Triflumizole as a Novel Lead Compound for Strigolactone Biosynthesis Inhibitor
Abstract
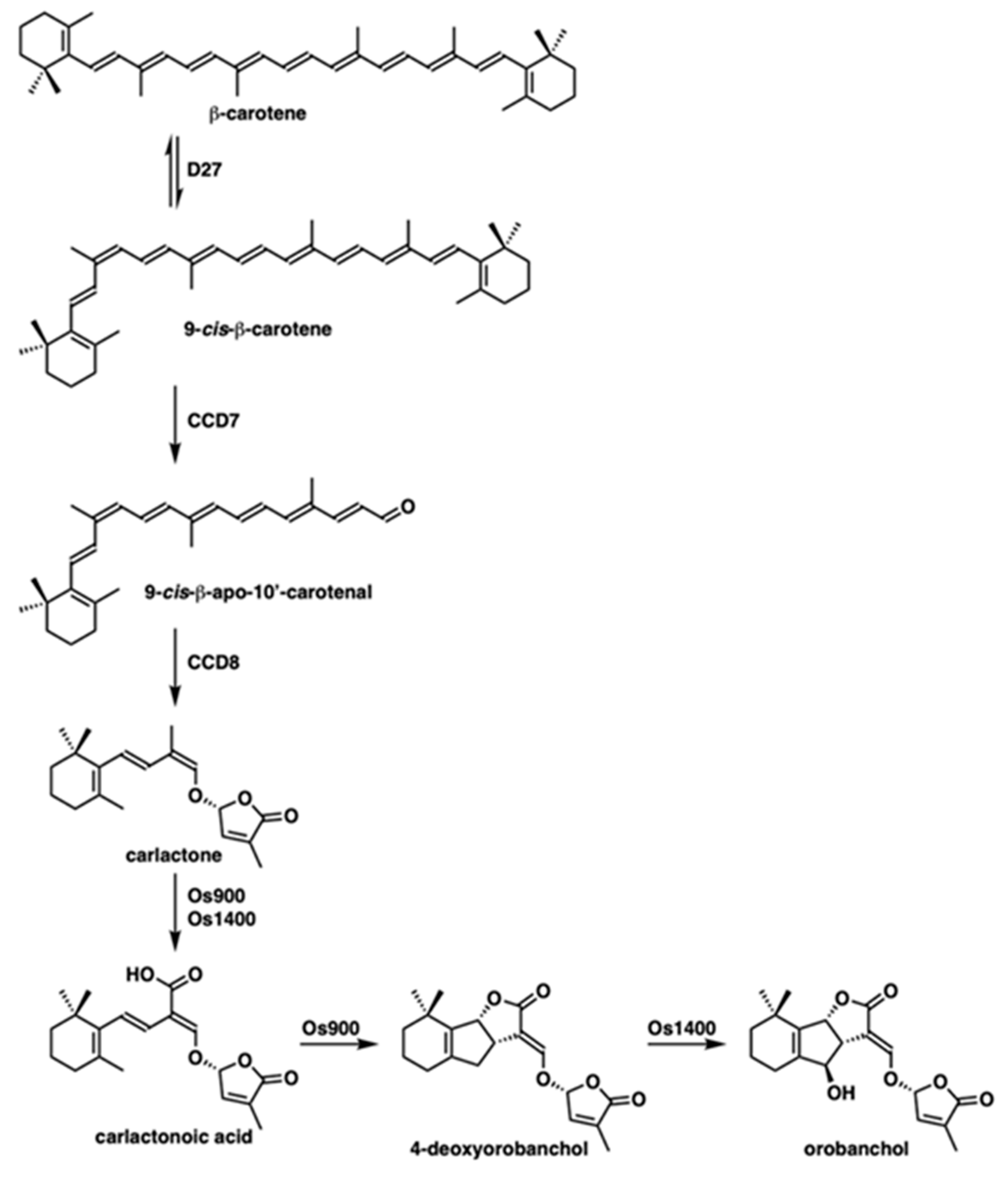
1. Introduction
2. Results
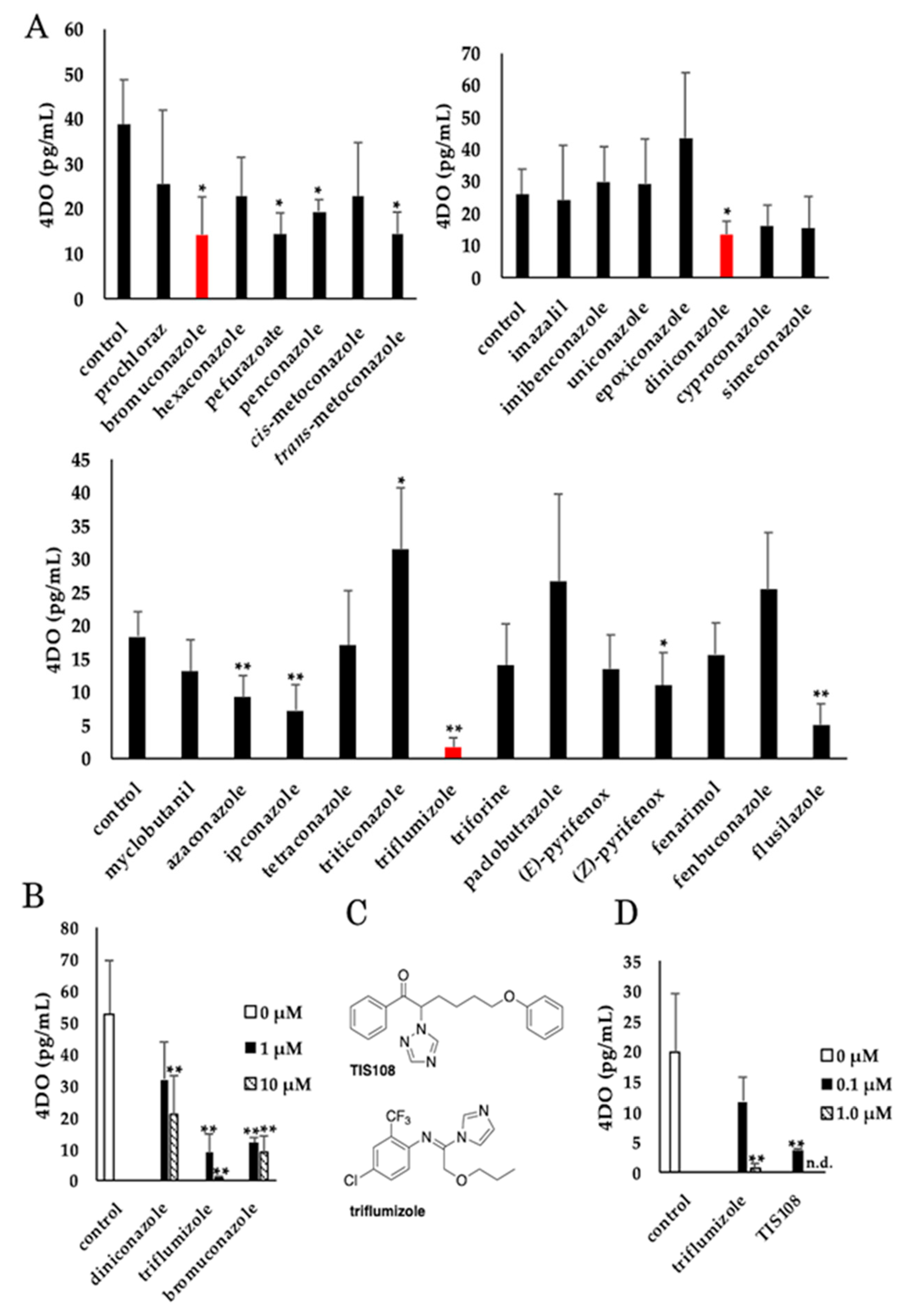
2.1. Screening for Compounds that Decrease 4DO Levels in Rice Root Exudates
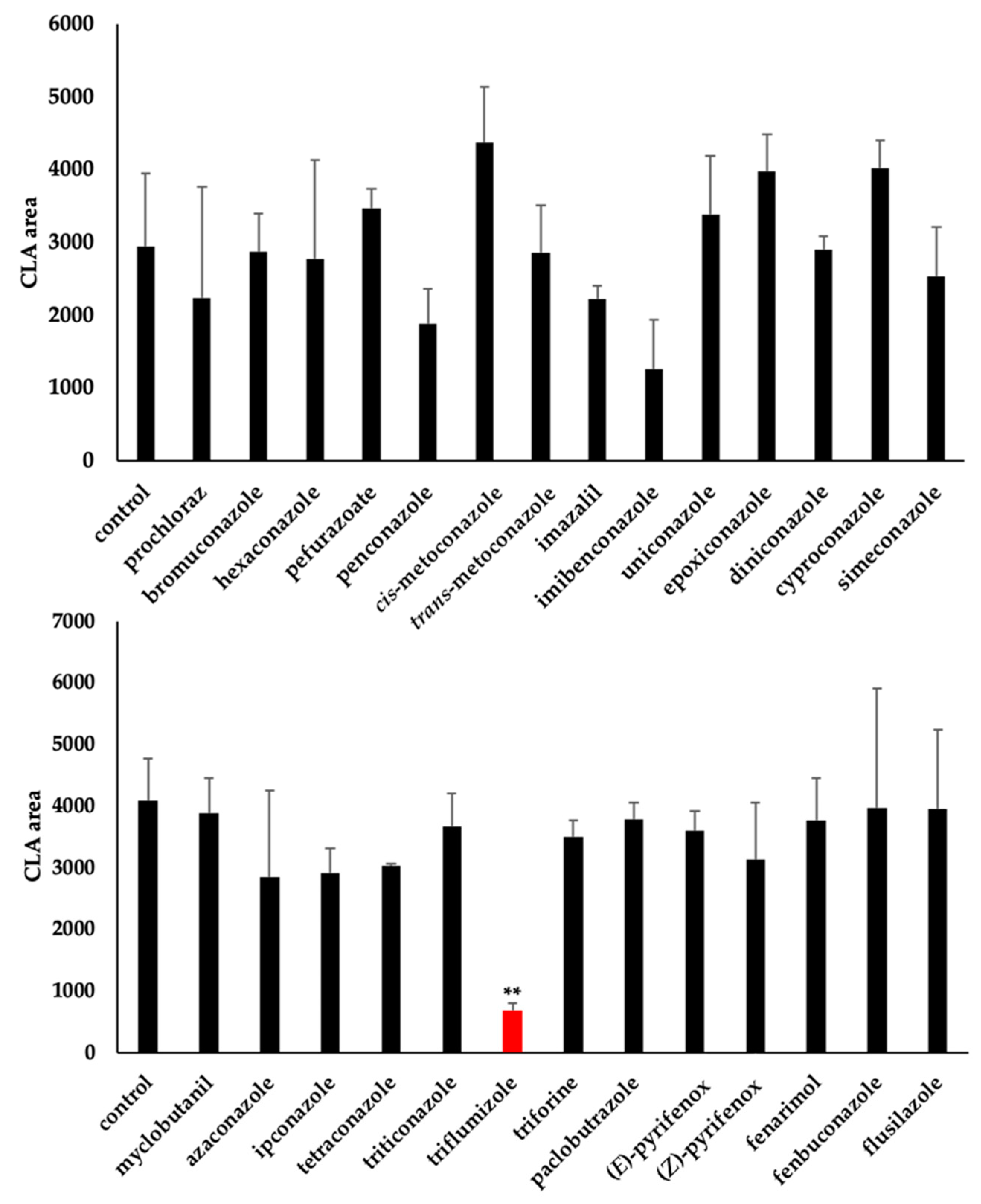
2.2. Os900 Inhibition Assay of Tested Compounds
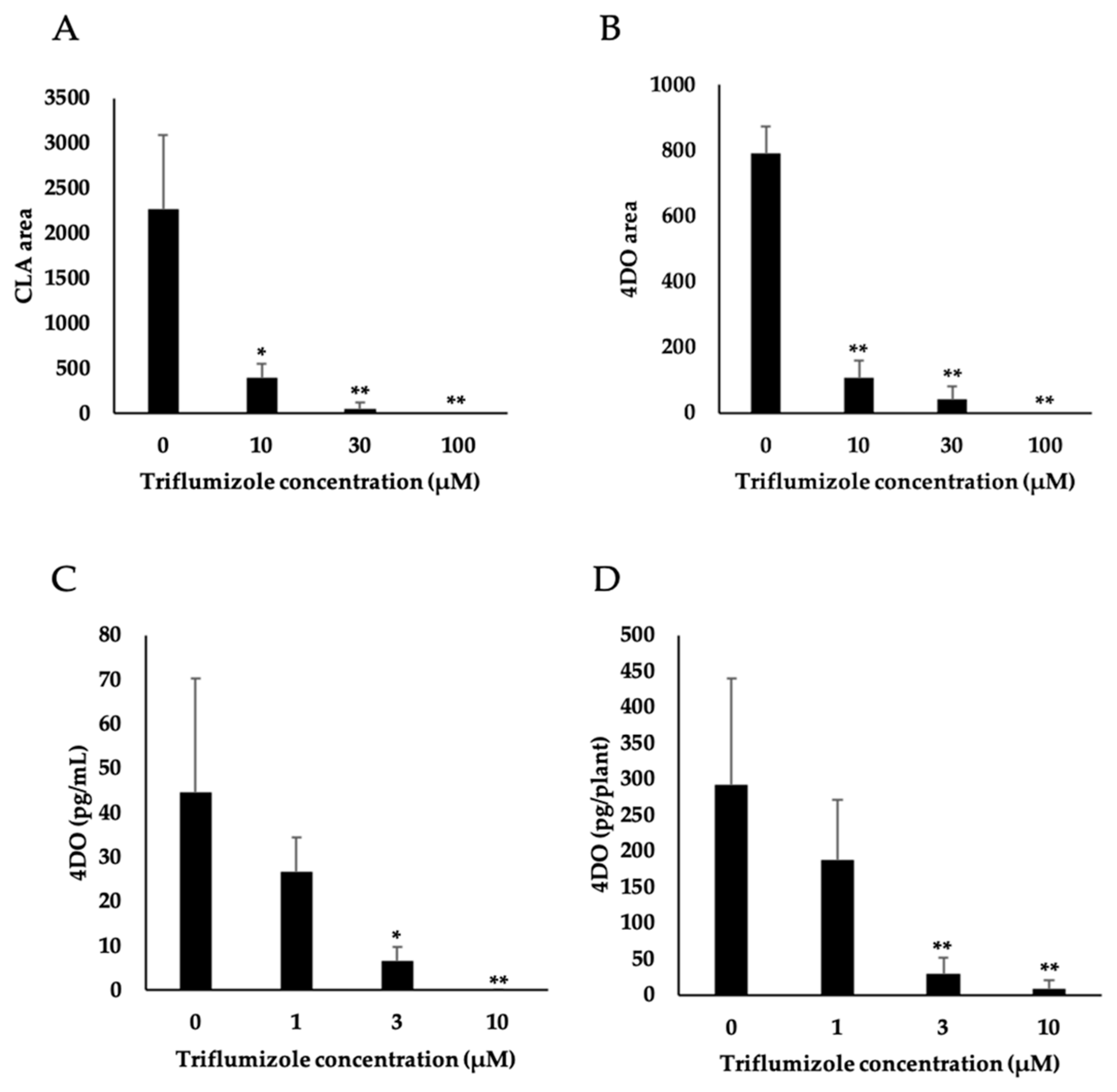
2.3. Effect of Triflumizole on SL Biosynthesis
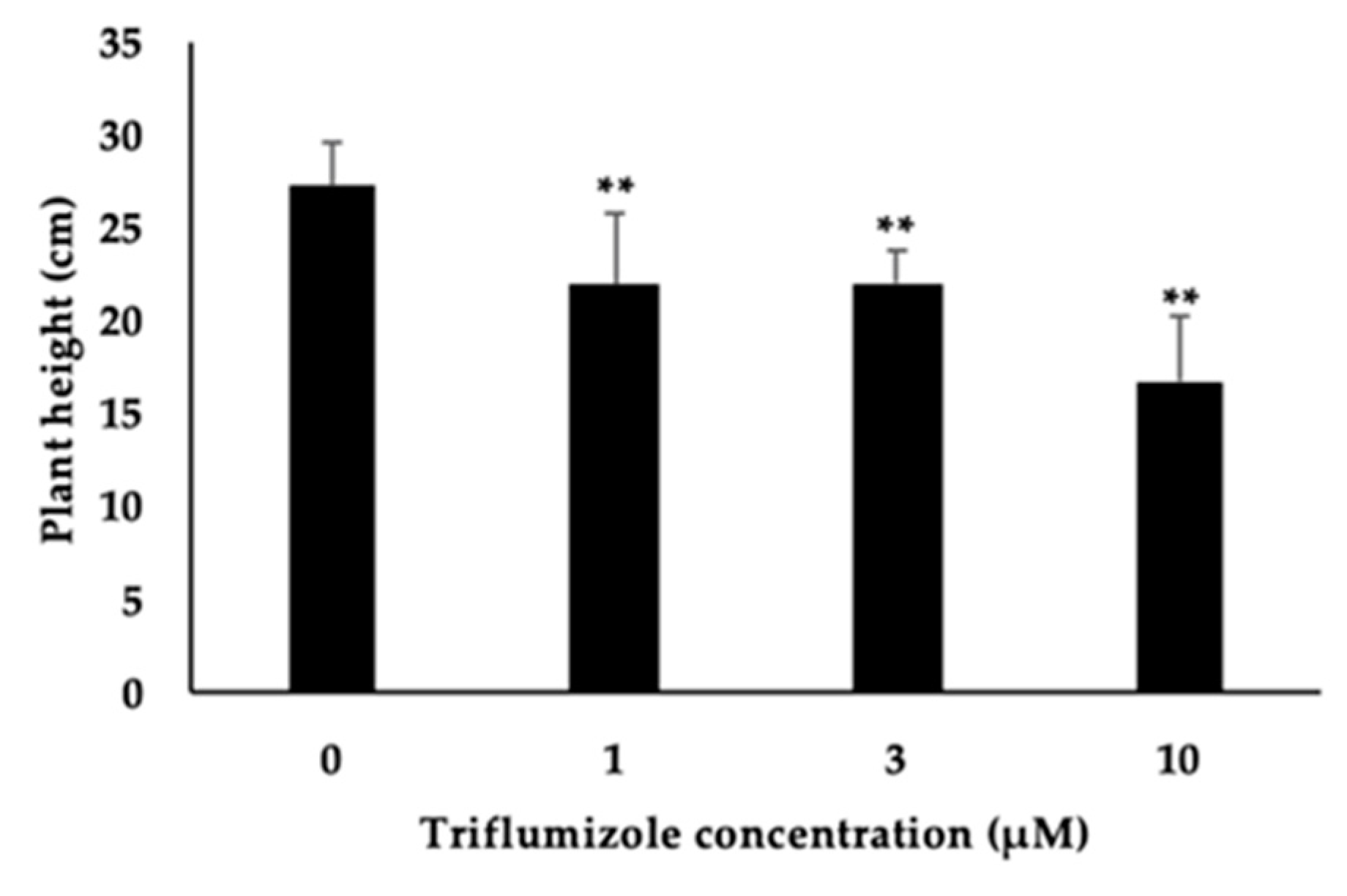
2.4. Side Effect of Triflumizole
2.5. Striga Germination Assay
3. Discussion
4. Materials and Methods
4.1. Rice Growth Condition
4.2. Quantification of the Endogenous 4DO Level
4.3. Examination of Inhibition of Os900
4.3.1. Conditions of the Enzyme Assay
4.3.2. LC-MS/MS Analysis of CLA Level
4.4. Striga Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Omoarelojie, L.O.; Kulkarni, M.G.; Finnie, J.F.; Van Staden, J. Strigolactones and their crosstalk with other phytohormones. Ann. Bot. 2019, 124, 749–767. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Lopez-Obando, M.; Ligerot, Y.; Bonhomme, S.; Boyer, F.D.; Rameau, C. Strigolactone biosynthesis and signaling in plant development. Development 2015, 142, 3615–3619. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of Witchweed (Striga lutea Lour.): Isolation and Properties of a Potent Stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef]
