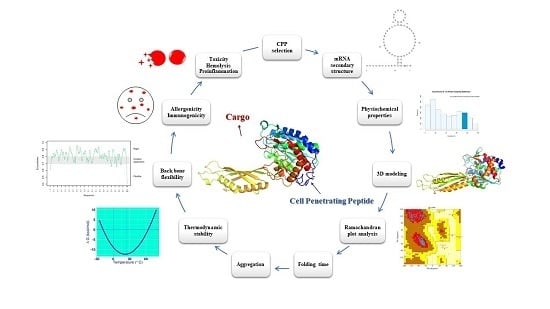
Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo Molecule
Abstract
1. Introduction
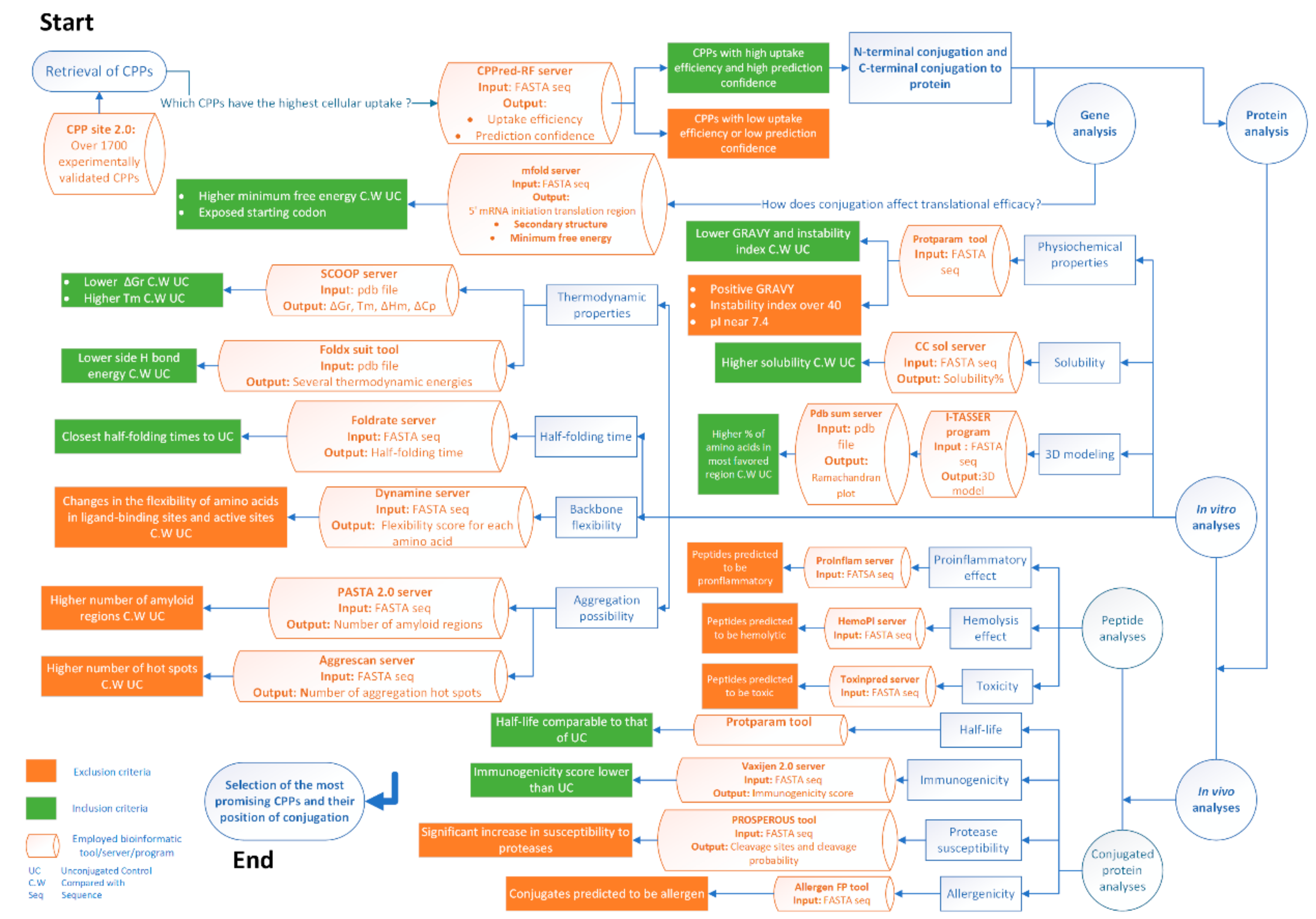
2. Results and Discussions
2.1. Primary Dataset and Penetration Prediction of CPPs
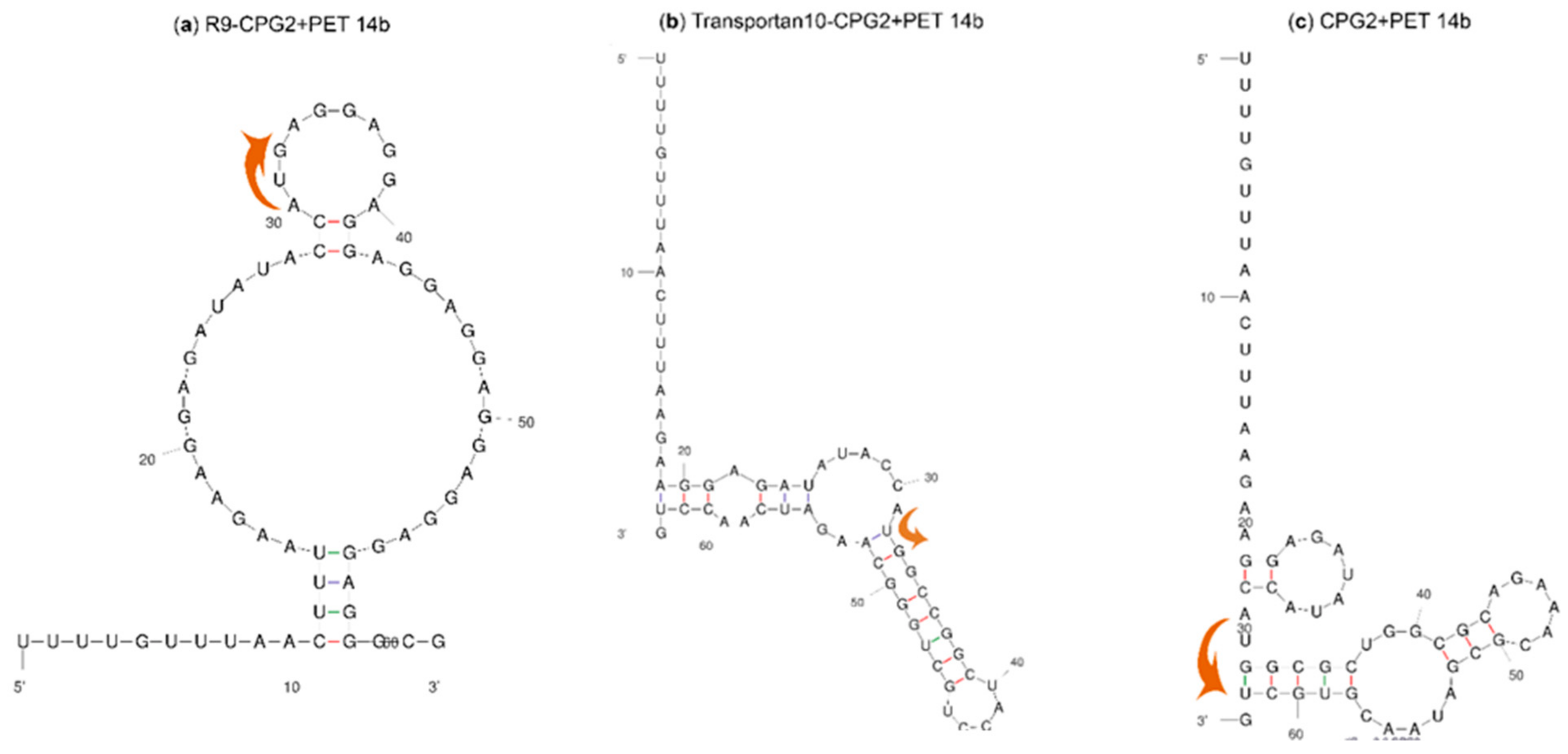
2.2. mRNA Secondary Structure Prediction of cpp-cpg2/cpg2-cpp Conjugates
2.3. Physiochemical Properties of CPPs and CPP-CPG2/CPG2-CPP Conjugates
2.4. The Solubility of CPP-CPG2 and CPG2-CPP Conjugates
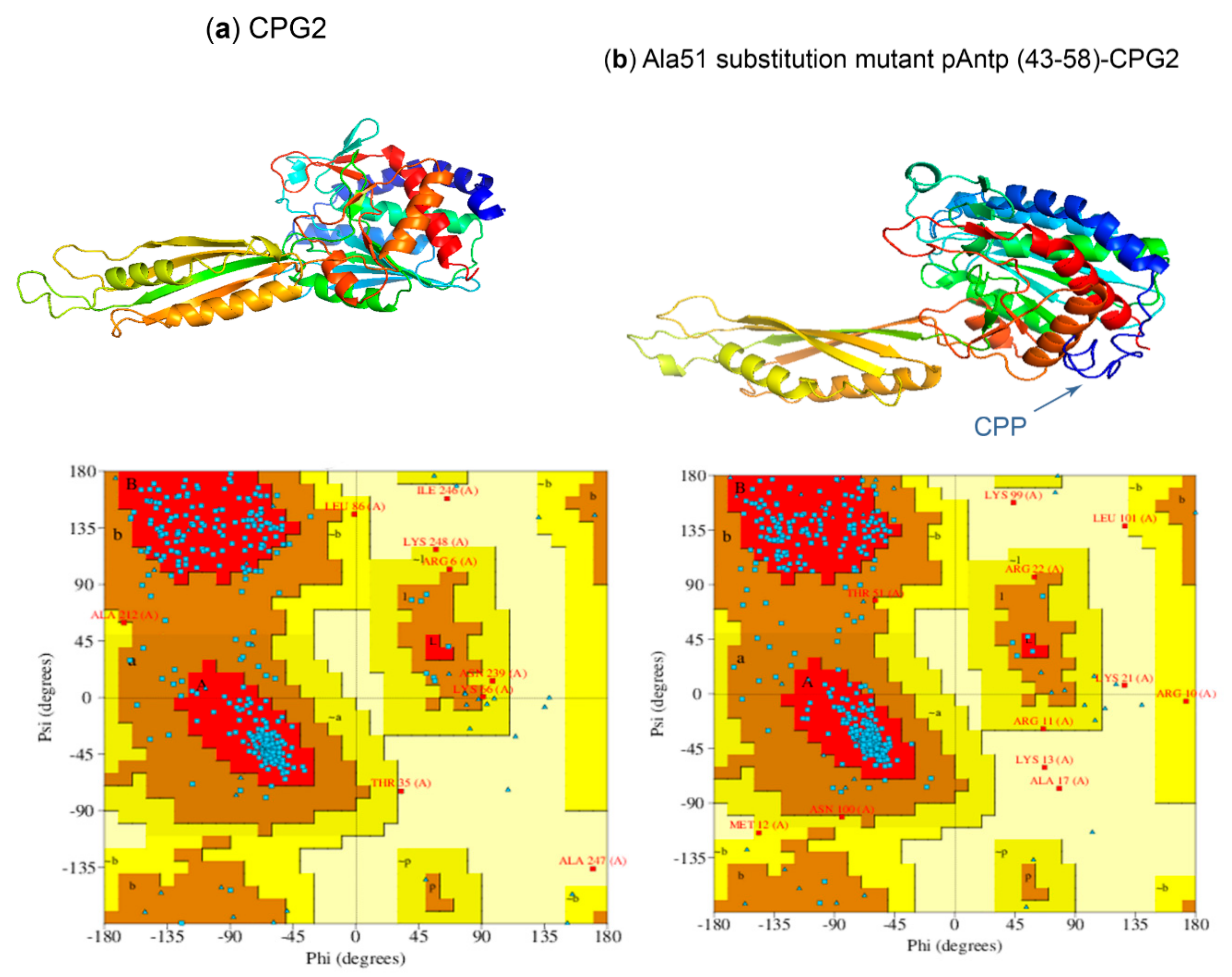
2.5. Three-Dimensional Modeling of CPP-CPG2 and CPG2-CPP Conjugates
2.6. Thermodynamic Characteristics of CPP-CPG2 and CPG2-CPP Conjugates
2.7. Prediction of the Aggregation Possibility of CPP-CPG2 and CPG2-CPP Conjugates
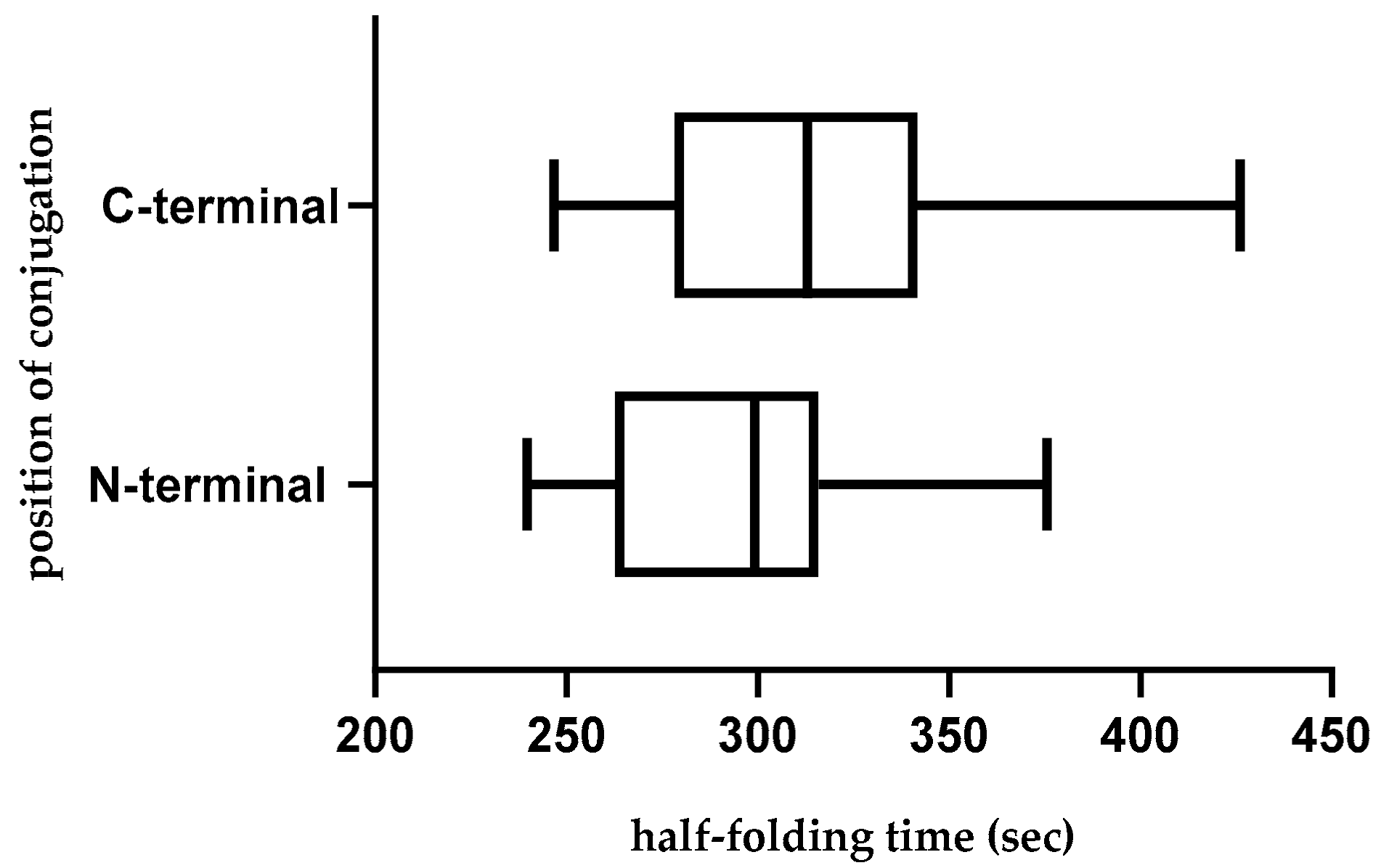
2.8. Folding Rate and Backbone Flexibility of CPP-CPG2 and CPG2-CPP Conjugates
2.9. Further Analyses for In Vivo Applications
2.9.1. Analyses of CPP-CPG2 and CPG2-CPP Conjugates
2.9.2. Analyses of top CPPs for In Vivo Application
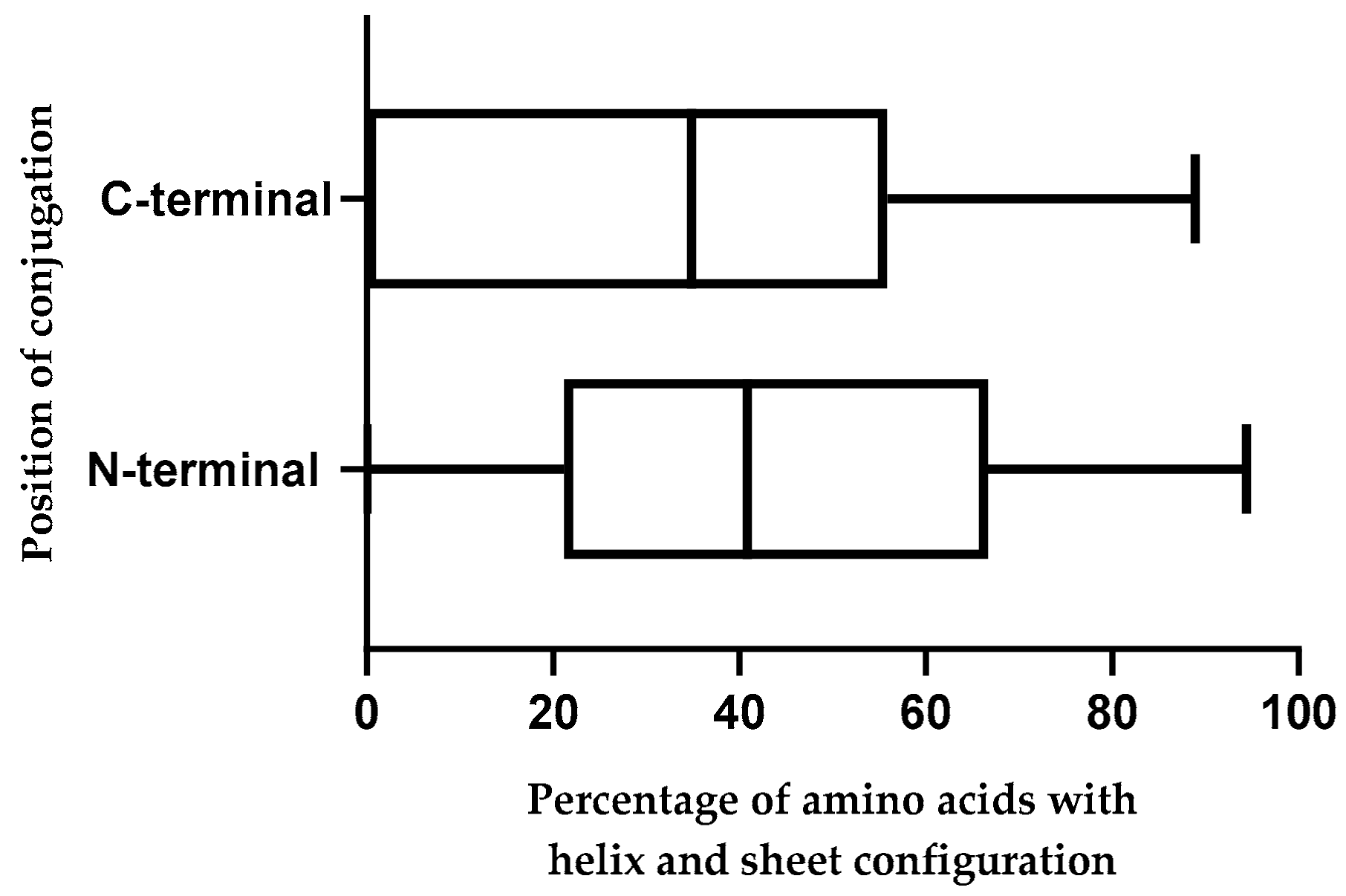
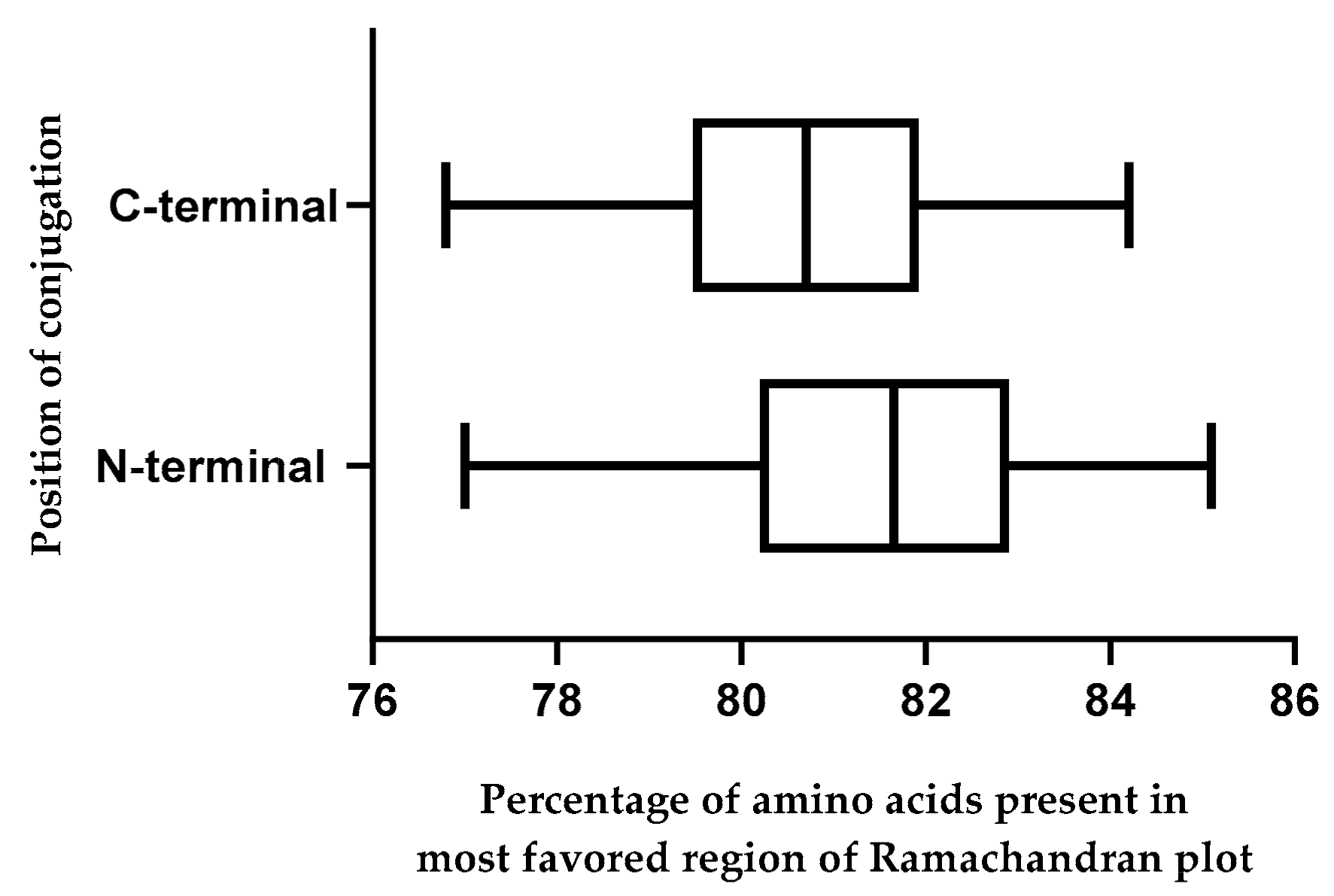
2.10. Effect of Position of Conjugations on CPP-CPG2 and CPG2-CPP Conjugates
2.11. Most Promising CPP Candidates to Design CPP-CPG2/CPG2-CPP Conjugates
2.12. Analysis of Susceptibility to Human Proteases
2.13. Limitations of the Current Study
3. Materials and Methods
3.1. Primary Dataset Collection
3.2. Penetration Prediction of CPPs
3.3. mRNA Secondary Structure Prediction of cpg2 and cpp-cpg2/cpg2-cpp Conjugates
3.4. Prediction of Physiochemical Properties of CPPs and CPP-CPG2/CPG2-CPP Conjugates
3.5. Solubility Prediction of CPG2 and CPP-CPG2/CPG2-CPP Conjugates
3.6. Modeling 3D Structures of CPG2 and CPP-CPG2/CPG2-CPP Conjugates
3.7. Thermodynamic Quantities of CPG2 and CPP-CPG2/CPG2-CPP Conjugates
3.8. Prediction of Aggregation Possibility of CPG2 and CPP-CPG2/CPG2-CPP Conjugates
3.9. Folding Rate of CPG2 and CPP-CPG2/CPG2-CPP Conjugates
3.10. Backbone Flexibility of CPG2 and CPP-CPG2/CPG2-CPP Conjugates
3.11. Further Analyses for In Vivo Application
3.11.1. Analyses of CPP-CPG2 and CPG2-CPP Conjugates
3.11.2. Analyses of CPPs
3.12. Evaluation of CPP-CPG2/CPG2-CPP Conjugates Susceptibility to Human Proteases
3.13. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sebbage, V. Cell-penetrating peptides and their therapeutic applications. Biosci. Horiz. 2009, 2, 64–72. [Google Scholar] [CrossRef]
- Ramsey, J.D.; Flynn, N.H. Cell-penetrating peptides transport therapeutics into cells. Pharmacol. Ther. 2015, 154, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef]
- Borrelli, A.; Tornesello, A.; Tornesello, M.; Buonaguro, F. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules 2018, 23, 295. [Google Scholar] [CrossRef]
- Kauffman, W.B.; Fuselier, T.; He, J.; Wimley, W.C. Mechanism Matters: A Taxonomy of Cell Penetrating Peptides. Trends Biochem. Sci. 2015, 40, 749–764. [Google Scholar] [CrossRef]
- Kristensen, M.; Birch, D.; Mørck Nielsen, H. Applications and challenges for use of cell-penetrating peptides as delivery vectors for peptide and protein cargos. Int. J. Mol. Sci. 2016, 17, 185. [Google Scholar] [CrossRef]
- Keller, A.-A.; Mussbach, F.; Breitling, R.; Hemmerich, P.; Schaefer, B.; Lorkowski, S.; Reissmann, S. Relationships between cargo, cell penetrating peptides and cell type for uptake of non-covalent complexes into live cells. Pharmaceuticals (Basel) 2013, 6, 184–203. [Google Scholar] [CrossRef]
- Herce, H.D.; Deng, W.; Helma, J.; Leonhardt, H.; Cardoso, M.C. Visualization and targeted disruption of protein interactions in living cells. Nat. Commun. 2013, 4, 2660. [Google Scholar] [CrossRef]
- Koshman, Y.E.; Waters, S.B.; Walker, L.A.; Los, T.; Tombe, P.d.; Goldspink, P.H.; Russell, B. Delivery and visualization of proteins conjugated to quantum dots in cardiac myocytes. J. Mol. Cell. Cardiol. 2008, 45, 853–856. [Google Scholar] [CrossRef]
- Liang, J.F.; Yang, V.C. Insulin-cell penetrating peptide hybrids with improved intestinal absorption efficiency. Biochem. Biophys. Res. Commun. 2005, 335, 734–738. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Fu, R.; Rao, P.; Weller, R.; Bradshaw, J.; Liu, S. Can the Cellular Internalization of Cargo Proteins Be Enhanced by Fusing a Tat Peptide in the Center of Proteins? A Fluorescence Study. J. Pharm. Sci. 2018, 107, 879–886. [Google Scholar] [CrossRef]
- Patel, S.G.; Sayers, E.J.; He, L.; Narayan, R.; Williams, T.L.; Mills, E.M.; Allemann, R.K.; Luk, L.Y.P.; Jones, A.T.; Tsai, Y.-H. Cell-penetrating peptide sequence and modification dependent uptake and subcellular distribution of green florescent protein in different cell lines. Sci. Rep. 2019, 9, 6298. [Google Scholar] [CrossRef] [PubMed]
- Courtois, F.; Schneider, C.P.; Agrawal, N.J.; Trout, B.L. Rational Design of Biobetters with Enhanced Stability. J. Pharm. Sci. 2015, 104, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Griswold, K.E.; Bailey-Kellogg, C. Design and engineering of deimmunized biotherapeutics. Curr. Opin. Struct. Biol. 2016, 39, 79–88. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.; Gautam, A. CPPsite 2.0: A repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res. 2016, 44, D1098–D1103. [Google Scholar] [CrossRef]
- Bolhassani, A.; Jafarzade, B.S.; Mardani, G. In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides 2017, 87, 50–63. [Google Scholar] [CrossRef]
- Hajiebrahimi, A.; Owji, H.; Hemmati, S. Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome 2017, 60, 797–814. [Google Scholar] [CrossRef]
- Courtois, F.; Agrawal, N.J.; Lauer, T.M.; Trout, B.L. Rational design of therapeutic mAbs against aggregation through protein engineering and incorporation of glycosylation motifs applied to bevacizumab. mAbs 2016, 8, 99–112. [Google Scholar] [CrossRef]
- Schwartz, S.; Borner, K.; Muller, K.; Martus, P.; Fischer, L.; Korfel, A.; Auton, T.; Thiel, E. Glucarpidase (carboxypeptidase g2) intervention in adult and elderly cancer patients with renal dysfunction and delayed methotrexate elimination after high-dose methotrexate therapy. Oncologist 2007, 12, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Widemann, B.C.; Sung, E.; Anderson, L.; Salzer, W.L.; Balis, F.M.; Monitjo, K.S.; McCully, C.; Hawkins, M.; Adamson, P.C. Pharmacokinetics and metabolism of the methotrexate metabolite 2, 4-diamino-N(10)-methylpteroic acid. J. Pharmacol. Expe. Ther. 2000, 294, 894–901. [Google Scholar]
- Green, J.M. Glucarpidase to combat toxic levels of methotrexate in patients. Ther. Clin. Risk Manag. 2012, 8, 403–413. [Google Scholar] [CrossRef]
- Souza, C.; Pellosi, D.S.; Tedesco, A.C. Prodrugs for targeted cancer therapy. Expert Rev. Anticancer Ther. 2019. [Google Scholar] [CrossRef]
- Hedley, D.; Ogilvie, L.; Springer, C. Carboxypeptidase G2-based gene-directed enzyme–prodrug therapy: A new weapon in the GDEPT armoury. Nat. Rev. Cancer 2007, 7, 870. [Google Scholar] [CrossRef] [PubMed]
- Jeyaharan, D.; Brackstone, C.; Schouten, J.; Davis, P.; Dixon, A.M. Characterisation of the Carboxypeptidase G2 Catalytic Site and Design of New Inhibitors for Cancer Therapy. ChemBioChem 2018, 19, 1959–1968. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Balis, F.M.; O’Brien, M.M.; Schmiegelow, K.; Pauley, J.L.; Bleyer, A.; Widemann, B.C.; Askenazi, D.; Bergeron, S.; Shirali, A.; et al. Consensus Guideline for Use of Glucarpidase in Patients with High-Dose Methotrexate Induced Acute Kidney Injury and Delayed Methotrexate Clearance. Oncologist 2018, 23, 52–61. [Google Scholar] [CrossRef]
- Sadeghian, I.; Khalvati, B.; Ghasemi, Y.; Hemmati, S. TAT-mediated intracellular delivery of carboxypeptidase G2 protects against methotrexate-induced cell death in HepG2 cells. Toxicol. Appl. Pharmacol. 2018, 346, 9–18. [Google Scholar] [CrossRef]
- Jha, D.; Mishra, R.; Gottschalk, S.; Wiesmuller, K.H.; Ugurbil, K.; Maier, M.E.; Engelmann, J. CyLoP-1: A novel cysteine-rich cell-penetrating peptide for cytosolic delivery of cargoes. Bioconjug. Chem. 2011, 22, 319–328. [Google Scholar] [CrossRef]
- Jones, S.W.; Christison, R.; Bundell, K.; Voyce, C.J.; Brockbank, S.M.; Newham, P.; Lindsay, M.A. Characterisation of cell-penetrating peptide-mediated peptide delivery. Br. J. Pharmacol. 2005, 145, 1093–1102. [Google Scholar] [CrossRef]
