NMR Study of the Secondary Structure and Biopharmaceutical Formulation of an Active Branched Antimicrobial Peptide
Abstract
:1. Introduction
2. Results and Discussion
2.1. NMR Characterization of SET-M33 Peptide
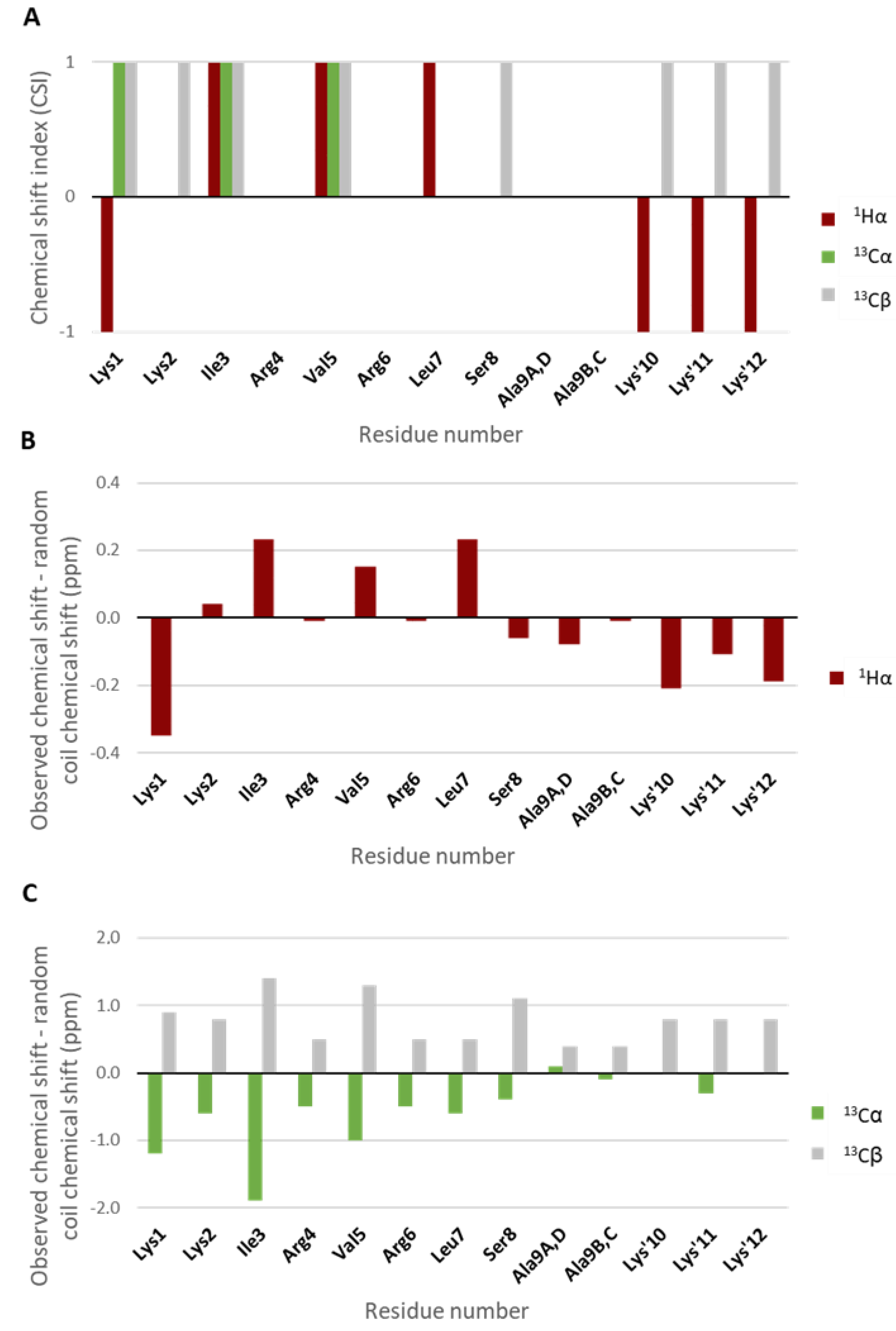
2.2. Assessment of Secondary Structure Elements
2.3. TFA/Chloride Counter Ion Exchange
2.4. MIC Determination
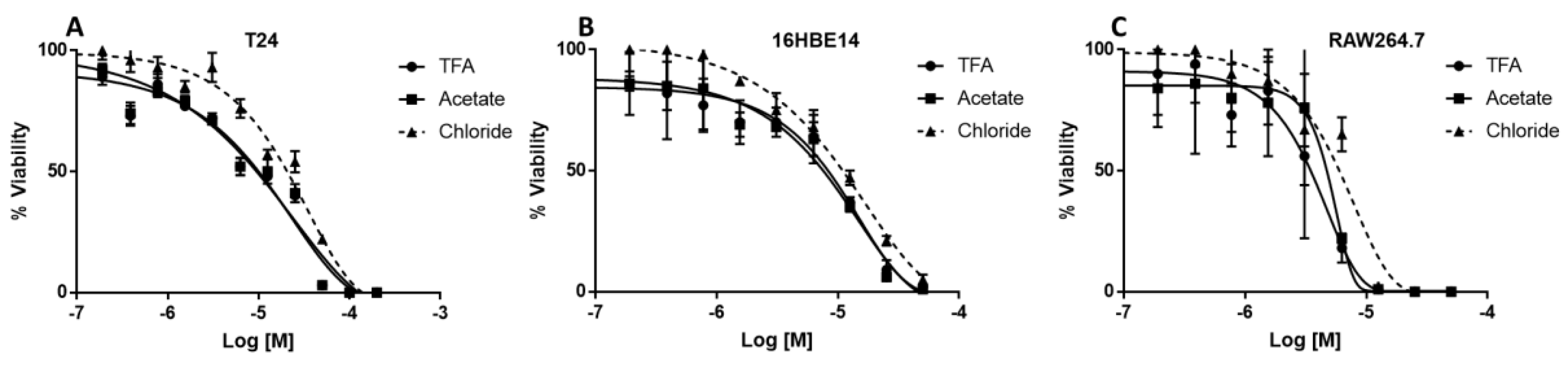
2.5. Toxicity of Different SET-M33 Forms for Human Cells
2.6. Acute Toxicity in Mice
3. Discussion
4. Materials and Methods
4.1. M33 Peptide Synthesis
4.2. NMR
4.3. TFA/Acetate Ion-exchange
4.4. TFA/Chloride Ion-exchange
4.5. Antimicrobial Susceptibility of Bacterial Isolates – MIC Assay
4.6. Cytotoxicity
4.7. Acute Toxicity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pini, A.; Giuliani, A.; Falciani, C.; Runci, Y.; Ricci, C.; Lelli, B.; Malossi, M.; Neri, P.; Rossolini, G.M.; Bracci, L. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob. Agents. Ch. 2005, 49, 2665–2672. [Google Scholar] [CrossRef]
- Bracci, L.; Falciani, C.; Lelli, B.; Lozzi, L.; Runci, Y.; Pini, A.; De Montis, M.G.; Tagliamonte, A.; Neri, P. Synthetic peptides in the form of dendrimers become resistant to protease activity. J. Biol. Chem. 2003, 278, 46590–46595. [Google Scholar] [CrossRef]