- Spallek, T.; Mutuku, M.; Shirasu, K. The genus Striga: A witch profile. Mol. Plant. Pathol. 2013, 14, 861–869. [Google Scholar] [CrossRef]
- Ejeta, D. Breeding for Resistance in Sorghum: Exploitation of an Intricate Host-Parasite Biology. Crop. Sci. 2007, 47, S216–S227. [Google Scholar] [CrossRef]
- Rademacher, W. Plant Growth Regulators: Backgrounds and Uses in Plant Production. J. Plant Growth Regul. 2015, 34, 845–872. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H.I.; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 18084–18089. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Mori, N.; Sato, T.; Yoda, A.; Xie, X.; Okamoto, M.; Iwanaga, M.; Ohnishi, T.; Nishiwaki, H.; Asami, T.; et al. Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol. 2018, 218, 1522–1533. [Google Scholar] [CrossRef] [PubMed]
- Neunzig, I.; Drăgan, C.A.; Widjaja, M.; Schwaninger, A.E.; Peters, F.T.; Maurer, H.H.; Bureik, M. Whole-cell biotransformation assay for investigation of the human drug metabolizing enzyme CYP3A7. Biochim. Biophys. Acta 2011, 1814, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gao, B.; He, Z.; Li, L.; Zhang, Q.; Kaziem, A.E.; Wang, M. Stereoselective bioactivity of the chiral triazole fungicide prothioconazole and its metabolite. Pestic. Biochem. Physiol. 2019, 160, 112–118. [Google Scholar] [CrossRef]