- Fischer, P.M.; Zhelev, N.Z.; Wang, S.; Melville, J.E.; Fahraeus, R.; Lane, D.P. Structure-activity relationship of truncated and substituted analogues of the intracellular delivery vector Penetratin. J. Pept. Res. 2000, 55, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Kilk, K.; Langel, U. Predicting cell-penetrating peptides. Adv. Drug Deliv. Rev. 2008, 60, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Calvet, S.; Trembleau, A.; Brunissen, A.; Chassaing, G.; Prochiantz, A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996, 271, 18188–18193. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Kaneto, H.; Weir, G.C.; Bonner-Weir, S. PDX-1 protein containing its own antennapedia-like protein transduction domain can transduce pancreatic duct and islet cells. Diabetes 2003, 52, 1732–1737. [Google Scholar] [CrossRef]
- Kondo, E.; Saito, K.; Tashiro, Y.; Kamide, K.; Uno, S.; Furuya, T.; Mashita, M.; Nakajima, K.; Tsumuraya, T.; Kobayashi, N.; et al. Tumour lineage-homing cell-penetrating peptides as anticancer molecular delivery systems. Nat. Commun. 2012, 3, 951. [Google Scholar] [CrossRef]
- Kamide, K.; Nakakubo, H.; Uno, S.; Fukamizu, A. Isolation of novel cell-penetrating peptides from a random peptide library using in vitro virus and their modifications. Int. J. Mol. Med. 2010, 25, 41–51. [Google Scholar] [CrossRef][Green Version]
- Gomez, J.A.; Chen, J.; Ngo, J.; Hajkova, D.; Yeh, I.J.; Gama, V.; Miyagi, M.; Matsuyama, S. Cell-Penetrating Penta-Peptides (CPP5s): Measurement of Cell Entry and Protein-Transduction Activity. Pharmaceuticals (Basel) 2010, 3, 3594–3613. [Google Scholar] [CrossRef]
- Elmquist, A.; Hansen, M.; Langel, U. Structure-activity relationship study of the cell-penetrating peptide pVEC. Biochim. Biophys. Acta 2006, 1758, 721–729. [Google Scholar] [CrossRef]
- Cohen-Avrahami, M.; Shames, A.I.; Ottaviani, M.F.; Aserin, A.; Garti, N. HIV-TAT enhances the transdermal delivery of NSAID drugs from liquid crystalline mesophases. J. Phys. Chem. B 2014, 118, 6277–6287. [Google Scholar] [CrossRef]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef]
- Nakase, I.; Hirose, H.; Tanaka, G.; Tadokoro, A.; Kobayashi, S.; Takeuchi, T.; Futaki, S. Cell-surface accumulation of flock house virus-derived peptide leads to efficient internalization via macropinocytosis. Mol. Ther. 2009, 17, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Wender, P.A.; Mitchell, D.J.; Pattabiraman, K.; Pelkey, E.T.; Steinman, L.; Rothbard, J.B. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13003–13008. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.J.; El-Andaloussi, S.; Holm, T.; Mae, M.; Janes, J.; Maimets, T.; Langel, U. Characterization of a novel cytotoxic cell-penetrating peptide derived from p14ARF protein. Mol. Ther. 2008, 16, 115–123. [Google Scholar] [CrossRef]
- El-Andaloussi, S.; Johansson, H.J.; Holm, T.; Langel, U. A novel cell-penetrating peptide, M918, for efficient delivery of proteins and peptide nucleic acids. Mol. Ther. 2007, 15, 1820–1826. [Google Scholar] [CrossRef]
- Scheller, A.; Oehlke, J.; Wiesner, B.; Dathe, M.; Krause, E.; Beyermann, M.; Melzig, M.; Bienert, M. Structural requirements for cellular uptake of alpha-helical amphipathic peptides. J. Pept. Sci. 1999, 5, 185–194. [Google Scholar] [CrossRef]
- Fernandez-Carneado, J.; Kogan, M.J.; Pujals, S.; Giralt, E. Amphipathic peptides and drug delivery. Biopolymers 2004, 76, 196–203. [Google Scholar] [CrossRef]
- Shinde, A.; Feher, K.M.; Hu, C.; Slowinska, K. Peptide internalization enabled by folding: Triple helical cell-penetrating peptides. J. Pept. Sci. 2015, 21, 77–84. [Google Scholar] [CrossRef]
- Eiriksdottir, E.; Konate, K.; Langel, U.; Divita, G.; Deshayes, S. Secondary structure of cell-penetrating peptides controls membrane interaction and insertion. Biochim. Biophys. Acta 2010, 1798, 1119–1128. [Google Scholar] [CrossRef]
- Zhu, F.; Liu, F.; Wu, B.; He, B. Efficient extracellular expression of metalloprotease for Z-aspartame synthesis. J. Agric. Food Chem. 2016, 64, 9631–9638. [Google Scholar] [CrossRef]
- Owji, H.; Hemmati, S. A comprehensive in silico characterization of bacterial signal peptides for the excretory production of Anabaena variabilis phenylalanine ammonia lyase in Escherichia coli. 3 Biotech 2018, 8, 488. [Google Scholar] [CrossRef]
- Kramer, R.M.; Shende, V.R.; Motl, N.; Pace, C.N.; Scholtz, J.M. Toward a molecular understanding of protein solubility: Increased negative surface charge correlates with increased solubility. Biophys. J. 2012, 102, 1907–1915. [Google Scholar] [CrossRef]
- Dubikovskaya, E.A.; Thorne, S.H.; Pillow, T.H.; Contag, C.H.; Wender, P.A. Overcoming multidrug resistance of small-molecule therapeutics through conjugation with releasable octaarginine transporters. Proc. Natl. Acad. Sci. USA 2008, 105, 12128–12133. [Google Scholar] [CrossRef]
- Duan, Z.; Chen, C.; Qin, J.; Liu, Q.; Wang, Q.; Xu, X.; Wang, J. Cell-penetrating peptide conjugates to enhance the antitumor effect of paclitaxel on drug-resistant lung cancer. Drug Deliv. 2017, 24, 752–764. [Google Scholar] [CrossRef]
- Kristensen, M.; de Groot, A.M.; Berthelsen, J.; Franzyk, H.; Sijts, A.; Nielsen, H.M. Conjugation of cell-penetrating peptides to parathyroid hormone affects its structure, potency, and transepithelial permeation. Bioconjug. Chem. 2015, 26, 477–488. [Google Scholar] [CrossRef]
- Rahmatabadi, S.S.; Sadeghian, I.; Ghasemi, Y.; Sakhteman, A.; Hemmati, S. Identification and characterization of a sterically robust phenylalanine ammonia-lyase among 481 natural isoforms through association of in silico and in vitro studies. Enzyme Microb. Technol. 2019, 122, 36–54. [Google Scholar] [CrossRef]
- Sadeghian, I.; Rezaie, Z.; Rahmatabadi, S.S.; Hemmati, S. Biochemical insights into a novel thermo/organo tolerant bilirubin oxidase from Thermosediminibacter oceani and its application in dye decolorization. Process Biochem. 2019. [Google Scholar] [CrossRef]
- Edgeworth, M.J.; Phillips, J.J.; Lowe, D.C.; Kippen, A.D.; Higazi, D.R.; Scrivens, J.H. Global and local conformation of human IgG antibody variants rationalizes loss of thermodynamic stability. Angew. Chem. Int. Ed. Engl. 2015, 54, 15156–15159. [Google Scholar] [CrossRef]
- Pucci, F.; Rooman, M. Stability curve prediction of homologous proteins using temperature-dependent statistical potentials. PLoS Comput. Biol. 2014, 10, e1003689. [Google Scholar] [CrossRef]
- Ahmed, A.B.; Kajava, A.V. Breaking the amyloidogenicity code: Methods to predict amyloids from amino acid sequence. FEBS Lett. 2013, 587, 1089–1095. [Google Scholar] [CrossRef]
- Wang, W. Instability, stabilization, and formulation of liquid protein pharmaceuticals. Int. J. Pharm. 1999, 185, 129–188. [Google Scholar] [CrossRef]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef]
- DuBay, K.F.; Pawar, A.P.; Chiti, F.; Zurdo, J.; Dobson, C.M.; Vendruscolo, M. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J. Mol. Biol. 2004, 341, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of” hot spots” of aggregation in polypeptides. BMC Bioinformatics 2007, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics 2009, 26, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Echols, N.; Milburn, D.; Gerstein, M. MolMovDB: Analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 2003, 31, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.J.; Benson, M.L.; Smith, R.D.; Carlson, H.A. Inherent versus induced protein flexibility: Comparisons within and between apo and holo structures. PLoS Comput. Biol. 2019, 15, e1006705. [Google Scholar] [CrossRef]
- Hemmati, S.; Schneider, B.; Schmidt, T.J.; Federolf, K.; Alfermann, A.W.; Fuss, E. Justicidin B 7-hydroxylase, a cytochrome P450 monooxygenase from cell cultures of Linum perenne Himmelszelt involved in the biosynthesis of diphyllin. Phytochemistry 2007, 68, 2736–2743. [Google Scholar] [CrossRef]
- Rizzuti, M.; Nizzardo, M.; Zanetta, C.; Ramirez, A.; Corti, S. Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov. Today 2015, 20, 76–85. [Google Scholar] [CrossRef]
- Dupont, E.; Prochiantz, A.; Joliot, A. Penetratin story: An overview. In Cell-Penetrating Peptides; Langel, Ü, Ed.; Cell-Penetrating Peptides. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; Volume 1324, pp. 29–37. [Google Scholar] [CrossRef]
- Kalafatovic, D.; Giralt, E. Cell-penetrating peptides: Design strategies beyond primary structure and amphipathicity. Molecules 2017, 22, 1929. [Google Scholar] [CrossRef]
