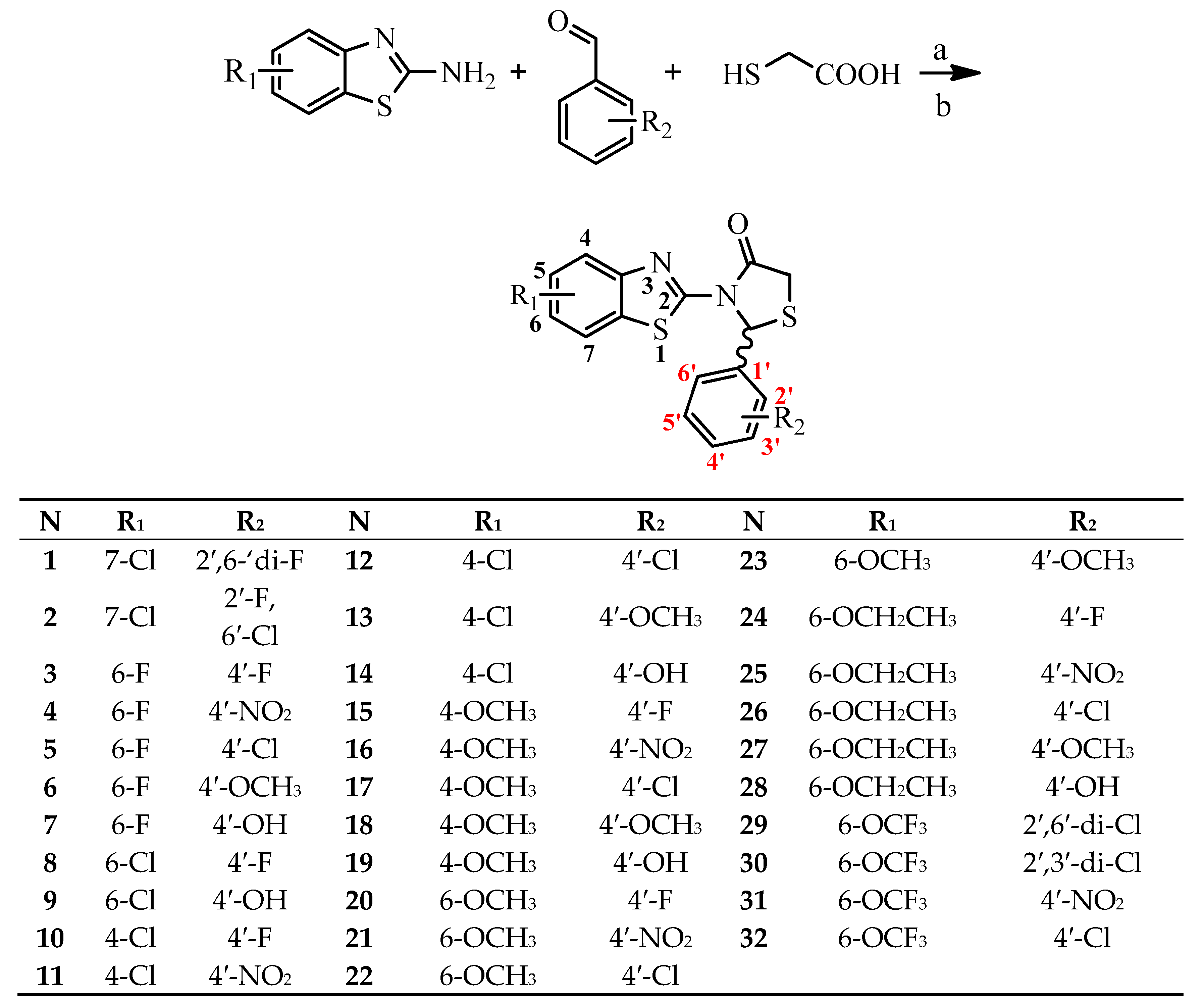
Abstract
Background: HIV is the causative agent of Acquired Immunodeficiency Syndrome (AIDS), an infectious disease with increasing incidence worldwide. Non-nucleoside reverse transcriptase inhibitors (NNRTIs) play an important role in the treatment of AIDS. Although, many compounds are already being used as anti-HIV drugs, research for the development of new inhibitors continues as the virus develops resistant strains. Methods: The best features of available NNRTIs were taken into account for the design of novel inhibitors. PASS (Prediction of activity spectra for substances) prediction program and molecular docking studies for the selection of designed compounds were used for the synthesis. Compounds were synthesized using conventional and microwave irradiation methods and HIV RT inhibitory action was evaluated by colorimetric photometric immunoassay. Results: The evaluation of HIV-1 RT inhibitory activity revealed that seven compounds have significantly lower ΙC50 values than nevirapine (0.3 μΜ). It was observed that the activity of compounds depends not only on the nature of substituent and it position in benzothiazole ring but also on the nature and position of substituents in benzene ring. Conclusion: Twenty four of the tested compounds exhibited inhibitory action lower than 4 μΜ. Seven of them showed better activity than nevirapine, while three of the compounds exhibited IC50 values lower than 5 nM. Two compounds 9 and 10 exhibited very good inhibitory activity with IC50 1 nM.