- Min, Y.K.; Asami, T.; Fujioka, S.; Murofushi, N.; Yamaguchi, I.; Yoshida, S. New lead compounds for brassinosteroid biosynthesis inhibitors. Bioorg. Med. Chem. Lett. 1999, 9, 425–430. [Google Scholar] [CrossRef]
- Sasaki, E.; Ogura, T.; Takei, K.; Kojima, M.; Kitahata, N.; Sakakibara, H.; Asami, T.; Shimada, Y. Uniconazole, a cytochrome P450 inhibitor, inhibits trans-zeatin biosynthesis in Arabidopsis. Phytochemistry 2013, 87, 30–38. [Google Scholar] [CrossRef]
- Takeuchi, J.; Okamoto, M.; Mega, R.; Kanno, Y.; Ohnishi, T.; Seo, M.; Todoroki, Y. Abscinazole-E3M, a practical inhibitor of abscisic acid 8’-hydroxylase for improving drought tolerance. Sci. Rep. 2016, 6, 37060. [Google Scholar] [CrossRef]
- Ito, S.; Kitahata, N.; Umehara, M.; Hanada, A.; Kato, A.; Ueno, K.; Mashiguchi, K.; Kyozuka, J.; Yoneyama, K.; Yamaguchi, S.; et al. A new lead chemical for strigolactone biosynthesis inhibitors. Plant. Cell. Physiol. 2010, 51, 1143–1150. [Google Scholar] [CrossRef]
- Ito, S.; Umehara, M.; Hanada, A.; Kitahata, N.; Hayase, H.; Yamaguchi, S.; Asami, T. Effects of triazole derivatives on strigolactone levels and growth retardation in rice. PLoS ONE 2011, 6, e21723. [Google Scholar] [CrossRef]