- Falciani, C.; Lozzi, L.; Pini, A.; Corti, F.; Fabbrini, M.; Bernini, A.; Lelli, B.; Niccolai, N.; Bracci, L. Molecular basis of branched peptides resistance to enzyme proteolysis. Chem. Biol. Drug. Des. 2007, 69, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Pini, A.; Falciani, C.; Bracci, L. Branched peptides as therapeutics. Curr. Protein. Pept. Sci. 2008, 9, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, J.; Falciani, C.; Bracci, L.; Pini, A. Branched peptides as bioactive molecules for drug design. J. Pept. Sci. 2018, 110, e24089. [Google Scholar] [CrossRef]
- Pini, A.; Falciani, C.; Mantengoli, E.; Bindi, S.; Brunetti, J.; Iozzi, S.; Rossolini, G.M.; Bracci, L. A novel tetrabranched antimicrobial peptide that neutralizes bacterial lipopolysaccharide and prevents septic shock in vivo. FASEB. J. 2010, 24, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, J.; Falciani, C.; Roscia, G.; Pollini, S.; Bindi, S.; Scali, S.; Arrieta, U.C.; Gómez-Vallejo, V.; Quercini, L.; Ibba, E.; et al. In vitro and in vivo efficacy, toxicity, bio-distribution and resistance selection of a novel antibacterial drug candidate. Sci. Rep. 2016, 6, 26077. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, J.; Roscia, G.; Lampronti, I.; Gambari, R.; Quercini, L.; Falciani, C.; Bracci, L.; Pini, A. Immunomodulatory and Anti-inflammatory Activity in Vitro and in Vivo of a Novel Antimicrobial Candidate. J. Biol. Chem. 2016, 291, 25742–25748. [Google Scholar] [CrossRef]
- Pini, A.; Lozzi, L.; Bernini, A.; Brunetti, J.; Falciani, C.; Scali, S.; Bindi, S.; Di Maggio, T.; Rossolini, G.M.; Niccolai, N.; et al. Efficacy and toxicity of the antimicrobial peptide M33 produced with different counter-ions. Amino. Acids. 2012, 43, 467–473. [Google Scholar] [CrossRef]
- Roux, S.; Zékri, E.; Rousseau, B.; Paternostre, M.; Cintrat, J.C.; Fay, N. Elimination and exchange of trifluoroacetate counter-ion from cationic peptides: A critical evaluation of different approaches. J. Pept. Sci. 2008, 14, 354–359. [Google Scholar] [CrossRef]
- Andrushchenko, V.V.; Vogel, H.J.; Prenner, E.J. Optimization of the hydrochloric acid concentration used for trifluoroacetate removal from synthetic peptides. J. Pept. Sci. 2007, 13, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.A.; Loudet, C.; Gröbner, G.; Dufourc, E.J. Pro-apoptotic bax-alpha 1 