- Gomez, J.A.; Gama, V.; Yoshida, T.; Sun, W.; Hayes, P.; Leskov, K.; Boothman, D.; Matsuyama, S. Bax-inhibiting peptides derived from Ku70 and cell-penetrating pentapeptides. Biochem. Soc. Trans. 2007, 35, 797–801. [Google Scholar] [CrossRef]
- Li, Y.; Yokota, T.; Gama, V.; Yoshida, T.; Gomez, J.A.; Ishikawa, K.; Sasaguri, H.; Cohen, H.Y.; Sinclair, D.A.; Mizusawa, H.; et al. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ. 2007, 14, 2058–2067. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; WuWong, D.J.; Wong, S.; Matsuyama, M.; Matsuyama, S. Pharmacological inhibition of Bax-induced cell death: Bax-inhibiting peptides and small compounds inhibiting Bax. Exp. Biol. Med. 2019, 244, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Böttger, R.; Hoffmann, R.; Knappe, D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PloS ONE 2017, 12, e0178943. [Google Scholar] [CrossRef] [PubMed]
- Yachnin, B.J.; Khare, S.D. Engineering carboxypeptidase G2 circular permutations for the design of an autoinhibited enzyme. Protein Eng. Des. Sel. 2017, 30, 321–331. [Google Scholar] [CrossRef]
- AlQahtani, A.D.; Al-mansoori, L.; Bashraheel, S.S.; Rashidi, F.B.; Al-Yafei, A.; Elsinga, P.; Domling, A.; Goda, S.K. Production of “biobetter” glucarpidase variants to improve drug detoxification and antibody directed enzyme prodrug therapy for cancer treatment. Eur. J. Pharm. Sci. 2019, 127, 79–91. [Google Scholar] [CrossRef]
- AlQahtani, A.D.; O’Connor, D.; Domling, A.; Goda, S.K. Strategies for the production of long-acting therapeutics and efficient drug delivery for cancer treatment. Biomed. Pharmacother. 2019, 113, 108750. [Google Scholar] [CrossRef]
- Erazo-Oliveras, A.; Muthukrishnan, N.; Baker, R.; Wang, T.-Y.; Pellois, J.-P. Improving the Endosomal Escape of Cell-Penetrating Peptides and Their Cargos: Strategies and Challenges. Pharmaceuticals 2012, 5, 1177–1209. [Google Scholar] [CrossRef]
- Rydström, A.; Deshayes, S.; Konate, K.; Crombez, L.; Padari, K.; Boukhaddaoui, H.; Aldrian, G.; Pooga, M.; Divita, G. Direct translocation as major cellular uptake for CADY self-assembling peptide-based nanoparticles. PloS ONE 2011, 6, e25924. [Google Scholar] [CrossRef]
- Hayashi, Y.; Yamauchi, J.; Khalil, I.A.; Kajimoto, K.; Akita, H.; Harashima, H. Cell penetrating peptide-mediated systemic siRNA delivery to the liver. Int. J. Pharm. 2011, 419, 308–313. [Google Scholar] [CrossRef]
- El-Sayed, N.S.; Shirazi, A.N.; Sajid, M.I.; Park, S.E.; Parang, K.; Tiwari, R.K. Synthesis and Antiproliferative Activities of Conjugates of Paclitaxel and Camptothecin with a Cyclic Cell-Penetrating Peptide. Molecules 2019, 24, 1427. [Google Scholar] [CrossRef]
- Shirazi, A.N.; Mozaffari, S.; Sherpa, R.T.; Tiwari, R.; Parang, K. Efficient Intracellular Delivery of Cell-Impermeable Cargo Molecules by Peptides Containing Tryptophan and Histidine. Molecules 2018, 23, 1536. [Google Scholar] [CrossRef]
- Najjar, K.; Erazo-Oliveras, A.; Brock, D.J.; Wang, T.-Y.; Pellois, J.-P. An l- to d-Amino Acid Conversion in an Endosomolytic Analog of the Cell-penetrating Peptide TAT Influences Proteolytic Stability, Endocytic Uptake, and Endosomal Escape. J. Biol. Chem. 2017, 292, 847–861. [Google Scholar] [CrossRef]
- Wei, L.; Xing, P.; Su, R.; Shi, G.; Ma, Z.S.; Zou, Q. CPPred-RF: A Sequence-based Predictor for Identifying Cell-Penetrating Peptides and Their Uptake Efficiency. J. Proteome Res. 2017, 16, 2044–2053. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, 1st ed.; Walker, J.M., Ed.; Humana Press: Hatfield, UK, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Agostini, F.; Vendruscolo, M.; Tartaglia, G.G. Sequence-based prediction of protein solubility. J. Mol. Biol. 2012, 421, 237–241. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008, 9, 40. [Google Scholar] [CrossRef]
- Laskowski, R.A. Enhancing the functional annotation of PDB structures in PDBsum using key figures extracted from the literature. Bioinformatics 2007, 23, 1824–1827. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Pucci, F.; Kwasigroch, J.M.; Rooman, M. SCooP: An accurate and fast predictor of protein stability curves as a function of temperature. Bioinformatics 2017, 33, 3415–3422. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Walsh, I.; Seno, F.; Tosatto, S.C.; Trovato, A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014, 42, W301–W307. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. FoldRate: A web-server for predicting protein folding rates from primary sequence. Open Bioinforma. J. 2009, 3, 31–50. [Google Scholar] [CrossRef]
- Cilia, E.; Pancsa, R.; Tompa, P.; Lenaerts, T.; Vranken, W.F. The DynaMine webserver: Predicting protein dynamics from sequence. Nucleic Acids Res. 2014, 42, W264–W270. [Google Scholar] [CrossRef]
- Dimitrov, I.; Naneva, L.; Doytchinova, I.; Bangov, I. AllergenFP: Allergenicity prediction by descriptor fingerprints. Bioinformatics 2013, 30, 846–851. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 2007, 8, 4. [Google Scholar] [CrossRef]
- Bachmair, A.; Finley, D.; Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 1986, 234, 179–186. [Google Scholar] [CrossRef]
- Varshavsky, A. The N-end rule pathway of protein degradation. Genes Cells 1997, 2, 13–28. [Google Scholar] [CrossRef]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.; Open Source Drug Discovery Consortium. In silico approach for predicting toxicity of peptides and proteins. PloS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Gupta, S.; Madhu, M.K.; Sharma, A.K.; Sharma, V.K. ProInflam: A webserver for the prediction of proinflammatory antigenicity of peptides and proteins. J. Transl. Med. 2016, 14, 178. [Google Scholar] [CrossRef]
- Song, J.; Li, F.; Leier, A.; Marquez-Lago, T.T.; Akutsu, T.; Haffari, G.; Chou, K.-C.; Webb, G.I.; Pike, R.N. PROSPERous: High-throughput prediction of substrate cleavage sites for 90 proteases with improved accuracy. Bioinformatics 2017, 34, 684–687. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Li, C.; Marquez-Lago, T.T.; Leier, A.; Rawlings, N.D.; Haffari, G.; Revote, J.; Akutsu, T.; Chou, K.-C. Twenty years of bioinformatics research for protease-specific substrate and cleavage site prediction: A comprehensive revisit and benchmarking of existing methods. Brief. Bioinform. 2018, bby077. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2015, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
| Peptide No. | CPPs’ Names | Amino Acid Sequence | Cell-Penetrating or Not | Prediction Confidence 1 | Uptake Efficiency | Prediction Confidence 2 |
|---|---|---|---|---|---|---|
| 1 | Transportan 10 (TP10) | AGYLLGKINLKALAALAKKIL | Cell-penetrating | 0.98 | High | 1.00 |
| 2 | Ala43 substitution mutant of pAntp (43–58) | AQIKIWFQNRRMKWKK | Cell-penetrating | 0.95 | High | 0.96 |
| 3 | Crot (27–39) derivative 1 | CRFRFKCCKK | Cell-penetrating | 0.96 | High | 0.98 |
| 4 | Crot (27–39) derivative 2 | CRFRWKCCKK | Cell-penetrating | 0.96 | High | 0.99 |
| 5 | Crot (27–39) derivative 3 | CRWRFKCCKK | Cell-penetrating | 0.96 | High | 1.00 |
| 6 | CyLoP–1 | CRWRWKCCKK | Cell-penetrating | 0.95 | High | 1.00 |
| 7 | Crot (27–39) derivative 4 | CRWRWKCGCKK | Cell-penetrating | 0.92 | High | 0.99 |
| 8 | Crot (27–39) derivative 5 | DCRWRWKCCKK | Cell-penetrating | 0.82 | High | 0.99 |
| 9 | pAntp (49–58) | FQNRRMKWKK | Cell-penetrating | 0.84 | High | 0.91 |
| 10 | Tat (48–60) | GRKKRRQRRRPPQ | Cell-penetrating | 0.97 | High | 0.94 |
| 11 | pAntp (45–58) | IKIWFQNRRMKWKK | Cell-penetrating | 0.93 | High | 0.91 |
| 12 | Bip15 | IPMLK | Cell-penetrating | 0.56 | High | 0.92 |
| 13 | pAntp (47–58) | IWFQNRRMKWKK | Cell-penetrating | 0.89 | High | 0.97 |
| 14 | II | KALAALLKKLAKLLAALK | Cell-penetrating | 1.00 | High | 0.93 |
| 15 | Crot (27–39) derivative 6 | KCCKWRWRCK | Cell-penetrating | 0.95 | High | 0.94 |
| 16 | Crot (27–39) derivative 7 | KCGCRWRWKCGCKK | Cell-penetrating | 0.95 | High | 0.90 |
| 17 | Crot (27–39) derivative 8 | KCRWRWKCCKK | Cell-penetrating | 0.95 | High | 0.98 |
| 18 | Crot (27–39) derivative 9 | KDCRWRWKCCKK | Cell-penetrating | 0.78 | High | 0.99 |
| 19 | pAntp (46–58) | KIWFQNRRMKWKK | Cell-penetrating | 0.93 | High | 0.96 |
| 20 | 7 | KLWMRWWSPTTRRYG | Cell-penetrating | 0.98 | High | 0.93 |
| 21 | No.14–2 | KLWMRWYSATTRRYG | Cell-penetrating | 0.98 | High | 0.97 |
| 22 | No.14 | KLWMRWYSPTTRRYG | Cell-penetrating | 0.98 | High | 0.96 |
| 23 | No.14–7 | KLWMRWYSPWTRRYG | Cell-penetrating | 0.96 | High | 0.92 |