- Kawada, K.; Takahashi, I.; Arai, M.; Sasaki, Y.; Asami, T.; Yajima, S.; Ito, S. Synthesis and Biological Evaluation of Novel Triazole Derivatives as Strigolactone Biosynthesis Inhibitors. J. Agric. Food Chem. 2019, 67, 6143–6149. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Umehara, M.; Hanada, A.; Yamaguchi, S.; Asami, T. Tebuconazole derivatives are potent inhibitors of strigolactone biosynthesis. J. Pestic. Sci. 2013, 38, 147–151. [Google Scholar] [CrossRef]
- Harrison, P.J.; Newgas, S.A.; Descombes, F.; Shepherd, S.A.; Thompson, A.J.; Bugg, T.D. Biochemical characterization and selective inhibition of β-carotene cis-trans isomerase D27 and carotenoid cleavage dioxygenase CCD8 on the strigolactone biosynthetic pathway. FEBS J. 2015, 282, 3986–4000. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Xie, X.; Kusumoto, D.; Sekimoto, H.; Sugimoto, Y.; Takeuchi, Y. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 2007, 227, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, A.; Ikura, K.; Katsuura, K.; Hashimoto, S.; Nakata, A. Quantitative Structure-Activity Relationships, Conformational Analyses and Computer Graphics Study of Triflumizole Analogs, Fungicidal N-(1-Imidazol-1-ylalkylidene)anilines. J. Pestic. Sci. 1989, 14, 23–37. [Google Scholar] [CrossRef]
- Asami, T.; Mizutani, M.; Shimada, Y.; Goda, H.; Kitahata, N.; Sekimata, K.; Han, S.Y.; Fujioka, S.; Takatsuto, S.; Sakata, K.; et al. Triadimefon, a fungicidal triazole-type P450 inhibitor, induces brassinosteroid deficiency-like phenotypes in plants and binds to DWF4 protein in the brassinosteroid biosynthesis pathway. Biochem. J. 2003, 369, 71–76. [Google Scholar] [CrossRef]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef]