synthesis and evidence for beta-sheet to alpha helix conformational change as triggered by negatively charged lipid membranes. J. Pept. Sci. 2007, 13, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Bussat, M.C.; Klinquer- Hamour, C.; Goetsch, L.; Aubry, J.P.; Champion, T.; Julien, E.; Haeuw, J.F.; Bonnefoy, J.Y.; Corvaia, N. Stability and CTL activity of N-terminal glutamic acid containing peptides. J. Pept. Res. 2001, 57, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR of Proteins and Nucleic Acids, 1st ed.; Wiley: Hoboken, NJ, USA, 1986; pp. 1–292. [Google Scholar]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochem 1992, 31, 1647–1651. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994, 4, 171–180. [Google Scholar] [CrossRef]
- Van der Weide, H.; Brunetti, J.; Pini, A.; Bracci, L.; Ambrosini, C.; Lupetti, P.; Paccagnini, E.; Gentile, M.; Bernini, A.; Niccolai, N.; et al. Investigations into the killing activity of an antimicrobial peptide active against extensively antibiotic-resistant K. pneumoniae and P. aeruginosa. BBA-Biomembranes 2017, 1859, 1796–1804. [Google Scholar] [CrossRef]
- Sikora, K.; Neubauer, D.; Jaśkiewicz, M.; Kamysz, W. Citropin 1.1 Trifluoroacetate to Chloride Counter-Ion Exchange in HCl-Saturated Organic Solutions: An Alternative Approach. Int. J. Pept. Res. Ther. 2018, 24, 265–270. [Google Scholar] [CrossRef]
- Cornish, J.; Callon, K.E.; Lin, C.Q.X.; Xiao, C.L.; Mulvey, T.B.; Cooper, G.J.S.; Reid, I.R. Trifluoroacetate, a contaminant in purified proteins, inhibits proliferation of osteoblasts and chondrocytes. Am. J. Phisyol. 1999, 277, E779–E783. [Google Scholar] [CrossRef]
- Zhang, O.; Kay, L.E.; Olivier, J.P.; Forman-Kay, J.D. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR. 1994, 4, 845–858. [Google Scholar] [CrossRef]
- Little, M.J.; Aubry, N.; Beaudoin, M.E.; Goudreau, N.; La Plante, S.R. Quantifying trifluoroacetic acid as counterion in drug discovery by 19F NMR and capillary electrophoresis. J. Pharm. Biomed. Anal. 2007, 43, 1324–1330. [Google Scholar] [CrossRef]
- Preiss, A.; Kruppa, J.; Buschmann, J.; Mügge, C. The determination of trifluoroacetic acid in rat milk samples by 19F-NMR spectroscopy and capillary gas chromatography. J. Pharm. Biomed. Anal. 1998, 16, 1381–1385. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compound SET-M33 are available from the authors. |
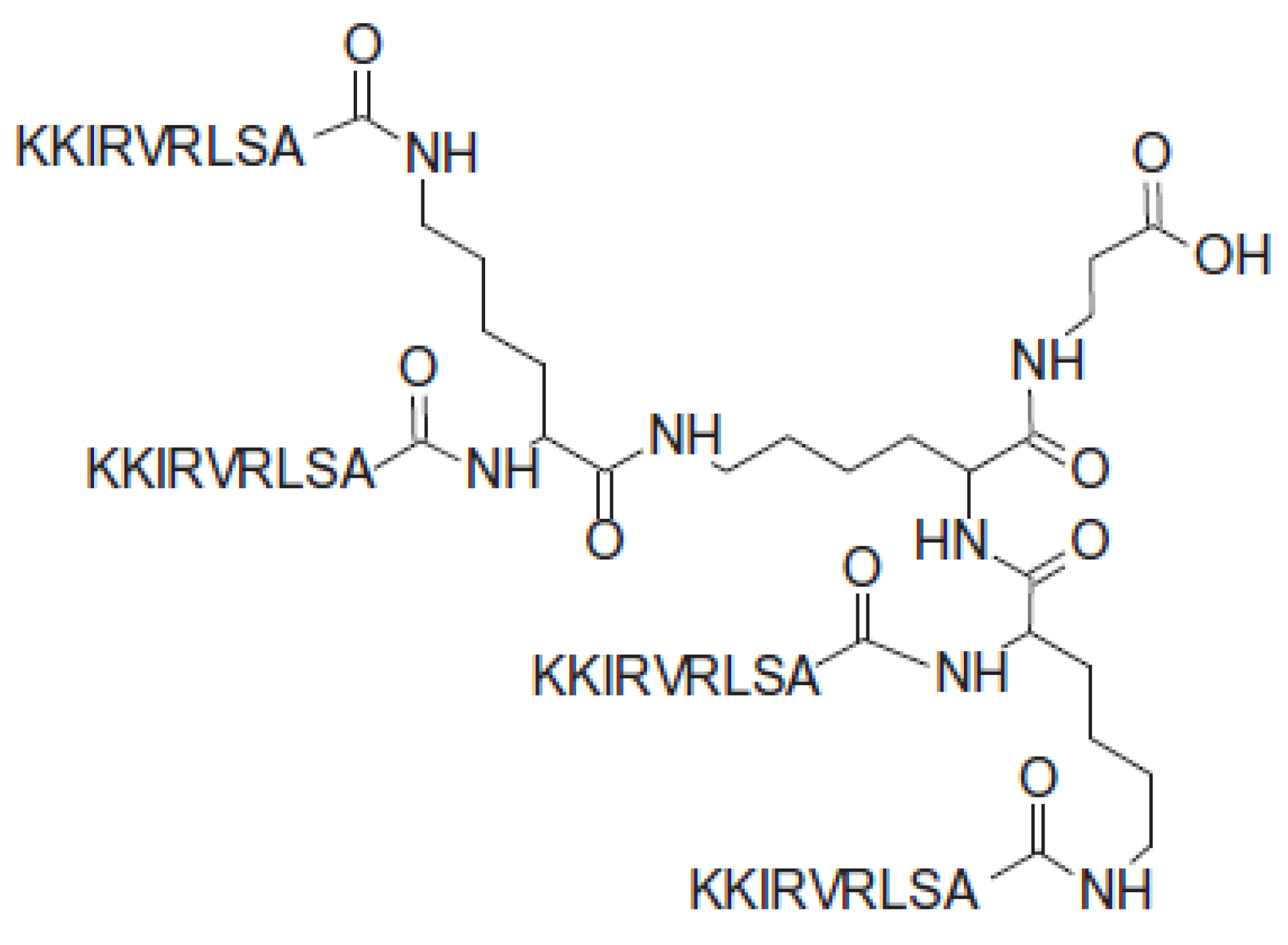