1. Introduction
Acute Immunodeficiency Syndrome (AIDS) is a result of the infection and destruction of T-lymphocytes by the human immunodeficiency virus (HIV). Over 60 million people are infected with the virus placing AIDS as the fourth cause of death worldwide [1,2,3].
Non-nucleoside reverse transcriptase inhibitors, as a part of Highly Active Antiretroviral Therapy (HAART), remain in the first line of fighting against AIDS. However, research for the development of novel NNRT inhibitors still continues as the virus develops resistant strains, limiting their use.
Among the approved by FDA compounds are several heterocyclic compounds such as benzoxazin-2-one (etravirine), dipirydo [1,4] diazepine-6-one (etravirin) [4], piperazine and indolyl (delaverdine) moieties [5,6]. Except of these drugs many other molecules have been found to display RT inhibitory action [7,8,9,10,11,12,13], among which are: benzothiazine dioxides [14], N1,N3-disubstituted uracils [15], 6-arylmethyl-substituted S-DABOs [16], adamantyl-substituted thiazolidine-4-ones [17,18] and many others [19,20,21,22,23,24,25,26,27]. The 2,3-diaryl-thiazolidin-4-one scaffold appeared as a selective NNRTI [28,29]. Modeling studies on the HIV-1 RT inhibitory activity of 2,3-diaryl-thiazolidin-4-ones showed the importance of overall hydrophobicity of the analogues and the presence of a heteroaryl system over the 3-aryl moiety for better HIV-1 RT inhibitory activity [30,31,32,33,34].
Computational methods are an important part of the drug design process and they are presently used to get a deeper understanding into the drug-enzyme interactions.
In this study, taking into account the best features of available NNRTIs a large number of novel 2,3-diaryl-thiazolidin-4ones were designed. The most promising compounds were selected with the aid of Molecular docking studies and prediction of spectra of biological activity for compounds by computer program PASS, synthesized and evaluated.
2. Results and Discussion
2.1. Rational Design of the Compounds
It is known that HIV-1 reverse transcriptase (HIV-1 RT) has a flexible structure that contains two binding domains. The active site characterized by the catalytic triad of Asp110, Asp185 and Asp186 residues, where the deoxynucleoside triphosphates are bound by the physiological process, and an allosteric site of NNRTIs about 10 Å away from the active site.
The NNRTIs binding pocket is hydrophobic, as most of its amino acids are hydrophobic (Pro59, Leu100, Val106, Val179, Leu234 and Pro236) and five of them are aromatic (Tyr181, Tyr188, Phe227, Trp229 and Tyr232). Certain hydrophilic amino acids such as Lys101, Lys103, Ser105, Asp132 and Glu224 are also present [35]. For the binding of NNRTIs to the reverse transcriptase enzyme, their ability to adopt a configuration characterized as a “butterfly orientation” is needed [36]. Aromatic rings are usually present in both wings of “batterfly” conformation. A heeterocyclic ring or other hydrogen donor/acceptor groups stabilized the complex via hydrogen bond formation, usually with Lys101.