- Kloer, D.P.; Ruch, S.; Al-Babili, S.; Beyer, P.; Schulz, G.E. The structure of a retinal-forming carotenoid oxygenase. Science 2005, 308, 267–269. [Google Scholar] [CrossRef]
- Kloer, D.P.; Schulz, G.E. Structural and biological aspects of carotenoid cleavage. Cell. Mol. Life Sci. 2006, 63, 2291–2303. [Google Scholar] [CrossRef]
- Cui, H.; Wang, Y.; Qin, S. Genomewide analysis of carotenoid cleavage dioxygenases in unicellular and filamentous cyanobacteria. Comp. Funct. Genom. 2012, 2012, 164690. [Google Scholar] [CrossRef]
- Harrison, P.J.; Bugg, T.D. Enzymology of the carotenoid cleavage dioxygenases: Reaction mechanisms, inhibition and biochemical roles. Arch. Biochem. Biophys. 2014, 544, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Sheldon, C.C.; Olive, M.R.; Walker, A.R.; Zeevaart, J.A.; Peacock, W.J.; Dennis, E.S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 1998, 95, 9019–9024. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Chandler, P.M.; Poole, A.; Dennis, E.S.; Peacock, W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 2065–2070. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, A.; Fujioka, S. Studies on biosynthesis of brassinosteroids. Biosci. Biotechnol. Biochem. 1997, 61, 757–762. [Google Scholar]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef]