| 24 | Crot (27–39) | KMDCRWRWKCCKK | Cell-penetrating | 0.80 | High | 0.94 |
| 25 | Crot (27–39) derivative 10 | KMDCRWRWKCKK | Cell-penetrating | 0.78 | High | 0.95 |
| 26 | Crot (27–39) derivative 11 | KMDCRWRWKCSKK | Cell-penetrating | 0.82 | High | 0.95 |
| 27 | Crot (27–39) derivative 12 | KMDCRWRWKSCKK | Cell-penetrating | 0.83 | High | 0.95 |
| 28 | pVEC mutant 1 | LLIILRARIRKQAHAHSK | Cell-penetrating | 0.98 | High | 0.90 |
| 29 | pVEC mutant 2 | LLIILRRAIRKQAHAHSK | Cell-penetrating | 0.98 | High | 0.95 |
| 30 | pVEC mutant 3 | LLIILRRRIRAQAHAHSK | Cell-penetrating | 0.98 | High | 0.94 |
| 31 | Crot (27–39) derivative 13 | MDCRWRWKCCKK | Cell-penetrating | 0.79 | High | 0.93 |
| 32 | ARF (1-22) | MVRRFLVTLRIRRACGPPRVRV | Cell-penetrating | 0.88 | High | 0.93 |
| 33 | M918 | MVTVLFRRLRIRRACGPPRVRV | Cell-penetrating | 0.90 | High | 0.94 |
| 34 | pAntp (51–58) | NRRMKWKK | Cell-penetrating | 0.90 | High | 0.91 |
| 35 | pAntp (44–58) | QIKIWFQNRRMKWKK | Cell-penetrating | 0.96 | High | 0.92 |
| 36 | pAntp (50–8) | QNRRMKWKK | Cell-penetrating | 0.88 | High | 0.96 |
| 37 | Ala44 substitution mutant of pAntp (43–58) | RAIKIWFQNRRMKWKK | Cell-penetrating | 1.00 | High | 0.99 |
| 38 | PDX -1-PTD | RHIKIWFQNRRMKWKK | Cell-penetrating | 0.99 | High | 0.92 |
| 39 | No.14–25 | RLFMRFYSPTTRRYG | Cell-penetrating | 0.95 | High | 0.93 |
| 40 | No.14–17 | RLWMRWASPTTRRYG | Cell-penetrating | 0.99 | High | 0.96 |
| 41 | No.14–18 | RLWMRWYAPTTRRYG | Cell-penetrating | 0.98 | High | 0.98 |
| 42 | No.14–20 | RLWMRWYSPATRRYG | Cell-penetrating | 0.99 | High | 1.00 |
| 43 | No.14–21 | RLWMRWYSPTARRYG | Cell-penetrating | 0.99 | High | 1.00 |
| 44 | No.14–35 | RLWMRWYSPTTRRYA | Cell-penetrating | 0.98 | High | 0.98 |
| 45 | No.14–1 | RLWMRWYSPTTRRYG | Cell-penetrating | 0.99 | High | 0.98 |
| 46 | 30 | RLYMRYYSPTTRRYG | Cell-penetrating | 0.97 | High | 0.93 |
| 47 | Ala45 substitution mutant of pAntp (43–58) | RQAKIWFQNRRMKWKK | Cell-penetrating | 0.98 | High | 0.98 |
| 48 | Ala46 substitution mutant of pAntp (43–58) | RQIAIWFQNRRMKWKK | Cell-penetrating | 0.98 | High | 0.91 |
| 49 | Ala47 substitution mutant of pAntp (43–58) | RQIKAWFQNRRMKWKK | Cell-penetrating | 0.99 | High | 0.99 |
| 50 | Ala48 substitution mutant of pAntp (43–58) | RQIKIAFQNRRMKWKK | Cell-penetrating | 1.00 | High | 0.94 |
| 51 | Ala49 substitution mutant of pAntp (43–58) | RQIKIWAQNRRMKWKK | Cell-penetrating | 1.00 | High | 0.98 |
| 52 | Ala50 substitution mutant of pAntp (43–58) | RQIKIWFANRRMKWKK | Cell-penetrating | 0.99 | High | 0.99 |
| 53 | pAntpHD (Pro50) | RQIKIWFPNRRMKWKK | Cell-penetrating | 0.99 | High | 0.96 |
| 54 | Ala51 substitution mutant of pAntp (43–58) | RQIKIWFQARRMKWKK | Cell-penetrating | 0.99 | High | 0.94 |
| 55 | Ala52 substitution mutant of pAntp (43–58) | RQIKIWFQNARMKWKK | Cell-penetrating | 0.95 | High | 0.92 |
| 56 | Met-Arg | RQIKIWFQNMRRKWKK | Cell-penetrating | 1.00 | High | 0.93 |
| 57 | Ala54 substitution mutant of pAntp (43–58) | RQIKIWFQNRRAKWKK | Cell-penetrating | 0.99 | High | 0.97 |
| 58 | Penetratin | RQIKIWFQNRRMKWKK | Cell-penetrating | 1.00 | High | 0.97 |
| 59 | Retro - Tat (57–49) | RRRQRRKKR | Cell-penetrating | 1.00 | High | 0.90 |
| 60 | R6 | RRRRRR | Cell-penetrating | 1.00 | High | 0.91 |
| 61 | R9 | RRRRRRRRR | Cell-penetrating | 1.00 | High | 0.91 |
| 62 | Crot (27–39) derivative 14 | RWRWKCCKK | Cell-penetrating | 0.91 | High | 0.97 |
| 63 | Crot (27–39) derivative 15 | SRWRWKCCKK | Cell-penetrating | 0.94 | High | 0.93 |
| 64 | Rev (34–50) | TRQARRNRRRRWRERQR | Cell-penetrating | 0.98 | High | 0.90 |
| 65 | HIV-1 Rev (34–50) | TRQARRNRRRRWRERQRGC | Cell-penetrating | 0.96 | High | 0.90 |
| 66 | Bip6 | VPALK | Cell-penetrating | 0.74 | High | 0.96 |
| 67 | Bip1 | VPMLK | Cell-penetrating | 0.57 | High | 0.96 |
| 68 | Bip2 | VPTLK | Cell-penetrating | 0.67 | High | 0.99 |
| 69 | Bip16 | VPTLQ | Cell-penetrating | 0.60 | High | 0.91 |
| 70 | pAntp (48–58) | WFQNRRMKWKK | Cell-penetrating | 0.84 | High | 0.97 |
| Category | Names of CPPs | Experimental Uptake Efficiency | References |
|---|---|---|---|
| Crot (27–39) and its derivatives | CyLoP-1 | High, higher than D-Tat peptide, penetratin, and D-R8 | [29] |
| Crot (27–39) | 78% of CyLoP1 | ||
| Crot (27–39) derivative 1 | 63% of CyLoP1 | ||
| Crot (27–39) derivative 2 | 66% of CyLoP1 | ||
| Crot (27–39) derivative 3 | 61% of CyLoP1 | ||
| Crot (27–39) derivative 4 | 59% of CyLoP1 | ||
| Crot (27–39) derivative 5 | 47% of CyLoP1 | ||
| Crot (27–39) derivative 6 | 42% of CyLoP1 | ||
| Crot (27–39) derivative 7 | 75% of CyLoP1 | ||
| Crot (27–39) derivative 8 | 39% of CyLoP1 | ||
| Crot (27–39) derivative 9 | 26% of CyLoP1 | ||
| Crot (27–39) derivative 10 | 79% of CyLoP1 | ||
| Crot (27–39) derivative 11 | 58% of CyLoP1 | ||
| Crot (27–39) derivative 12 | 29% of CyLoP1 | ||
| Crot (27–39) derivative 13 | 83% of CyLoP1 | ||
| Crot (27–39) derivative 14 | 37% of CyLoP1 | ||
| Crot (27–39) derivative 15 | 46% of CyLoP1 | ||
| Penetratin and its derivatives | Penetratin/pAntp (43–58) | Lower than polyarginines but higher than Tat peptide and transportan | [30] |
| pAntp (44–58) | 85% of pAntp (43–58) | [31] | |
| pAntp (45–58) | 95% of pAntp (43–58) | ||
| pAntp (46–58) | 65% of pAntp (43–58) | ||
| pAntp (47–58) | 50% of pAntp (43–58) | ||
| pAntp (48–58) | 55% of pAntp (43–58) | ||
| pAntp (49–58) | 65% of pAntp (43–58) | ||
| pAntp (50–58) | 60% of pAntp (43–58) | ||
| pAntp (51–58) | 60% of pAntp (43–58) | ||
| Ala43 substitution mutant of pAntp (43–58) | 90% of pAntp (43–58) | ||
| Ala44 substitution mutant of pAntp (43–58) | 65% of (pAntp) (43–58) | ||
| Ala45 substitution mutant of pAntp (43–58) | 80% of pAntp (43–58) | ||
| Ala46 substitution mutant of pAntp (43–58) | 50% of pAntp (43–58) | ||
| Ala47 substitution mutant of pAntp (43–58) | 55% of pAntp (43–58) | ||
| Ala48 substitution mutant of pAntp (43–58) | 65% of pAntp (43–58) | ||
| Ala49 substitution mutant of pAntp (43–58) | 90% of pAntp (43–58) | ||
| Ala50 substitution mutant of pAntp (43–58) | 90% of pAntp (43–58) | ||
| Ala51 substitution mutant of pAntp (43–58) | 60% of pAntp (43–58) | ||
| Ala52 substitution mutant of pAntp (43–58) | 45% of pAntp (43–58) | ||
| Ala54 substitution mutant of pAntp (43–58) | 90% of pAntp (43–58) | ||
| Met-Arg | Not available | [32] | |
| pAntpHD (Pro50) | High Comparable to pAntp (43–58) | [33] | |
| CPP derived from PDX-1 protein | PDX-1-PTD | Not available | [34] |
| Tumor lineage-homing CPPs | 7 | Not available | [35] |
| Peptide 14 and its derivatives | No.14 | Higher than Tat peptide | [36] |
| No.14–1 | Higher than Tat and peptide No. 14 | ||
| No.14–2 | Higher than Tat peptide | ||
| No.14–7 | Higher than Tat and peptide No. 14 | ||
| No.14–17 | Equal to Peptide No. 14 | ||
| No.14–18 | Higher than Peptide 14 and equal to peptide 14–1 | ||
| No.14–20 | Higher than Peptide 14 and equal to peptide 14–1 | ||
| No.14–21 | Higher than Peptide 14 and equal to peptide 14–1 | ||
| No.14–25 | Equal to peptide 14 | ||
| No.14–30 | Higher than peptide 14 and lower than peptide 14–1 | ||
| No.14–35 | Higher than Peptide 14 and equal to peptide 14–1 | ||
| Cell-penetrating penta peptides (CPP5s) | Bip1 | High About 97% of KLPVM | [37] |
| Bip2 | About 61% of KLPVM) | ||
| Bip6 | About 71% of KLPVM | ||
| Bip15 | Not available | ||
| Bip16 | About 70% KLPVM | ||
| pVEC (CPP derived from murine vascular endothelial cadherin) | pVEC mutant 1 | Comparable to pVEC | [38] |
| pVEC mutant 2 | Higher than pVEC | ||
| pVEC mutant 3 | Comparable to pVEC | ||
| Tat peptide and its derivatives | Tat (48–60) | High Lower than polyarginine and penetratin but equal to transportan | [30,39] |
| Rev (34–50) | High Comparable to Tat (48–60) | [40] | |
| HIV-1 Rev (34–50) | 2.5-6.6 times more than Tat (48–60) | [41] | |
| Retro - Tat (57–49) | High Compared to Tat (49–57) | [42] | |
| Polyarginines | R6 | Lower than R9 | [42] |
| R9 | High | ||
| Transportan peptide derivative | Transportan 10 (TP10) | High | [43] |
| CPPs derived from tumor suppressor protein p14ARF | ARF(1–22) | High Comparable to TP10 | [43] |
| M918 | Higher than Penetratin | [44] | |
| α-helical amphipathic CPPs | II | High | [45] |
| Peptide Number | CPPs’ Names | N-Terminal Conjugation with CPG2 | C-Terminal Conjugation with CPG2 | ||
|---|---|---|---|---|---|