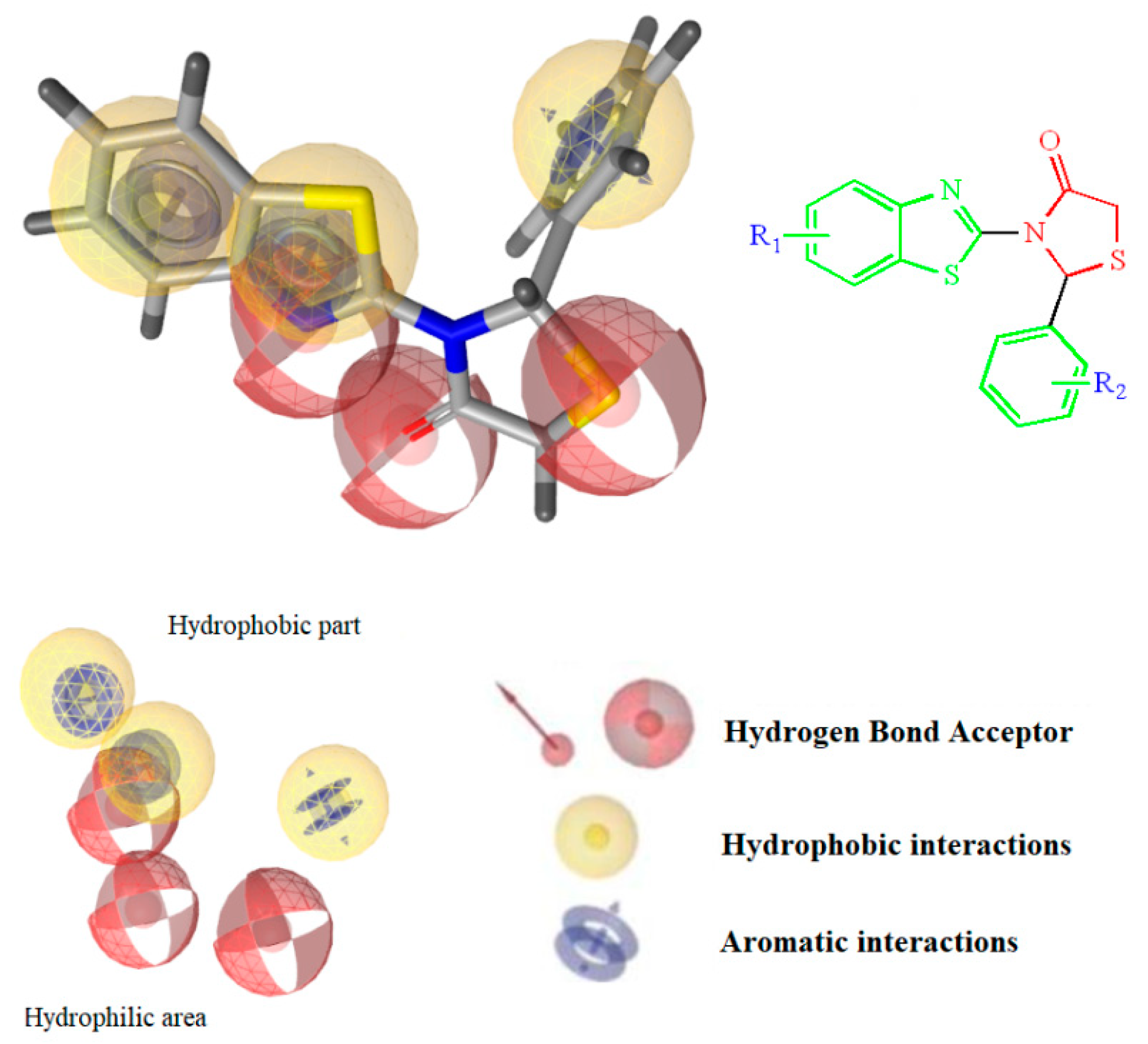
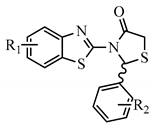
The design of the compounds was based on the presence of the above characteristics and consist of a benzothiazole ring connected via thiazolidinone moiety with a phenyl ring. The benzothiazole and the phenyl ring serve as the wings of the ‘’butterfly’’ confirmation, forming hydrothobic interactions, while the thiazolidinone moiety involves in the formation of hydrogen bond interactions especially with Lys101 (Figure 1). Different substituents at the benzothiazole and phenyl rings were used to determinate the compounds with the best predicted activity.