- Zhang, A.; Xie, X.; Liu, W. Enantioselective separation and phytotoxicity on rice seedlings of paclobutrazol. J. Agric. Food Chem. 2011, 59, 4300–4305. [Google Scholar] [CrossRef]
- Li, W.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Tian, C.; Yin, X.; Li, K.; Yang, Y.; Guo, J.; Miao, Y.; et al. Negative Roles of Strigolactone-Related SMXL6, 7 and 8 Proteins in Drought Resistance in Arabidopsis. Biomolecules 2020, 10, 607. [Google Scholar] [CrossRef]
- Ito, S.; Yamagami, D.; Asami, T. Effects of gibberellin and strigolactone on rice tiller bud growth. J. Pestic. Sci. 2018, 43, 220–223. [Google Scholar] [CrossRef]
- Ueno, K.; Hanada, A.; Yamaguchi, S.; Asami, T. Preparation of multideuterated 5-deoxystrigol for use as an internal standard for quantitative LC/MS. J. Label. Compd. Radiopharm. 2010, 53, 763–766. [Google Scholar] [CrossRef]
- Ito, S.; Yamagami, D.; Umehara, M.; Hanada, A.; Yoshida, S.; Sasaki, Y.; Yajima, S.; Kyozuka, J.; Ueguchi-Tanaka, M.; Matsuoka, M.; et al. Regulation of Strigolactone Biosynthesis by Gibberellin Signaling. Plant Physiol. 