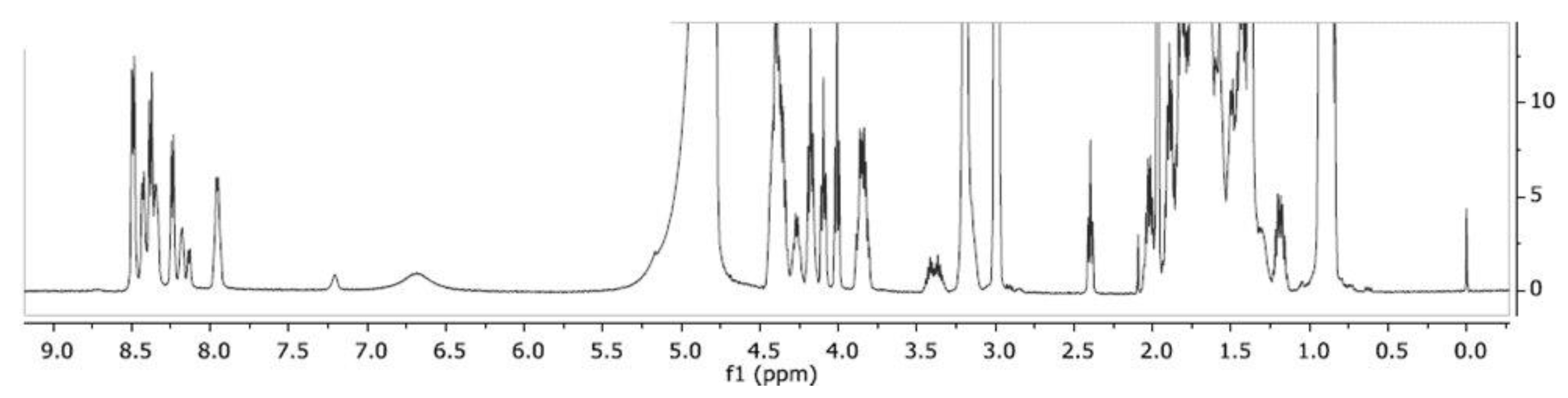
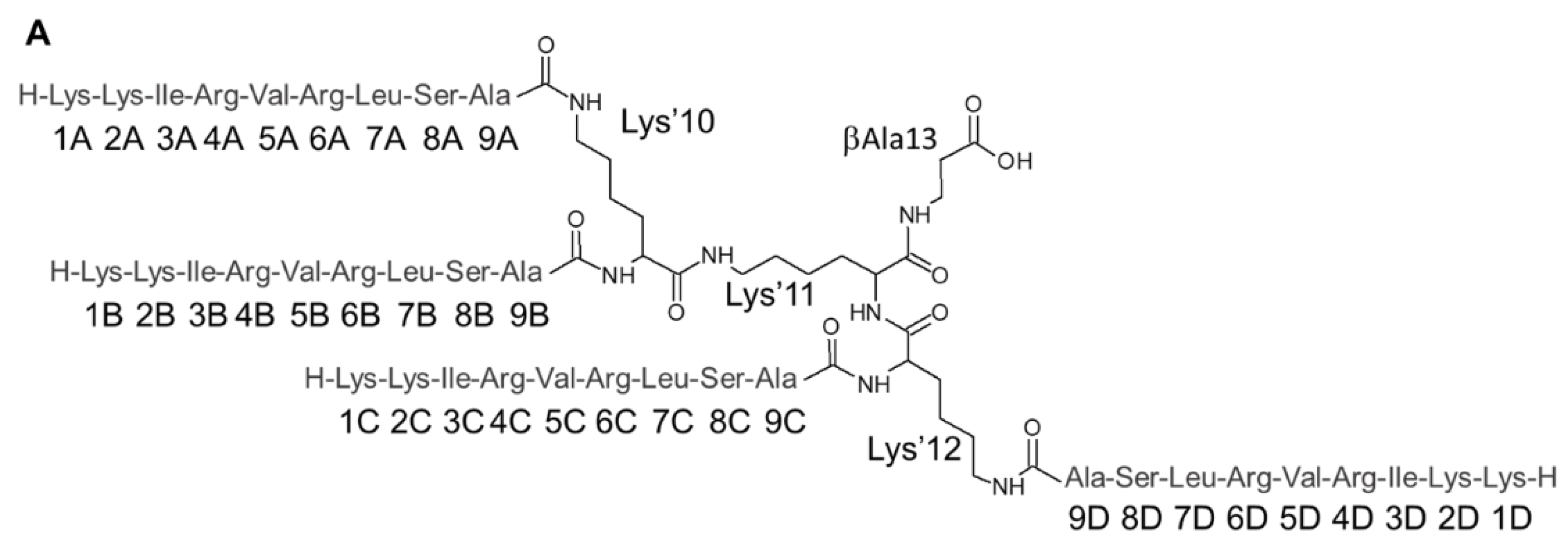
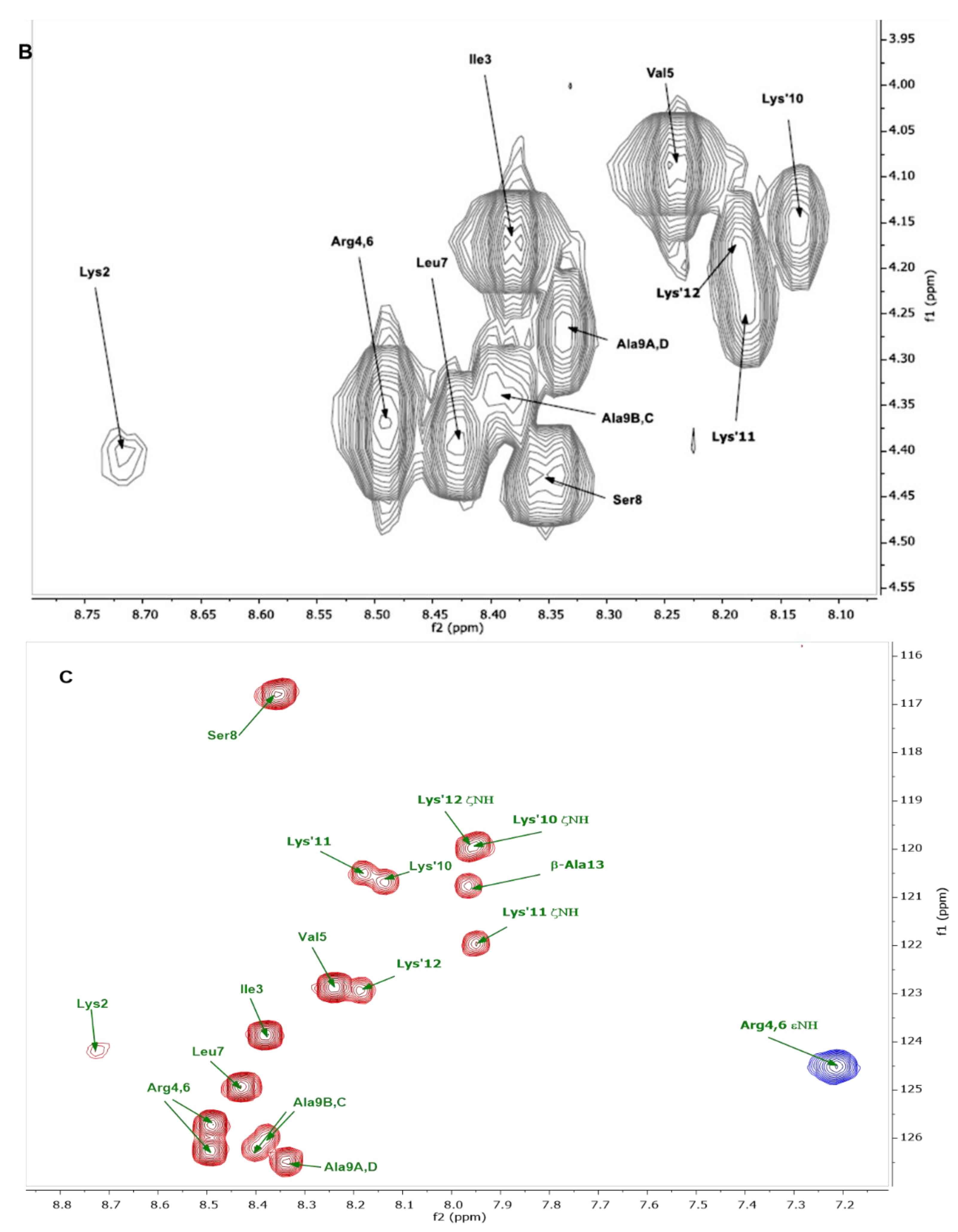

| Residue | 1HN | 15N | 1Hα | 13Cα | 1Hβ | 13Cβ | Others |
|---|---|---|---|---|---|---|---|
| Lys1A-D | NO | NO | 4.01 | 55.5 | 1.89 | 33.2 | γ: 1.43/23.8; δ: 1.69/29.1; ε: 2.99/41.9 |
| Lys2A-D | 8.72 | 124.2 | 4.40 | 56.1 | 1.76 | 33.1 | γ: 1.40/24.7; δ: 1.40/24.7; ε: 2.99/41.9 |
| Ile3A-D | 8.38 | 123.9 | 4.18 | 60.7 | 1.83 | 38.9 | γ1: 1.19, 1.47/27.1; γ2: 0.89/17.3; δ1: 0.86/12.6 |
| Arg4A-D | 8.49 | 125.71 | 4.37 | 55.8 | 1.75, 1.82 | 30.8 | γ:1.58, 1.64/27.0; δ: 3.20/43.3; ε: 7.21/84.5 (15N) |
| Val5A-D | 8.24 | 122.9 | 4.10 | 62.0 | 2.03 | 33.0 | γ1: 0.92/20.8; γ2:0.94/20.5 |
| Arg6A-D | 8.49 | 126.31 | 4.37 | 55.8 | 1.75, 1.82 | 30.8 | γ:1.58, 1.64/27.0; δ: 3.20/43.3; ε: 7.21/84.5 (15N) |
| Leu7A-D | 8.43 | 124.9 | 4.40 | 55.1 | 1.59, 1.64 | 42.4 | γ: 1.64/26.8; δ1: 0.93/24.8; δ2: 0.87/23.3 |
| Ser8A-D | 8.35 | 116.8 | 4.44 | 57.9 | 3.85 | 63.8 | |
| Ala9A,D | 8.34 | 126.5 | 4.27 | 52.6 | 1.38 | 19.4 | |
| Ala9B,C | 8.40, 8.38 | 126.2, 126.0 | 4.34 | 52.4 | 1.38 | 19.4 | |
| Lys’10 | 8.13 | 120.7 | 4.15 | 56.7 | 1.76 | 33.1 | γ: 1.32/25.0; δ: 1.50/30.5; ε: 3.18/41.8; ζ: 7.95/119.9 (15N) |