| Secondary Structure after Conjugation with CPG2 (C: coil, S: Sheet, H: Helix) | Percentage of Amino Acids with Helix and Sheet Configuration | Secondary Structure after Conjugation with CPG2 (C: coil, S: sheet, H: helix) | Percentage of Amino Acids with Helix and Sheet Configuration | ||
| 1 | Transportan 10 (TP10) | CCSSSSSCHHHHCCCCCHHHC | 57.1 | CCCCCCCCCCHHHHHHHHHCC | 42.9 |
| 2 | Ala43 substitution mutant of pAntp (43–58) | CCHHHHHHHCCCCCHH | 56.2 | HHHHHHHHHHHHHHCC | 87.5 |
| 3 | Crot (27–39) derivative 1 | CSSSSSHHHH | 90.0 | CCCSSSSCCC | 40.0 |
| 4 | Crot (27–39) derivative 2 | CSSSSSSHHH | 90.0 | CCCSSSSCCC | 40.0 |
| 5 | Crot (27–39) derivative 3 | CSSSSSSCHH | 80.0 | CCHHHSSCCC | 50.0 |
| 6 | CyLoP-1 | CSSSHHHHHH | 90.0 | CCCCCCCCCC | 0.0 |
| 7 | Crot (27–39) derivative 4 | CCCCCCCCCCH | 9.1 | CCCCCCCCCCC | 0.0 |
| 8 | Crot (27–39) derivative 5 | CCSSSSHHHHH | 81.8 | CHHHHHHCCCC | 54.5 |
| 9 | pAntp (49–58) | CCCCHHHHHH | 60.0 | CCCCCCCCCC | 0.0 |
| 10 | Tat (48–60) | CCCCCCCCCCCCC | 0.0 | CCCCCCCCCCCCC | 0.0 |
| 11 | pAntp (45–58) | CCSSSCCCCCCCHH | 35.7 | CCCCCCCCCCCCCC | 0.0 |
| 12 | Bip15 | CCCCC | 0.0 | CCCCC | 0.0 |
| 13 | pAntp (47–58) | CCCCCCCCCCHH | 16.7 | CCCCCCCCCCCC | 0.0 |
| 14 | II | CHHHHHHHHHHHHHHHHH | 94.4 | HHHHHHHHHHHHHHHHCC | 88.9 |
| 15 | Crot (27–39) derivative 6 | CCCCCCCCCC | 0.0 | CCCCCCCCCC | 0.0 |
| 16 | Crot (27–39) derivative 7 | CCCCCCCCCCCCCH | 7.1 | CCHHHHHHHCCCCC | 50.0 |
| 17 | Crot (27–39) derivative 8 | CCSSSSSCHHH | 27.3 | CCCCCCCCCCC | 0.0 |
| 18 | Crot (27–39) derivative 9 | CCCSSSSHHHHH | 41.7 | CCCCCCCHHCCC | 16.7 |
| 19 | pAntp (46–58) | CCCCCCCCCCCHH | 15.4 | CCCCCHHHHHHCC | 46.2 |
| 20 | 7 | CCCSSSCCCCCCCCH | 26.7 | CCCCCCCCCCCCCCC | 0.0 |
| 21 | No.14–2 | CHHHHHHHHHHHCCC | 73.3 | HHHCCCCHHHHCCCC | 46.7 |
| 22 | No.14 | CCSSSSSCCCCCCHH | 53.3 | CCCCCCCCCCCCCCC | 0.0 |
| 23 | No.14–7 | CCHHHHHCCCCHHHH | 60.0 | CCCCCCCCCCCCCCC | 0.0 |
| 24 | Crot (27–39) | CCCSSSSSHHHHH | 76.9 | CCCCCHHHHHCCC | 38.5 |
| 25 | Crot (27–39) derivative 10 | CCCCSSHHHHHH | 66.7 | CCCCCCCCCCCC | 0.0 |
| 26 | Crot (27–39) derivative 11 | CCCCCCSSCCCHH | 30.8 | CCCCCCCCCCCCC | 0.0 |
| 27 | Crot (27–39) derivative 12 | CCCSSSSHHHHHH | 76.9 | CCCCHHHHHHCCC | 46.2 |
| 28 | pVEC mutant 1 | CHHHHHHHHHCCCCCCCH | 55.5 | CCHHHHHHHHHHHHCCCC | 66.7 |
| 29 | pVEC mutant 2 | CCSSSHHHHHCCCCCCCC | 44.4 | HHHHHHHHHHHCCCCCCC | 61.1 |
| 30 | pVEC mutant 3 | CSSSHHHHHHHCCCCCCH | 61.1 | CCHHHHHHHHHHHCCCCC | 61.1 |
| 31 | Crot (27–39) derivative 13 | CCCSSSSHHHHH | 75.0 | CCCCHHHHHCCC | 41.7 |
| 32 | ARF(1–22) | CCCHHHHHHHHHHHHCCCCCCC | 54.5 | HHHHHHHHHHHCCCCCCCCCCC | 50.0 |
| 33 | M918 | CCSSHHHHHHHHHHCCCCCCCC | 54.5 | CCHHHHHHHHCCCCCCCCCCCC | 36.4 |
| 34 | pAntp (51–58) | CCCCCCHH | 25.0 | CCCCCCCC | 0.0 |
| 35 | pAntp (44–58) | CCCHHHHCCCCCCHH | 40.0 | HHHHHHHHHCCCCCC | 60.0 |
| 36 | pAntp (50–58) | CCCCCCCHH | 22.2 | CCCCCCCCC | 0.0 |
| 37 | Ala44 substitution mutant of pAntp (43–58) | CCCCHHHCCCCCCCHH | 31.2 | HHHHHHHHCCHHHCCC | 68.8 |
| 38 | PDX -1-PTD | CCCHHHHCCCCCCCHH | 37.5 | HHHHHHHHHHHCCCCC | 68.8 |
| 39 | No.14–25 | CSSSSSCCCHHHHHH | 73.3 | CHHCCCCCCCCCCCC | 13.3 |
| 40 | No.14–17 | CSSSSCCCCCCHHHH | 53.3 | HHHHCCCCCCCCCCC | 26.7 |
| 41 | No.14–18 | CCSSSSSCCCCCCCC | 33.3 | HHHHHHHCCCCCCCC | 46.7 |
| 42 | No.14–20 | CCCCCCCCCCCCCCH | 6.7 | CCCCCCCCCCCCCCC | 0.0 |
| 43 | No.14–21 | CCSSSSSSCCCHHHH | 66.7 | HHHHHHHHHHHHHCC | 86.7 |
| 44 | No.14–1135 | CCSSSSSCCCCCCCC | 33.3 | CCCCCCCCCCCCCCC | 0.0 |
| 45 | No.14–1 | CCCSSSCCCCCCCHH | 33.3 | CCCCCCCCCCCCCCC | 0.0 |
| 46 | 30 | CCCSSSCCCHHHHCC | 46.7 | CCCCCCCCCCCCCCC | 0.0 |
| 47 | Ala45 substitution mutant of pAntp (43–58) | CCCSSSSCCCCCCCHH | 37.5 | HHHHHHHHHHHHHCCC | 81.3 |
| 48 | Ala46 substitution mutant of pAntp (43–58) | CCSSSSSCCCCCCCHH | 43.7 | HHHHHHHCCCHHHHCC | 68.8 |
| 49 | Ala47 substitution mutant of pAntp (43–58) | CCHHHCCCCCCCCCHH | 31.2 | HHHHHHHHHHCCCCCC | 62.5 |
| 50 | Ala48 substitution mutant of pAntp (43–58) | CCHHHHHHHCCCCCCC | 43.7 | CCCCHHHCHHHHHCCC | 50.0 |
| 51 | Ala49 substitution mutant of pAntp (43–58) | CCCSSCCCCCCCCCHH | 25 | HHCCHHHHCCCHHCCC | 50.0 |
| 52 | Ala50 substitution mutant of pAntp (43–58) | CCSSSSSCCCCCCCHH | 43.7 | HHHHHHHHHHHHCCCC | 87.5 |
| 53 | pAntpHD (Pro50) | CCCCCCCCCCCCCCHH | 12.5 | HHHHHHCCCCHHHCCC | 68.8 |
| 54 | Ala51 substitution mutant of pAntp (43–58) | CCCSSSSCCCCCCCCH | 31.2 | CCCCCCHHHHHHHHCC | 50.0 |
| 55 | Ala52 substitution mutant of pAntp(43–58) | CCHHHHHHHHHHHCCC | 68.7 | CCCCCHHHCCHHHCCC | 37.5 |
| 56 | Met-Arg | CCHHHHHHHHHHHHHH | 87.5 | HHHHHHHHHHHHHHCC | 87.5 |
| 57 | Ala54 substitution mutant of pAntp(43–58) | CSSSHHHCCCCCCCHH | 50.0 | HHHHHHHHCCHHHHCC | 75.0 |
| 58 | Penetratin | CCSSSSSCCCCCCCCH | 37.5 | HHHHHHHHHHHHHHCC | 87.5 |
| 59 | Retro - Tat (57–49) | CCCCCCCCC | 0.0 | CCCCCCCCC | 0.0 |
| 60 | R6 | CCHHHH | 66.7 | CCCCCC | 0.0 |
| 61 | R9 | CCCCCCCCC | 0.0 | CCCCHHCCC | 22.2 |
| 62 | Crot (27–39) derivative 14 | CSSHHHHHH | 88.9 | CCCSSSCCC | 33.3 |
| 63 | Crot (27–39) derivative 15 | CCSSSHHHHH | 80.0 | CCCCCCCCCC | 0.0 |
| 64 | Rev (34–50) | CCCCCCCCCCCCCCCCC | 0.0 | CCCCCCCCCCCCCCCCC | 0.0 |
| 65 | HIV-1 Rev (34–50) | CCCCCCCCCHHHHHHHCCC | 36.8 | CCCCCCCCCCCCCCCCCCC | 0.0 |
| 66 | Bip6 | CCCCC | 0.0 | CCCCC | 0.0 |
| 67 | Bip1 | CCCCC | 0.0 | CCCCC | 0.0 |
| 68 | Bip2 | CCCCC | 0.0 | CCCCC | 0.0 |
| 69 | Bip16 | CCCCC | 0.0 | CCCCC | 0.0 |
| 70 | pAntp (48–58) | CCCCCCCCCHH | 18.2 | CCCCCCCCCCC | 0.0 |
| Peptide Number | Name of the Conjugate | Number of Amino Acids | Mw (Da) | pI | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|
| 1 | Transportan 10 (TP10)-CPG2 | 414 | 43860.19 | 8.37 | 23.99 | 97.20 | −0.116 |
| 2 | Ala43 substitution mutant of pAntp (43-58)-CPG2 | 409 | 43839.07 | 8.82 | 25.71 | 91.69 | −0.218 |
| 3 | Crot (27–39) derevative 1-CPG2 | 403 | 42996.11 | 8.48 | 24.58 | 90.87 | −0.187 |
| 4 | Crot (27–39) derevative 2-CPG2 | 403 | 43035.15 | 8.48 | 26.37 | 90.87 | −0.196 |
| 5 | Crot (27–39) derevative 3-CPG2 | 403 | 43035.15 | 8.48 | 26.00 | 90.87 | −0.196 |
| 6 | CyLoP-1-CPG2 | 403 | 43074.18 | 8.48 | 27.79 | 90.87 | −0.205 |
| 7 | Crot (27–39) derevative 4-CPG2 | 404 | 43131.24 | 8.48 | 27.75 | 90.64 | −0.206 |
| 8 | Crot (27–39) derevative 5-CPG2 | 404 | 43189.27 | 8.21 | 27.75 | 90.64 | −0.214 |
| 9 | pAntp (49–58)-CPG2 | 403 | 43099.16 | 8.65 | 26.37 | 90.87 | −0.228 |
| 10 | Tat (48–60)-CPG2 | 406 | 43396.46 | 9.08 | 31.77 | 90.20 | −0.279 |
| 11 | pAntp (45–58)-CPG2 | 407 | 43639.86 | 8.82 | 25.79 | 91.89 | −0.215 |
| 12 | Bip15–CPG2 | 398 | 42278.25 | 6.56 | 24.66 | 93.97 | −0.159 |
| 13 | pAntp (47–58)-CPG2 | 405 | 43398.53 | 8.65 | 26.29 | 91.38 | −0.218 |
| 14 | II-CPG2 | 411 | 43554.90 | 8.64 | 23.83 | 97.20 | −0.121 |
| 15 | Crot (27–39) derevative 6-CPG2 | 403 | 43074.18 | 8.48 | 26.37 | 90.87 | −0.205 |
| 16 | Crot (27–39) derevative 7-CPG2 | 407 | 43419.60 | 8.61 | 27.62 | 89.98 | −0.209 |
| 17 | Crot (27–39) derevative 8-CPG2 | 404 | 43202.36 | 8.66 | 27.75 | 90.64 | −0.215 |
| 18 | Crot (27–39) derevative 9-CPG2 | 405 | 43317.45 | 8.47 | 27.70 | 90.42 | −0.223 |
| 19 | pAntp (46–58)-CPG2 | 406 | 43526.70 | 8.82 | 26.04 | 91.16 | −0.227 |
| 20 | 7-CPG2 | 408 | 43701.80 | 8.39 | 28.82 | 90.71 | −0.215 |
| 21 | No.14–2-CPG2 | 408 | 43652.73 | 8.38 | 27.74 | 90.96 | −0.208 |
| 22 | No.14-CPG2 | 408 | 43678.77 | 8.38 | 28.82 | 90.71 | −0.216 |
| 23 | No.14–7-CPG2 | 408 | 43763.88 | 8.38 | 28.38 | 90.71 | −0.216 |
| 24 | Crot (27–39)-CPG2 | 406 | 43448.64 | 8.47 | 28.46 | 90.20 | −0.217 |
| 25 | Crot (27–39) derevative 10-CPG2 | 405 | 43345.50 | 8.53 | 28.51 | 90.42 | −0.224 |
| 26 | Crot (27–39) derevative 11-CPG2 | 406 | 43432.58 | 8.53 | 28.46 | 90.20 | −0.226 |
| 27 | Crot (27–39) derevative 12-CPG2 | 406 | 43432.58 | 8.53 | 29.27 | 90.20 | −0.226 |
| 28 | pVEC mutant 1-CPG2 | 411 | 43802.04 | 8.66 | 25.76 | 95.52 | −0.169 |
| 29 | pVEC mutant 2-CPG2 | 411 | 43802.04 | 8.66 | 27.15 | 95.52 | −0.169 |
| 30 | pVEC mutant 3-CPG2 | 411 | 43830.05 | 8.67 | 27.97 | 95.52 | −0.171 |
| 31 | Crot (27–39) derevative 13-CPG2 | 405 | 43320.46 | 8.17 | 27.70 | 90.42 | −0.208 |
| 32 | ARF(1–22)-CPG2 | 415 | 44329.74 | 8.93 | 28.88 | 94.10 | −0.158 |
| 33 | M918-CPG2 | 415 | 44329.74 | 8.93 | 28.88 | 94.10 | −0.158 |
| 34 | pAntp (51–58)-CPG2 | 401 | 42823.85 | 8.65 | 26.45 | 91.32 | −0.227 |
| 35 | pAntp (44–58)-CPG2 | 408 | 43767.99 | 8.82 | 25.75 | 91.67 | −0.223 |
| 36 | pAntp (50–58)-CPG2 | 402 | 42951.98 | 8.65 | 26.41 | 91.09 | −0.235 |
| 37 | Ala44 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43867.13 | 8.95 | 25.71 | 91.69 | −0.221 |