2017, 174, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Seto, Y.; Sado, A.; Asami, K.; Hanada, A.; Umehara, M.; Akiyama, K.; Yamaguchi, S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 2014, 111, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Ueyama, T. Production of (+)-5-deoxystrigol by Lotus japonicus root culture. Phytochemistry 2008, 69, 212–217. [Google Scholar] [CrossRef]

Sample Availability: All tested compounds were purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawada, K.; Uchida, Y.; Takahashi, I.; Nomura, T.; Sasaki, Y.; Asami, T.; Yajima, S.; Ito, S. Triflumizole as a Novel Lead Compound for Strigolactone Biosynthesis Inhibitor. Molecules 2020, 25, 5525. https://doi.org/10.3390/molecules25235525
Kawada K, Uchida Y, Takahashi I, Nomura T, Sasaki Y, Asami T, Yajima S, Ito S. Triflumizole as a Novel Lead Compound for Strigolactone Biosynthesis Inhibitor. Molecules. 2020; 25(23):5525. https://doi.org/10.3390/molecules25235525
Chicago/Turabian StyleKawada, Kojiro, Yuya Uchida, Ikuo Takahashi, Takahito Nomura, Yasuyuki Sasaki, Tadao Asami, Shunsuke Yajima, and Shinsaku Ito. 2020. "Triflumizole as a Novel Lead Compound for Strigolactone Biosynthesis Inhibitor" Molecules 25, no. 23: 5525. https://doi.org/10.3390/molecules25235525
APA StyleKawada, K., Uchida, Y., Takahashi, I., Nomura, T., Sasaki, Y., Asami, T., Yajima, S., & Ito, S. (2020). Triflumizole as a Novel Lead Compound for Strigolactone Biosynthesis Inhibitor. Molecules, 25(23), 5525. https://doi.org/10.3390/molecules25235525