| Lys’11 | 8.18 | 120.5 | 4.25 | 56.4 | 1.76 | 33.1 | γ: 1.32/25.0; δ: 1.50/30.5; ε: 3.18/41.8; ζ: 7.95/122.0 (15N) |
| Lys’12 | 8.19 | 122.9 | 4.17 | 56.7 | 1.76 | 33.1 | γ: 1.32, 1.40/25.0; δ: 1.50/30.5; ε: 3.18/41.8; ζ: 7.95/119.9 (15N) |
| β-Ala13 | 7.96 | 120.8 | 3.38 | 39.3 | 2.40 | 39.0 |
| Bacterial Species | MIC (μM) SET-M33 | |
|---|---|---|
| Acetate | Chloride | |
| E. coli TG-1 | 1.5 | 1.5 |
| P. aeruginosa PAO-1 | 3 | 1.5 |
| SET-M33 | EC50 [M] | ||
|---|---|---|---|
| Counter ion | T24 | 16HBE14o- | RAW264.7 |
| TFA | 1.363 × 10−5 | 1.105 × 10−5 | 3.846 × 10−6 |
| Acetate | 1.243 × 10−5 | 9.618 × 10−6 | 5.104 × 10−6 |
| Chloride | 2.260 × 10−5 | 1.034 × 10−5 | 6.125 × 10−6 |
| SET-M33 | 20 mg/kg | 25 mg/kg | 30 mg/kg |
|---|---|---|---|
| Counter ion | |||
| TFA | (-) | mild to severe | (-) |
| Acetate | not-observable [6] | not-observable | not-observable |
| Chloride | not-observable | (-) | not-observable |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castiglia, F.; Zevolini, F.; Riolo, G.; Brunetti, J.; De Lazzari, A.; Moretto, A.; Manetto, G.; Fragai, M.; Algotsson, J.; Evenäs, J.; et al. NMR Study of the Secondary Structure and Biopharmaceutical Formulation of an Active Branched Antimicrobial Peptide. Molecules 2019, 24, 4290. https://doi.org/10.3390/molecules24234290