| 38 | PDX -1-PTD-CPG2 | 409 | 43933.19 | 8.95 | 27.26 | 91.44 | −0.233 |
| 39 | No.14–25-CPG2 | 408 | 43628.71 | 8.39 | 27.26 | 90.71 | −0.199 |
| 40 | No.14–17-CPG2 | 408 | 43614.68 | 8.40 | 28.66 | 90.96 | −0.210 |
| 41 | No.14–18-CPG2 | 408 | 43690.78 | 8.39 | 29.00 | 90.96 | −0.211 |
| 42 | No.14–20-CPG2 | 408 | 43676.75 | 8.39 | 29.50 | 90.96 | −0.211 |
| 43 | No.14–21-CPG2 | 408 | 43676.75 | 8.39 | 29.02 | 90.96 | −0.211 |
| 44 | No.14–35-CPG2 | 408 | 43720.81 | 8.39 | 30.02 | 90.96 | −0.212 |
| 45 | No.14–1-CPG2 | 408 | 43706.78 | 8.39 | 29.02 | 90.71 | −0.217 |
| 46 | 30-CPG2 | 408 | 43660.71 | 8.37 | 27.65 | 90.71 | −0.219 |
| 47 | Ala45 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43882.10 | 8.95 | 26.39 | 90.73 | −0.240 |
| 48 | Ala46 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43867.09 | 8.83 | 26.60 | 91.69 | −0.220 |
| 49 | Ala47 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43882.10 | 8.95 | 26.39 | 90.73 | −0.240 |
| 50 | Ala48 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43809.00 | 8.95 | 26.19 | 91.69 | −0.227 |
| 51 | Ala49 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43848.08 | 8.95 | 25.82 | 91.69 | −0.236 |
| 52 | Ala50 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43867.13 | 8.95 | 26.19 | 91.69 | −0.221 |
| 53 | pAntpHD (Pro50)-CPG2 | 409 | 43893.17 | 8.95 | 26.66 | 91.44 | −0.229 |
| 54 | Ala51 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43881.16 | 8.95 | 26.19 | 91.69 | −0.221 |
| 55 | Ala52 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43839.07 | 8.82 | 24.78 | 91.69 | −0.218 |
| 56 | Met-Arg-CPG2 | 409 | 43924.18 | 8.95 | 26.00 | 91.44 | −0.233 |
| 57 | Ala54 substitution mutant of pAntp (43–58)-CPG2 | 409 | 43864.07 | 8.95 | 26.19 | 91.69 | −0.234 |
| 58 | Penetratin-CPG2 | 409 | 43924.18 | 8.95 | 26.19 | 91.44 | −0.233 |
| 59 | Retro - Tat (57–49)-CPG2 | 402 | 43017.05 | 9.08 | 30.55 | 91.09 | −0.264 |
| 60 | R6-CPG2 | 399 | 42632.57 | 8.86 | 32.28 | 91.78 | −0.238 |
| 61 | R9-CPG2 | 402 | 43101.13 | 9.19 | 36.38 | 91.09 | −0.269 |
| 62 | Crot (27–39) derevative 14-CPG2 | 402 | 42971.04 | 8.53 | 27.84 | 91.09 | −0.212 |
| 63 | Crot (27–39) derevative 15-CPG2 | 403 | 43058.12 | 8.52 | 28.27 | 90.87 | −0.214 |
| 64 | Rev (34–50)-CPG2 | 410 | 44115.2 | 9.18 | 32.92 | 89.56 | −0.309 |
| 65 | HIV-1 Rev (34–50)-CPG2 | 412 | 44275.39 | 9.13 | 32.60 | 89.13 | −0.302 |
| 66 | Bip6-CPG2 | 398 | 42204.11 | 6.55 | 25.89 | 93.97 | −0.160 |
| 67 | Bip1-CPG2 | 398 | 42264.22 | 6.55 | 25.22 | 93.72 | −0.159 |
| 68 | Bip2-CPG2 | 398 | 42234.13 | 6.55 | 25.41 | 93.72 | −0.166 |
| 69 | Bip16-CPG2 | 398 | 42234.09 | 6.22 | 26.44 | 93.72 | −0.165 |
| 70 | pAntp (48–58)-CPG2 | 404 | 43285.37 | 8.65 | 26.33 | 90.64 | −0.229 |
| CPG2 | 393 | 41695.44 | 6.22 | 25.33 | 93.18 | −0.173 |
| Peptide Number | Name of the Conjugate | The Solubility Percentage of CPP-CPG2 N-Terminal Conjugates | The Solubility Percentage of CPG2-CPP C-Terminal Conjugates |
|---|---|---|---|
| 1 | Transportan 10 (TP10)-CPG2 | 86% | 86% |
| 2 | Ala43 substitution mutant of pAntp (43–58)-CPG2 | 79% | 79% |
| 3 | Crot (27–39) derevative 1-CPG2 | 87% | 84% |
| 4 | Crot (27–39) derevative 2-CPG2 | 86% | 83% |
| 5 | Crot (27–39) derevative 3-CPG2 | 86% | 83% |
| 6 | CyLoP-1-CPG2 | 85% | 82% |
| 7 | Crot (27–39) derevative 4-CPG2 | 86% | 82% |
| 8 | Crot (27–39) derevative 5-CPG2 | 85% | 83% |
| 9 | pAntp (49–58)-CPG2 | 82% | 81% |
| 10 | Tat (48–60)-CPG2 | 79% | 82% |
| 11 | pAntp (45–58)-CPG2 | 80% | 79% |
| 12 | Bip15-CPG2 | 84% | 84% |
| 13 | pAntp (47–58)-CPG2 | 81% | 79% |
| 14 | II-CPG2 | 87% | 88% |
| 15 | Crot (27–39) derevative 6-CPG2 | 81% | 85% |
| 16 | Crot (27–39) derevative 7-CPG2 | 84% | 85% |
| 17 | Crot (27–39) derevative 8-CPG2 | 85% | 83% |
| 18 | Crot (27–39) derevative 9-CPG2 | 84% | 84% |
| 19 | pAntp (46–58)-CPG2 | 81% | 80% |
| 20 | 7-CPG2 | 70% | 74% |
| 21 | No.14–2-CPG2 | 70% | 74% |
| 22 | No.14-CPG2 | 71% | 75% |
| 23 | No.14–7-CPG2 | 70% | 74% |
| 24 | Crot (27–39)-CPG2 | 84% | 83% |
| 25 | Crot (27–39) derevative 10-CPG2 | 83% | 83% |
| 26 | Crot (27–39) derevative 11-CPG2 | 83% | 83% |
| 27 | Crot (27–39) derevative 12-CPG2 | 83% | 82% |
| 28 | pVEC mutant 1-CPG2 | 80% | 80% |
| 29 | pVEC mutant 2-CPG2 | 80% | 80% |
| 30 | pVEC mutant 3-CPG2 | 78% | 77% |
| 31 | Crot (27–39) derevative 13-CPG2 | 84% | 82% |
| 32 | ARF(1–22)-CPG2 | 75% | 76% |
| 33 | M918-CPG2 | 75% | 76% |
| 34 | pAntp (51–58)-CPG2 | 84% | 82% |
| 35 | pAntp (44–58)-CPG2 | 79% | 79% |
| 36 | pAntp (50–58)-CPG2 | 83% | 82% |
| 37 | Ala44 substitution mutant of pAntp (43–58)-CPG2 | 79% | 78% |
| 38 | PDX -1-PTD-CPG2 | 79% | 78% |
| 39 | No.14–25-CPG2 | 73% | 75% |
| 40 | No.14–17-CPG2 | 72% | 73% |
| 41 | No.14–18-CPG2 | 71% | 72% |
| 42 | No.14–20-CPG2 | 72% | 73% |
| 43 | No.14–21-CPG2 | 72% | 73% |
| 44 | No.14–35-CPG2 | 70% | 72% |
| 45 | No.14–1-CPG2 | 71% | 72% |
| 46 | 30-CPG2 | 72% | 74% |
| 47 | Ala45 substitution mutant of pAntp (43–58)-CPG2 | 79% | 78% |
| 48 | Ala46 substitution mutant of pAntp (43–58)-CPG2 | 78% | 76% |
| 49 | Ala47 substitution mutant of pAntp (43–58)-CPG2 | 80% | 78% |
| 50 | Ala48 substitution mutant of pAntp (43–58)-CPG2 | 81% | 80% |
| 51 | Ala49 substitution mutant of pAntp (43–58)-CPG2 | 80% | 78% |
| 52 | Ala50 substitution mutant of pAntp (43–58)-CPG2 | 79% | 78% |
| 53 | pAntpHD (Pro50)-CPG2 | 79% | 78% |
| 54 | Ala51 substitution mutant of pAntp (43–58)-CPG2 | 79% | 77% |
| 55 | Ala52 substitution mutant of pAntp (43–58)-CPG2 | 80% | 79% |
| 56 | Met-Arg-CPG2 | 79% | 78% |
| 57 | Ala54 substitution mutant of pAntp (43–58)-CPG2 | 80% | 79% |
| 58 | Penetratin-CPG2 | 79% | 78% |
| 59 | Retro - Tat (57–49)-CPG2 | 82% | 81% |
| 60 | R6-CPG2 | 80% | 82% |
| 61 | R9-CPG2 | 77% | 79% |
| 62 | Crot (27–39) derevative 14-CPG2 | 86% | 81% |
| 63 | Crot (27–39) derevative 15-CPG2 | 85% | 81% |
| 64 | Rev (34–50)-CPG2 | 73% | 74% |
| 65 | HIV-1 Rev (34–50)-CPG2 | 74% | 73% |
| 66 | Bip6-CPG2 | 84% | 85% |
| 67 | Bip1-CPG2 | 84% | 85% |
| 68 | Bip2-CPG2 | 84% | 85% |
| 69 | Bip16-CPG2 | 83% | 85% |
| 70 | pAntp (48–58)-CPG2 | 82% | 79% |
| CPG2 | 83% | 83% |
| Peptide Number | Name of the Conjugate | N-Terminal Conjugates | C-Terminal Conjugates | ||
|---|---|---|---|---|---|
| Tm (degree~C) | ΔGr (kcal.mol−1) | Tm (degree~C) | ΔGr (kcal.mol−1) | ||
| 1 | Transportan 10 (TP10)-CPG2 | 64.1 | −12.3 | 60.9 | −15.9 |
| 2 | Ala43 substitution mutant of pAntp (43-58)-CPG2 | 59.1 | −13.1 | 60.7 | −11.6 |
| 3 | Crot (27−39) derivative 1-CPG2 | 61.2 | −13.3 | 61.7 | −13.1 |
| 4 | Crot (27−39) derivative 2-CPG2 | 60.7 | −14.2 | 61.7 | −13.1 |
| 5 | Crot (27−39) derivative 3-CPG2 | 61.3 | −14.1 | 59.0 | −14.0 |
| 6 | CyLoP-1-CPG2 | 62.0 | −13.2 | 61.5 | −13.6 |
| 7 | Crot (27−39) derivative 4-CPG2 | 63.1 | −14.0 | 65.8 | −13.8 |
| 8 | Crot (27−39) derivative 5-CPG2 | 62.4 | −13.1 | 60.2 | −15.0 |
| 9 | pAntp (49−58)-CPG2 | 60.5 | −16.0 | 59.3 | −16.6 |
| 10 | Tat (48−60)-CPG2 | 60.1 | −13.4 | 68.0 | −13.6 |
| 11 | pAntp (45−58)-CPG2 | 59.1 | −16.1 | 60.0 | −14.1 |
| 12 | Bip15-CPG2 | 60.7 | −12.7 | 63.1 | −14.4 |
| 13 | pAntp (47−58)-CPG2 | 59.7 | −15.4 | 62.2 | −13.2 |
| 14 | II-CPG2 | 59.7 | −15.4 | 62.5 | −14.8 |
| 15 | Crot (27−39) derivative 6-CPG2 | 62.8 | −13.3 | 62.3 | −12.9 |
| 16 | Crot (27−39) derivative 7-CPG2 | 61.7 | −15.9 | 64.1 | −15.2 |
| 17 | Crot (27−39) derivative 8-CPG2 | 61.1 | −14.8 | 60.3 | −15.9 |
| 18 | Crot (27−39) derivative 9-CPG2 | 59.1 | −13.2 | 61.7 | −13.3 |
| 19 | pAntp (46−58)-CPG2 | 63.0 | −12.7 | 61.2 | −14.6 |
| 20 | 7-CPG2 | 62.7 | −12.8 | 60.2 | −12.8 |
| 21 | No.14−2-CPG2 | 61.9 | −15.1 | 59.6 | −13.6 |
| 22 | No.14-CPG2 | 64.7 | −15.4 | 59.8 | −14.8 |
| 23 | No.14−7-CPG2 | 63.6 | −14.4 | 63.0 | −12.6 |
| 24 | Crot (27−39)-CPG2 | 61.4 | −15.0 | 61.0 | −14.9 |
| 25 | Crot (27−39) derivative 10-CPG2 | 60.6 | −14.7 | 57.3 | −13.0 |
| 26 | Crot (27−39) derivative 11-CPG2 | 61.0 | −15.1 | 58.9 | −15.4 |
| 27 | Crot (27−39) derivative 12-CPG2 | 60.5 | −13.6 | 59.5 | −13.9 |
| 28 | pVEC mutant 1-CPG2 | 63.8 | −14.0 | 64.2 | −12.0 |
| 29 | pVEC mutant 2-CPG2 | 60.2 | −16.8 | 57.9 | −15.8 |
| 30 | pVEC mutant 3-CPG2 | 63.8 | −11.2 | 62.1 | −13.7 |
| 31 | Crot (27−39) derivative 13-CPG2 | 64.2 | −14.3 | 58.5 | −13.3 |
| 32 | ARF(1−22)-CPG2 | 61.0 | −16.3 | 65.4 | −12.4 |
| 33 | M918-CPG2 | 65.5 | −14.8 | 63.3 | −12.6 |