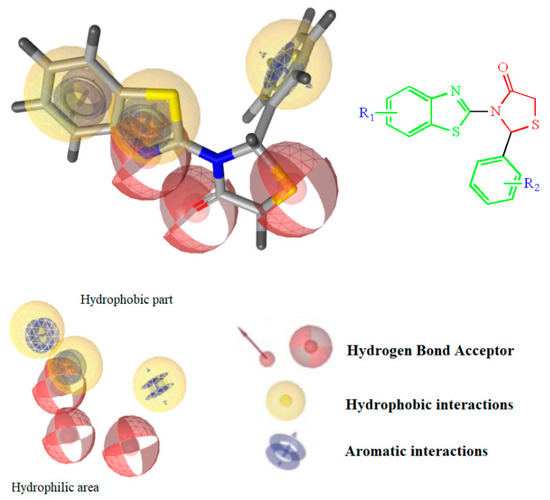
Figure 1.
Structure and Basic Characteristics of the Designed Compounds.
2.2. Computational Prediction of Anti-HIV Activity
PASS prediction of anti-HIV RT inhibitory activity was performed for several dozen designed molecules, from which thirty two were selected for synthesis and testing. Prediction was carried out using PASS online [37] version. For all selected compounds, anti-HIV inhibitory activity was predicted with Pa values in range of 0.297–0.704 (Table 1). The calculated Pa values for most of compounds were less than 0.5, indicating their relative novelty compared to the structures of the compounds from the PASS training set [38,39].

Table 1.
Molecular docking scores and PASS prediction of all the designed compounds.
2.3. Molecular Docking Prediction
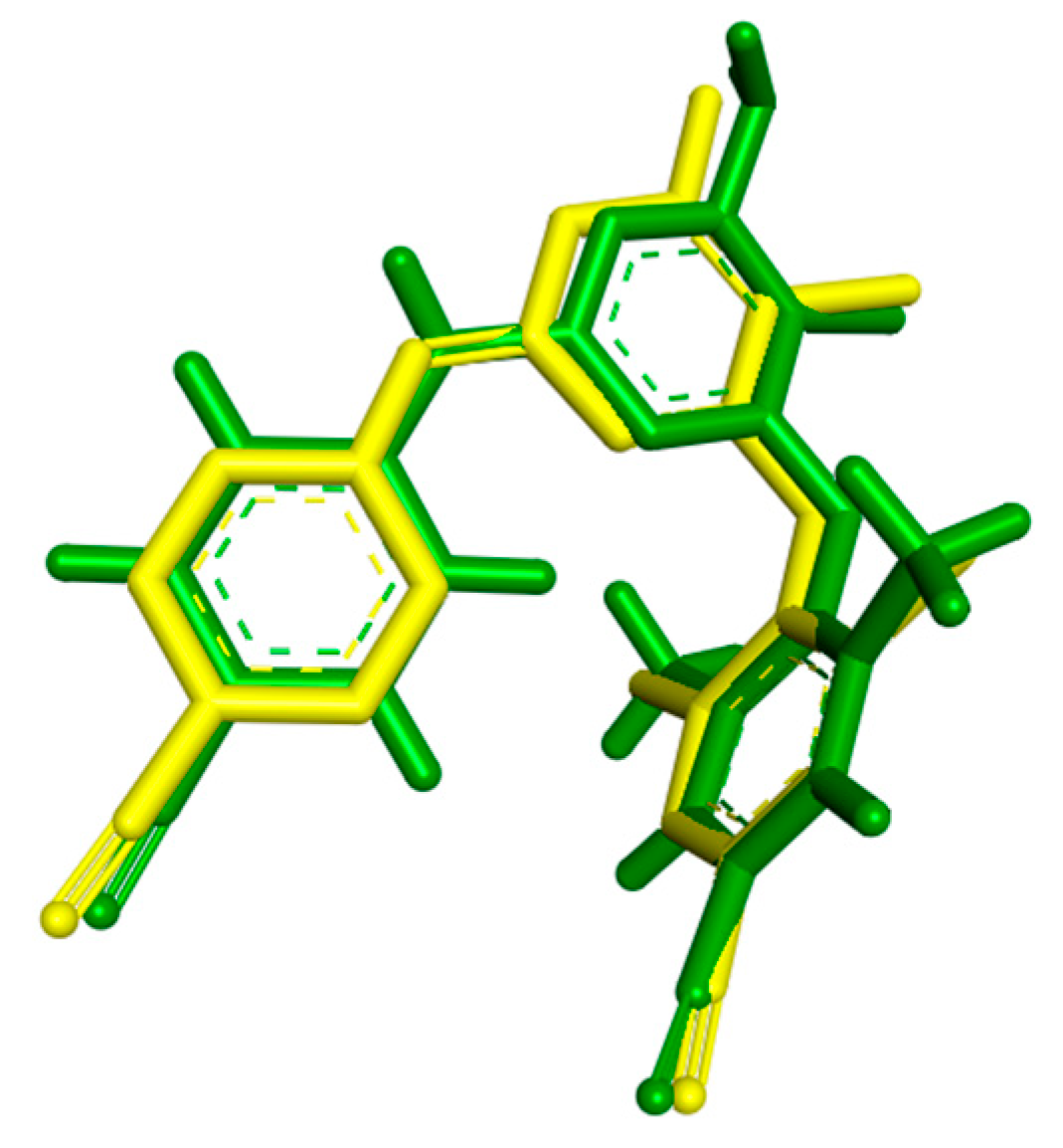
For the docking studies the enzyme of RT in complex with the inhibitor TMC 125, the most widely used in the literature, was selected (PDB code: 3MEC). At first stage, a validation test was performed to certify the program’s reliability. Firstly, the