Castiglia F, Zevolini F, Riolo G, Brunetti J, De Lazzari A, Moretto A, Manetto G, Fragai M, Algotsson J, Evenäs J, et al. NMR Study of the Secondary Structure and Biopharmaceutical Formulation of an Active Branched Antimicrobial Peptide. Molecules. 2019; 24(23):4290. https://doi.org/10.3390/molecules24234290
Chicago/Turabian StyleCastiglia, Francesca, Fabrizia Zevolini, Giulia Riolo, Jlenia Brunetti, Alessandra De Lazzari, Alberto Moretto, Giulia Manetto, Marco Fragai, Jenny Algotsson, Johan Evenäs, and et al. 2019. "NMR Study of the Secondary Structure and Biopharmaceutical Formulation of an Active Branched Antimicrobial Peptide" Molecules 24, no. 23: 4290. https://doi.org/10.3390/molecules24234290
APA StyleCastiglia, F., Zevolini, F., Riolo, G., Brunetti, J., De Lazzari, A., Moretto, A., Manetto, G., Fragai, M., Algotsson, J., Evenäs, J., Bracci, L., Pini, A., & Falciani, C. (2019). NMR Study of the Secondary Structure and Biopharmaceutical Formulation of an Active Branched Antimicrobial Peptide. Molecules, 24(23), 4290. https://doi.org/10.3390/molecules24234290