| 34 | pAntp (51−58)-CPG2 | 56.9 | −15.8 | 61.6 | −15.3 |
| 35 | pAntp (44−58)-CPG2 | 58.1 | −13.6 | 59.5 | −16.8 |
| 36 | pAntp (50−58)-CPG2 | 62.7 | −15.6 | 61.0 | −13.6 |
| 37 | Ala44 substitution mutant of pAntp (43−58)-CPG2 | 60.7 | −13.8 | 58.4 | −15.6 |
| 38 | PDX -1-PTD-CPG2 | 57.6 | −13.9 | 58.5 | −14.2 |
| 39 | No.14−25-CPG2 | 62.3 | −11.5 | 58.5 | −15.3 |
| 40 | No.14−17-CPG2 | 60.3 | −14.6 | 58.7 | −14.9 |
| 41 | No.14−18-CPG2 | 60.6 | −13.3 | 61.6 | −13.9 |
| 42 | No.14−20-CPG2 | 62.3 | −14.1 | 56.6 | −15.2 |
| 43 | No.14−21-CPG2 | 63.8 | −14.8 | 60.8 | −14.2 |
| 44 | No.14−35-CPG2 | 62.5 | −15.9 | 59.9 | −15.4 |
| 45 | No.14−1-CPG2 | 61.7 | −15.5 | 60.7 | −13.4 |
| 46 | 30-CPG2 | 62.8 | −13.8 | 58.3 | −15.0 |
| 47 | Ala45 substitution mutant of pAntp (43−58)-CPG2 | 60.2 | −14.0 | 59.7 | −16.3 |
| 48 | Ala46 substitution mutant of pAntp (43−58)-CPG2 | 63.2 | −12.9 | 60.4 | −14.6 |
| 49 | Ala47 substitution mutant of pAntp (43−58)-CPG2 | 60.6 | −13.6 | 61.3 | −13.0 |
| 50 | Ala48 substitution mutant of pAntp (43−58)-CPG2 | 62.2 | −14.0 | 57.1 | −16.1 |
| 51 | Ala49 substitution mutant of pAntp (43−58)-CPG2 | 61.1 | −14.6 | 60.7 | −14.7 |
| 52 | Ala50 substitution mutant of pAntp (43−58)-CPG2 | 60.8 | −13.3 | 59.5 | −15.3 |
| 53 | pAntpHD (Pro50)-CPG2 | 62.7 | −12.7 | 60.7 | −13.4 |
| 54 | Ala51 substitution mutant of pAntp (43−58)-CPG | 60.3 | −14.9 | 56.6 | −13.4 |
| 55 | Ala52 substitution mutant of pAntp (43−58)-CPG2 | 63.9 | −11.9 | 61.1 | −13.4 |
| 56 | Met-Arg-CPG2 | 64.2 | −14.3 | 59.6 | −14.3 |
| 57 | Ala54 substitution mutant of pAntp (43−58)-CPG2 | 60.4 | −14.7 | 59.5 | −14.9 |
| 58 | Penetratin-CPG2 | 60.4 | −15.1 | 60.0 | −12.5 |
| 59 | Retro - Tat (57−49)-CPG2 | 61.0 | −14.0 | 60.4 | −14.7 |
| 60 | R6-CPG2 | 58.7 | −14.3 | 62.0 | −14.4 |
| 61 | R9-CPG2 | 62.7 | −13.8 | 57.7 | −15.5 |
| 62 | Crot (27−39) derivative 14-CPG2 | 63.5 | −13.2 | 62.6 | −13.0 |
| 63 | Crot (27−39) derivative 15-CPG2 | 60.3 | −13.6 | 63.0 | −15.1 |
| 64 | Rev (34−50)-CPG2 | 59.1 | −16.4 | 58.5 | −16.0 |
| 65 | HIV-1 Rev (34−50)-CPG2 | 62.0 | −13.3 | 59.9 | −15.2 |
| 66 | Bip6-CPG2 | 62.6 | −15.0 | 62.6 | −13.9 |
| 67 | Bip1-CPG2 | 56.7 | −14.9 | 63.4 | −11.2 |
| 68 | Bip2-CPG2 | 60.8 | −13.7 | 60.5 | −13.0 |
| 69 | Bip16-CPG2 | 61.8 | −14.6 | 61.4 | −14.3 |
| 70 | pAntp (48−58)-CPG2 | 56.5 | −15.6 | 59.5 | −13.5 |
| CPG2 | 61.7 | −15.1 | 61.7 | −15.1 | |
| Peptide Number | Name of the Conjugate | N-Terminal Conjugates | C-Terminal Conjugates | ||
|---|---|---|---|---|---|
| ΔG side H bond (kcal.mol−1) | ΔGtotal (kcal.mol−1) | ΔG side H bond (kcal.mol−1) | ΔGtotal (kcal.mol−1) | ||
| 1 | Transportan 10 (TP10)-CPG2 | −129.40 | 225.54 | −109.75 | 280.94 |
| 2 | Ala43 substitution mutant of pAntp (43-58)-CPG2 | −122.58 | 276.34 | −116.80 | 290.77 |
| 3 | Crot (27–39) derivative 1-CPG2 | −124.28 | 212.60 | −116.83 | 231.74 |
| 4 | Crot (27–39) derivative 2-CPG2 | −118.09 | 244.42 | −116.83 | 231.74 |
| 5 | Crot (27–39) derivative 3-CPG2 | −124.84 | 201.96 | −116.51 | 221.44 |
| 6 | CyLoP-1-CPG2 | −114.78 | 222.25 | −121.71 | 237.72 |
| 7 | Crot (27–39) derivative 4-CPG2 | −118.05 | 240.24 | −117.87 | 258.05 |
| 8 | Crot (27–39) derivative 5-CPG2 | −111.72 | 242.79 | −122.19 | 226.79 |
| 9 | pAntp (49–58)-CPG2 | −117.91 | 213.28 | −128.23 | 192.93 |
| 10 | Tat (48–60)-CPG2 | −113.01 | 237.15 | −110.09 | 285.79 |
| 11 | pAntp (45–58)-CPG2 | −109.21 | 257.80 | −123.56 | 222.57 |
| 12 | Bip15-CPG2 | −111.99 | 219.08 | −110.39 | 206.12 |
| 13 | pAntp (47–58)-CPG2 | −115.32 | 251.89 | −119.12 | 257.36 |
| 14 | II-CPG2 | −118.36 | 220.02 | −120.90 | 269.15 |
| 15 | Crot (27–39) derivative 6-CPG2 | −122.01 | 240.34 | −112.90 | 247.76 |
| 16 | Crot (27–39) derivative 7-CPG2 | −114.80 | 218.64 | −104.19 | 264.74 |
| 17 | Crot (27–39) derivative 8-CPG2 | −113.66 | 231.91 | −118.05 | 228.78 |
| 18 | Crot (27–39) derivative 9-CPG2 | −129.44 | 213.50 | −116.83 | 261.88 |
| 19 | pAntp (46–58)-CPG2 | −129.66 | 234.61 | −122.16 | 220.00 |
| 20 | 7-CPG2 | −118.37 | 248.66 | −113.78 | 227.61 |
| 21 | No.14–2-CPG2 | −128.25 | 266.61 | −106.97 | 305.73 |
| 22 | No.14-CPG2 | −121.58 | 288.14 | −125.08 | 244.91 |
| 23 | No.14–7-CPG2 | −106.30 | 268.12 | −120.60 | 253.67 |
| 24 | Crot (27–39)-CPG2 | −121.32 | 239.73 | −121.87 | 211.89 |
| 25 | Crot (27–39) derivative 10-CPG2 | −123.15 | 236.04 | −119.18 | 232.36 |
| 26 | Crot (27–39) derivative 11-CPG2 | −117.73 | 240.33 | −120.16 | 223.44 |
| 27 | Crot (27–39) derivative 12-CPG2 | −119.24 | 236.96 | −125.03 | 239.37 |
| 28 | pVEC mutant 1-CPG2 | −122.35 | 279.77 | −112.86 | 279.81 |
| 29 | pVEC mutant 2-CPG2 | −118.54 | 275.22 | −114.38 | 236.84 |
| 30 | pVEC mutant 3-CPG2 | −123.99 | 238.03 | −118.77 | 231.76 |
| 31 | Crot (27–39) derivative 13-CPG2 | −117.23 | 274.19 | −124.87 | 208.41 |
| 32 | ARF(1–22)-CPG2 | −116.67 | 287.55 | −127.44 | 277.30 |
| 33 | M918-CPG2 | −100.79 | 353.34 | −120.74 | 304.20 |
| 34 | pAntp (51–58)-CPG2 | −120.11 | 213.01 | −119.48 | 227.21 |
| 35 | pAntp (44–58)-CPG2 | −120.79 | 271.81 | −113.18 | 237.39 |
| 36 | pAntp (50–58)-CPG2 | −117.81 | 219.53 | −116.73 | 245.44 |
| 37 | Ala44 substitution mutant of pAntp (43-58)-CPG2 | −131.75 | 260.06 | −107.05 | 248.14 |
| 38 | PDX -1-PTD-CPG2 | −118.12 | 230.27 | −123.45 | 254.56 |
| 39 | No.14–25-CPG2 | −117.33 | 259.77 | −111.95 | 223.28 |
| 40 | No.14–17-CPG2 | −122.74 | 201.85 | −124.49 | 287.89 |
| 41 | No.14–18-CPG2 | −110.20 | 279.65 | −111.53 | 235.99 |
| 42 | No.14–20-CPG2 | −113.46 | 286.37 | −120.35 | 235.61 |
| 43 | No.14–21-CPG2 | −123.65 | 306.20 | −103.28 | 271.59 |
| 44 | No.14–35-CPG2 | −118.98 | 316.52 | −127.82 | 288.57 |
| 45 | No.14–1-CPG2 | −117.56 | 288.64 | −111.92 | 232.73 |
| 46 | 30-CPG2 | −114.00 | 274.45 | −112.39 | 225.22 |
| 47 | Ala45 substitution mutant of pAntp (43–58)-CPG2 | −121.63 | 244.81 | −128.84 | 256.18 |
| 48 | Ala46 substitution mutant of pAntp (43–58)-CPG2 | −114.00 | 258.27 | −109.97 | 275.43 |
| 49 | Ala47 substitution mutant of pAntp (43–58)-CPG2 | −111.21 | 279.55 | −118.80 | 250.65 |
| 50 | Ala48 substitution mutant of pAntp (43–58)-CPG2 | −115.90 | 215.33 | −113.15 | 225.09 |
| 51 | Ala49 substitution mutant of pAntp (43–58)-CPG2 | −120.58 | 259.79 | −125.14 | 244.61 |
| 52 | Ala50 substitution mutant of pAntp (43–58)-CPG2 | −122.21 | 266.42 | −117.50 | 275.80 |
| 53 | pAntpHD (Pro50)-CPG2 | −116.73 | 247.08 | −113.98 | 279.80 |
| 54 | Ala51 substitution mutant of pAntp (43–58)-CPG2 | −120.61 | 256.74 | −117.29 | 277.38 |
| 55 | Ala52 substitution mutant of pAntp (43–58)-CPG2 | −114.28 | 273.55 | −127.58 | 224.55 |
| 56 | Met-Arg-CPG2 | −120.88 | 294.89 | −130.43 | 225.80 |
| 57 | Ala54 substitution mutant of pAntp (43–58)-CPG2 | −113.18 | 274.17 | −126.16 | 223.46 |
| 58 | Penetratin-CPG2 | −114.09 | 237.56 | −114.95 | 232.56 |
| 59 | Retro - Tat (57–49)-CPG2 | −104.71 | 239.28 | −115.58 | 209.58 |
| 60 | R6-CPG2 | −108.43 | 215.13 | −120.23 | 219.49 |
| 61 | R9-CPG2 | −126.43 | 207.16 | −138.39 | 218.37 |
| 62 | Crot (27–39) derivative 14-CPG2 | −116.15 | 187.59 | −117.38 | 233.60 |
| 63 | Crot (27–39) derivative 15-CPG2 | −109.48 | 259.98 | −112.67 | 226.84 |
| 64 | Rev (34–50)-CPG2 | −140.73 | 250.30 | −120.99 | 267.37 |
| 65 | HIV-1 Rev (34–50)-CPG2 | −123.81 | 241.35 | −120.42 | 237.54 |
| 66 | Bip6-CPG2 | −126.11 | 176.25 | −117.74 | 237.91 |
| 67 | Bip1-CPG2 | −120.74 | 163.93 | −125.52 | 199.54 |
| 68 | Bip2-CPG2 | −106.63 | 227.56 | −122.97 | 213.03 |
| 69 | Bip16-CPG2 | −118.69 | 199.30 | −126.85 | 187.15 |
| 70 | pAntp (48–58)-CPG2 | −121.70 | 239.55 | −125.92 | 230.92 |
| CPG2 | −124.91 | 203.85 | −124.91 | 203.85 | |
| Peptide Number | Name of the Conjugate | N-Terminal Conjugates | C-Terminal Conjugates |
|---|---|---|---|
| Predicted Half-Folding Time (sec) | Predicted Half-Folding Time (sec) | ||
| 1 | Transportan 10 (TP10)-CPG2 | 315.75 | 292.07 |
| 2 | Ala43 substitution mutant of pAntp (43-58)-CPG2 | 341.86 | 375.47 |
| 3 | Crot (27–39) derivative 1-CPG2 | 269.35 | 294.03 |
| 4 | Crot (27–39) derivative 2-CPG2 | 266.25 | 297.08 |
| 5 | Crot (27–39) derivative 3-CPG2 | 266.25 | 297.08 |
| 6 | CyLoP-1-CPG2 | 263.17 | 296.95 |
| 7 | Crot (27–39) derivative 4-CPG2 | 290.06 | 299.60 |
| 8 | Crot (27–39) derivative 5-CPG2 | 251.58 | 269.07 |
| 9 | pAntp (49–58)-CPG2 | 260.00 | 265.84 |
| 10 | Tat (48–60)-CPG2 | 250.01 | 261.29 |
| 11 | pAntp (45–58)-CPG2 | 326.14 | 336.71 |
| 12 | Bip15-CPG2 | 254.99 | 251.17 |
