 | [42] |
| CYP90D1 | CYTOCHROME P450 90D1 | AT3G13730 | [49] |
| CYP710A1 | CYTOCHROME P450 710A1 | AT2G34500 | [50] |
| CYP710A2 | CYTOCHROME P450 710A2 | AT2G34490 | [50] |
| DET2; DWF6 | DEETIOLATED 2 | AT2G38050 | [18,51] |
| DWF1; DIM; CBB1 | DWARF 1 | AT3G19820 | [35,36] |
| DWF4; CYP90B1 | DWARF 4 | AT3G50660 | [52] |
| DWF5 | DWARF 5 | AT1G50430 | [37] |
| DWF7; BUL1; STE1 | DWARF 7 | AT3G02580 | [38] |
| FK; HYD2 | FACKEL | AT3G52940 | [43,44] |
| HYD1 | HYDRA 1 | AT1G20050 | [45] |
| ROT3; CYP90C1 | ROTUNDIFOLIA 3 | AT4G36380 | [49,53] |
| SMT1 | STEROL METHYLTRANSFERASE 1 | AT5G13710 | [41] |
| SMT2 | STEROL METHYLTRANSFERASE 2 | AT1G20330 | [39] |
| SQE1 | SQUALENE EPOXIDASE1 | AT1G58440 | [54] |
| SQE2 | SQUALENE EPOXIDASE2 | AT2G22830 | [54] |
| SQE3 | SQUALENE EPOXIDASE3 | AT4G37760 | [54] |
| Compound | Type | Commercially Available | IC50 in µM 1 | Target; Kd in µM 2 |
|---|---|---|---|---|
| BRZ | Triazole | Yes | 0.58 ± 0.38 At [170]; 0.73 ± 0.13 At L 5d [235]; 0.77 At D 6 d [217] | DWF4; 1.05 [174] |
| BRZ2001 | Triazole | No | 0.55 At [219]; 0.33 At D 6 d [217] | DWF4 [221] |
| BRZ220 | Triazole | No | 0.61 ± 0.14 At [169] | DWF4 [168] |
| (2S,4R)-BRZ220 | Triazole | No | 0.01 C L 8 d [224]; 1.21 At D 5d [168] | DWF4; 0.22 [168] |
| YCZ-18 | Triazole | No | 0.10 ± 0.03 At [170]; 0.13 ± 0.03 At [169] | CYP90D1; 0.79 [73] |
| YCZ-14 | Triazole | No | 0.12 ± 0.04 At L 5 d [235] | CYP90D1 (?) |
| (2S,4R)-YCZ-2013 | Triazole | No | 0.024 ± 0.003 3 At [236] | CYP90D1 (?) |
| YCZ-FL | Triazole | No | 1.56 At D 5 d [234] | CYP90D1 (?) |
| Yucaizol | Triazole | No | 0.8 R L 10 d [163]; 0.045 ± 0.003 At D 5 d [169] | CYP90D1 (?) |
| Propiconazole | Triazole | Yes | - | CYP90D1; 0.76 [75] |
| Triadimefon | Triazole | Yes | Approximately 3 4 [175] | DWF4; 2.5 [175] |
| Imazalil | Imidazole | Yes | 3.44 At D 6 d [217] | - |
| Fenarimol | Pyrimidine | Yes | 1.8 ± 02 At D 5 d [74] | CYP90D1; 0.79 [74] |
| DPP4 | Pyrimidine | No | - | CYP90D1 (?) |
| 4-MA | Steroid | No | - | DET2; Ki 0.3 [76,77] |
| Finasteride | Steroid | Yes | - | DET2 [76] |
| AFA76 | Benzo[c]quinolizin-3-one | No | - | DET2 [76] |
| Spironolactone | Steroid | Yes | Approximately 30 5 [285] | - |
| Brassinopride | Amide | No | 17 At D [298] | - |
| Voriconazole | Triazole | Yes | 0.083 ± 0.019 C L 7 d 6 | CYP51 [78] |
| Lithium chloride | Salt | Yes | - | BIN2 [132,138] |
| Brassinolide-2,3-acetonide | Steroid | No | - | BRI1 [310] |
| Compounds 1, 2, 4 and 6 | Phenylfuran | No | Comp. 1, ID50: 5 nmol/plant RLI; Comp. 2, ID50: 3.2 nmol/plant RLI [313] | BRI1 [313,314] |
| Compound 14 7 | - | No | ID50: 0.63 nmol/plant RLI [313] | BRI1 [313] |
| NSBR1 | - | No | ED50: 0.8 nmol/plant RLI [312] | BRI1 [312] |
| Bikinin | Pyridine | Yes | EC50: 23.3 At L 6 d [177] | BIN2; 241 [70] |
| Methyliodobikinin | Pyridine | No | EC50: 6.8 At L 6 d [177] | BIN2 [177] |
| Brazide | Thiazole | No | - | BIN2 (?) |
| KM-01 | Natural compound | No | - | - |
| F1874-108 | - | No | - | BIN2 (?) [341] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozhon, W.; Akter, S.; Fernandez, A.; Poppenberger, B. Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction. Molecules 2019, 24, 4372. https://doi.org/10.3390/molecules24234372
Rozhon W, Akter S, Fernandez A, Poppenberger B. Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction. Molecules. 2019; 24(23):4372. https://doi.org/10.3390/molecules24234372
Chicago/Turabian StyleRozhon, Wilfried, Sonia Akter, Atiara Fernandez, and Brigitte Poppenberger. 2019. "Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction" Molecules 24, no. 23: 4372. https://doi.org/10.3390/molecules24234372
APA StyleRozhon, W., Akter, S., Fernandez, A., & Poppenberger, B. (2019). Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction. Molecules, 24(23), 4372. https://doi.org/10.3390/molecules24234372