| 13 | pAntp (47–58)-CPG2 | 312.02 | 325.60 |
| 14 | II-CPG2 | 282.10 | 260.72 |
| 15 | Crot (27–39) derivative 6-CPG2 | 272.11 | 300.16 |
| 16 | Crot (27–39) derivative 7-CPG2 | 300.50 | 310.30 |
| 17 | Crot (27–39) derivative 8-CPG2 | 262.57 | 286.80 |
| 18 | Crot (27–39) derivative 9-CPG2 | 251.02 | 271.45 |
| 19 | pAntp (46–58)-CPG2 | 314.64 | 324.84 |
| 20 | 7-CPG2 | 304.54 | 328.42 |
| 21 | No.14–2-CPG2 | 303.00 | 306.34 |
| 22 | No.14-CPG2 | 312.62 | 340.53 |
| 23 | No.14–7-CPG2 | 288.59 | 325.49 |
| 24 | Crot (27–39)-CPG2 | 262.67 | 283.81 |
| 25 | Crot (27–39) derivative 10-CPG2 | 263.85 | 297.45 |
| 26 | Crot (27–39) derivative 11-CPG2 | 287.51 | 303.33 |
| 27 | Crot (27–39) derivative 12-CPG2 | 281.35 | 296.93 |
| 28 | pVEC mutant 1-CPG2 | 318.66 | 318.66 |
| 29 | pVEC mutant 2-CPG2 | 308.37 | 318.66 |
| 30 | pVEC mutant 3-CPG2 | 331.94 | 310.97 |
| 31 | Crot (27–39) derivative 13-CPG2 | 257.41 | 310.97 |
| 32 | ARF(1–22)-CPG2 | 375.57 | 426.04 |
| 33 | M918-CPG2 | 371.56 | 371.56 |
| 34 | pAntp (51–58)-CPG2 | 246.89 | 371.56 |
| 35 | pAntp (44–58)-CPG2 | 341.31 | 359.69 |
| 36 | pAntp (50–58)-CPG2 | 253.19 | 371.56 |
| 37 | Ala44 substitution mutant of pAntp (43–58)-CPG2 | 339.30 | 371.56 |
| 38 | PDX -1-PTD-CPG2 | 354.49 | 371.56 |
| 39 | No.14–25-CPG2 | 302.06 | 325.92 |
| 40 | No.14–17-CPG2 | 286.95 | 325.92 |
| 41 | No.14–18-CPG2 | 316.39 | 325.92 |
| 42 | No.14–20-CPG2 | 300.29 | 325.92 |
| 43 | No.14–21-CPG2 | 303.60 | 341.55 |
| 44 | No.14–35-CPG2 | 309.58 | 341.55 |
| 45 | No.14–1-CPG2 | 315.26 | 341.55 |
| 46 | 30-CPG2 | 311.23 | 328.53 |
| 47 | Ala45 substitution mutant of pAntp (43–58)-CPG2 | 321.46 | 368.26 |
| 48 | Ala46 substitution mutant of pAntp (43–58)-CPG2 | 341.11 | 368.26 |
| 49 | Ala47 substitution mutant of pAntp (43–58)-CPG2 | 301.29 | 364.51 |
| 50 | Ala48 substitution mutant of pAntp (43–58)-CPG2 | 301.06 | 364.51 |
| 51 | Ala49 substitution mutant of pAntp (43–58)-CPG2 | 328.09 | 338.69 |
| 52 | Ala50 substitution mutant of pAntp (43–58)-CPG2 | 342.91 | 357.57 |
| 53 | pAntpHD (Pro50)-CPG2 | 324.61 | 342.27 |
| 54 | Ala51 substitution mutant of pAntp (43–58)-CPG2 | 342.47 | 342.27 |
| 55 | Ala52 substitution mutant of pAntp (43–58)-CPG2 | 300.29 | 364.03 |
| 56 | Met-Arg-CPG2 | 298.31 | 315.14 |
| 57 | Ala54 substitution mutant of pAntp (43–58)-CPG2 | 295.42 | 305.35 |
| 58 | Penetratin-CPG2 | 354.39 | 400.84 |
| 59 | Retro - Tat (57–49)-CPG2 | 243.94 | 246.69 |
| 60 | R6-CPG2 | 255.43 | 249.84 |
| 61 | R9-CPG2 | 269.07 | 249.03 |
| 62 | Crot (27–39) derivative 14-CPG2 | 258.50 | 276.26 |
| 63 | Crot (27–39) derivative 15-CPG2 | 255.15 | 263.85 |
| 64 | Rev (34–50)-CPG2 | 265.86 | 251.26 |
| 65 | HIV-1 Rev (34–50)-CPG2 | 276.22 | 261.16 |
| 66 | Bip6-CPG2 | 239.77 | 256.43 |
| 67 | Bip1-CPG2 | 256.15 | 259.01 |
| 68 | Bip2-CPG2 | 240.98 | 260.66 |
| 69 | Bip16-CPG2 | 242.18 | 262.04 |
| 70 | pAntp (48–58)-CPG2 | 300.95 | 278.92 |
| CPG2 | 229.89 | 229.89 |
| Peptide Number | CPPs’ Name | Immunogenicity of Respective CPP-CPG2 /CPG2-CPP Conjugate | Toxicity of CPPs | Hemolysis Potency of CPPs (PROB Score) | |
|---|---|---|---|---|---|
| N-Terminal Conjugates | C-Terminal Conjugates | ||||
| 1 | Transportan 10 (TP10) | 0.6620 | 0.6790 | non-toxin | 0.83 |
| 2 | Ala43 substitution mutant of pAntp (43–58) | 0.6874 | 0.6907 | non-toxin | 0.48 |
| 3 | Crot (27–39) derivative 1 | 0.7201 | 0.7267 | Toxin | 0.49 |
| 4 | Crot (27–39) derivative 2 | 0.7199 | 0.7263 | Toxin | 0.49 |
| 5 | Crot (27–39) derivative 3 | 0.7238 | 0.7319 | Toxin | 0.49 |
| 6 | CyLoP-1 | 0.7244 | 0.7323 | Toxin | 0.49 |
| 7 | Crot (27–39) derivative 4 | 0.7361 | 0.7403 | non-toxin | 0.49 |
| 8 | Crot (27–39) derivative 5 | 0.7276 | 0.7334 | Toxin | 0.49 |
| 9 | pAntp (49–58) | 0.6973 | 0.7017 | non-toxin | 0.48 |
| 10 | Tat (48–60) | 0.7208 | 0.7385 | non-toxin | 0.49 |
| 11 | pAntp (45–58) | 0.6910 | 0.6794 | non-toxin | 0.49 |
| 12 | Bip15 | 0.6929 | 0.6903 | non-toxin | 0.49 |
| 13 | pAntp (47–58) | 0.6855 | 0.6690 | non-toxin | 0.48 |
| 14 | II | 0.6466 | 0.6521 | non-toxin | 0.83 |
| 15 | Crot (27–39) derivative 6 | 0.7219 | 0.7300 | Toxin | 0.49 |
| 16 | Crot (27–39) derivative 7 | 0.7439 | 0.7410 | non-toxin | 0.49 |
| 17 | Crot (27–39) derivative 8 | 0.7267 | 0.7314 | Toxin | 0.49 |
| 18 | Crot (27–39) derivative 9 | 0.7263 | 0.7363 | Toxin | 0.49 |
| 19 | pAntp (46–58) | 0.6813 | 0.6764 | non-toxin | 0.49 |
| 20 | 7 | 0.6942 | 0.6961 | non-toxin | 0.49 |
| 21 | No.14–2 | 0.6961 | 0.6980 | non-toxin | 0.49 |
| 22 | No.14 | 0.6915 | 0.6934 | non-toxin | 0.49 |
| 23 | No.14–7 | 0.6844 | 0.6863 | non-toxin | 0.49 |
| 24 | Crot (27–39) | 0.7371 | 0.7409 | Toxin | 0.49 |
| 25 | Crot (27–39) derivative 10 | 0.7366 | 0.7388 | Toxin | 0.49 |
| 26 | Crot (27–39) derivative 11 | 0.7344 | 0.7406 | Toxin | 0.49 |
| 27 | Crot (27–39) derivative 12 | 0.7307 | 0.7327 | non-toxin | 0.49 |
| 28 | pVEC mutant 1 | 0.7006 | 0.6886 | non-toxin | 0.32 |
| 29 | pVEC mutant 2 | 0.6874 | 0.6755 | non-toxin | 0.32 |
| 30 | pVEC mutant 3 | 0.6901 | 0.6782 | non-toxin | 0.31 |
| 31 | Crot (27–39) derivative 13 | 0.7304 | 0.7344 | Toxin | 0.49 |
| 32 | ARF(1–22) | 0.6848 | 0.6917 | non-toxin | 0.48 |
| 33 | M918 | 0.6865 | 0.6893 | non-toxin | 0.48 |
| 34 | pAntp (51–58) | 0.7023 | 0.7142 | non-toxin | 0.49 |
| 35 | pAntp (44–58) | 0.6892 | 0.6913 | non-toxin | 0.49 |
| 36 | pAntp (50–58) | 0.7037 | 0.7153 | non-toxin | 0.49 |
| 37 | Ala44 substitution mutant of pAntp (43–58) | 0.6840 | 0.6866 | non-toxin | 0.48 |
| 38 | PDX -1-PTD | 0.6845 | 0.6861 | non-toxin | 0.49 |
| 39 | No.14–25 | 0.6910 | 0.6907 | non-toxin | 0.49 |
| 40 | No.14–17 | 0.6889 | 0.6900 | non-toxin | 0.49 |
| 41 | No.14–18 | 0.6927 | 0.6938 | non-toxin | 0.49 |
| 42 | No.14–20 | 0.6922 | 0.6933 | non-toxin | 0.48 |
| 43 | No.14–21 | 0.6949 | 0.6961 | non-toxin | 0.48 |
| 44 | No.14–35 | 0.6924 | 0.6938 | non-toxin | 0.49 |
| 45 | No.14–1 | 0.6929 | 0.6941 | non-toxin | 0.49 |
| 46 | 30 | 0.6981 | 0.7012 | non-toxin | 0.49 |
| 47 | Ala45 substitution mutant of pAntp (43–58) | 0.6741 | 0.6817 | non-toxin | 0.47 |
| 48 | Ala46 substitution mutant of pAntp (43–58) | 0.6756 | 0.6765 | non-toxin | 0.47 |
| 49 | Ala47 substitution mutant of pAntp (43–58) | 0.6740 | 0.6779 | non-toxin | 0.47 |
| 50 | Ala48 substitution mutant of pAntp (43–58) | 0.7007 | 0.7046 | non-toxin | 0.48 |
| 51 | Ala49 substitution mutant of pAntp (43–58) | 0.6784 | 0.6823 | non-toxin | 0.48 |
| 52 | Ala50 substitution mutant of pAntp (43–58) | 0.6827 | 0.6865 | non-toxin | 0.48 |
| 53 | pAntpHD (Pro50) | 0.6887 | 0.6926 | non-toxin | 0.49 |
| 54 | Ala51 substitution mutant of pAntp (43–58) | 0.6878 | 0.6916 | non-toxin | 0.48 |
| 55 | Ala52 substitution mutant of pAntp (43–58) | 0.6932 | 0.6970 | non-toxin | 0.48 |
| 56 | Met-Arg | 0.6792 | 0.6833 | non-toxin | 0.48 |
| 57 | Ala54 substitution mutant of pAntp (43–58) | 0.6791 | 0.6828 | non-toxin | 0.48 |
| 58 | Penetratin | 0.6820 | 0.6859 | non-toxin | 0.48 |
| 59 | Retro - Tat (57–49) | 0.7094 | 0.7274 | non-toxin | 0.17 |
| 60 | R6 | 0.6983 | 0.7184 | non-toxin | 0.49 |
| 61 | R9 | 0.7084 | 0.7284 | non-toxin | 0.49 |
| 62 | Crot (27–39) derivative 14 | 0.7263 | 0.7208 | Toxin | 0.49 |
| 63 | Crot (27–39) derivative 15 | 0.7182 | 0.7284 | Toxin | 0.49 |
| 64 | Rev (34–50) | 0.7032 | 0.7193 | non-toxin | 0.49 |
| 65 | HIV-1 Rev (34–50) | 0.7128 | 0.7213 | non-toxin | 0.48 |
| 66 | Bip6 | 0.6956 | 0.6980 | non-toxin | 0.49 |
| 67 | Bip1 | 0.6920 | 0.6924 | non-toxin | 0.49 |
| 68 | Bip2 | 0.6945 | 0.6984 | non-toxin | 0.49 |
| 69 | Bip16 | 0.6957 | 0.6968 | non-toxin | 0.49 |
| 70 | pAntp (48–58) | 0.6838 | 0.6695 | non-toxin | 0.47 |
| CPG2 | 0.7012 | 0.7012 | - | - | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behzadipour, Y.; Hemmati, S. Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo Molecule. Molecules 2019, 24, 4318. https://doi.org/10.3390/molecules24234318
Behzadipour Y, Hemmati S. Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo Molecule. Molecules. 2019; 24(23):4318. https://doi.org/10.3390/molecules24234318
Chicago/Turabian StyleBehzadipour, Yasaman, and Shiva Hemmati. 2019. "Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo Molecule" Molecules 24, no. 23: 4318. https://doi.org/10.3390/molecules24234318
APA StyleBehzadipour, Y., & Hemmati, S. (2019). Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo Molecule. Molecules, 24(23), 4318. https://doi.org/10.3390/molecules24234318