ligand TMC 125 (etravirine), in the conformation found in the crystal structure, was extracted and docked back into the corresponding binding pocket to determine the ability of Autodock to reproduce the orientation and position of the inhibitor observed in the crystal structure. According to docking results, the orientation of the docked TMC 125 inhibitor was very close to that found in the crystal structure (Figure 2). The low RMS deviation of 0.58 Å between the docked and the crystal ligand is an indication of the very good alignment of the experimental and calculated positions.
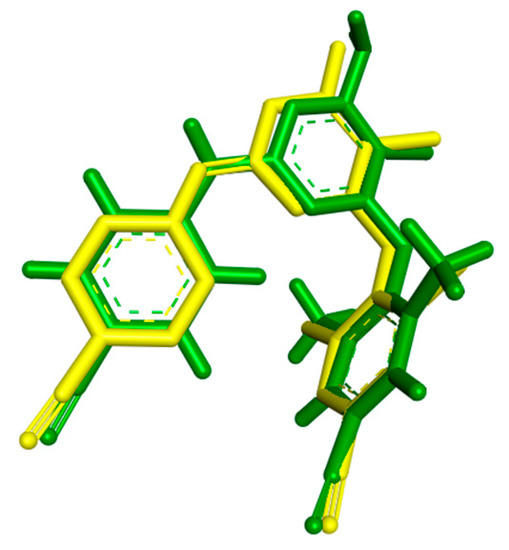
Figure 2.
Conformation of TMC 125-crystal Structure (green) as Compared to the Docked Conformation of TMC 125 (yellow).
All designed compounds and the reference drug nevirapine were docked to the HIV-1 reverse transcriptase enzyme using the same parameters (Table 1). Both the (S) and (R) isomer of each compound were docked separately. It was observed that the (R) isomer exhibited proportionaly decreased free binding energy for each compound [40,41,42].
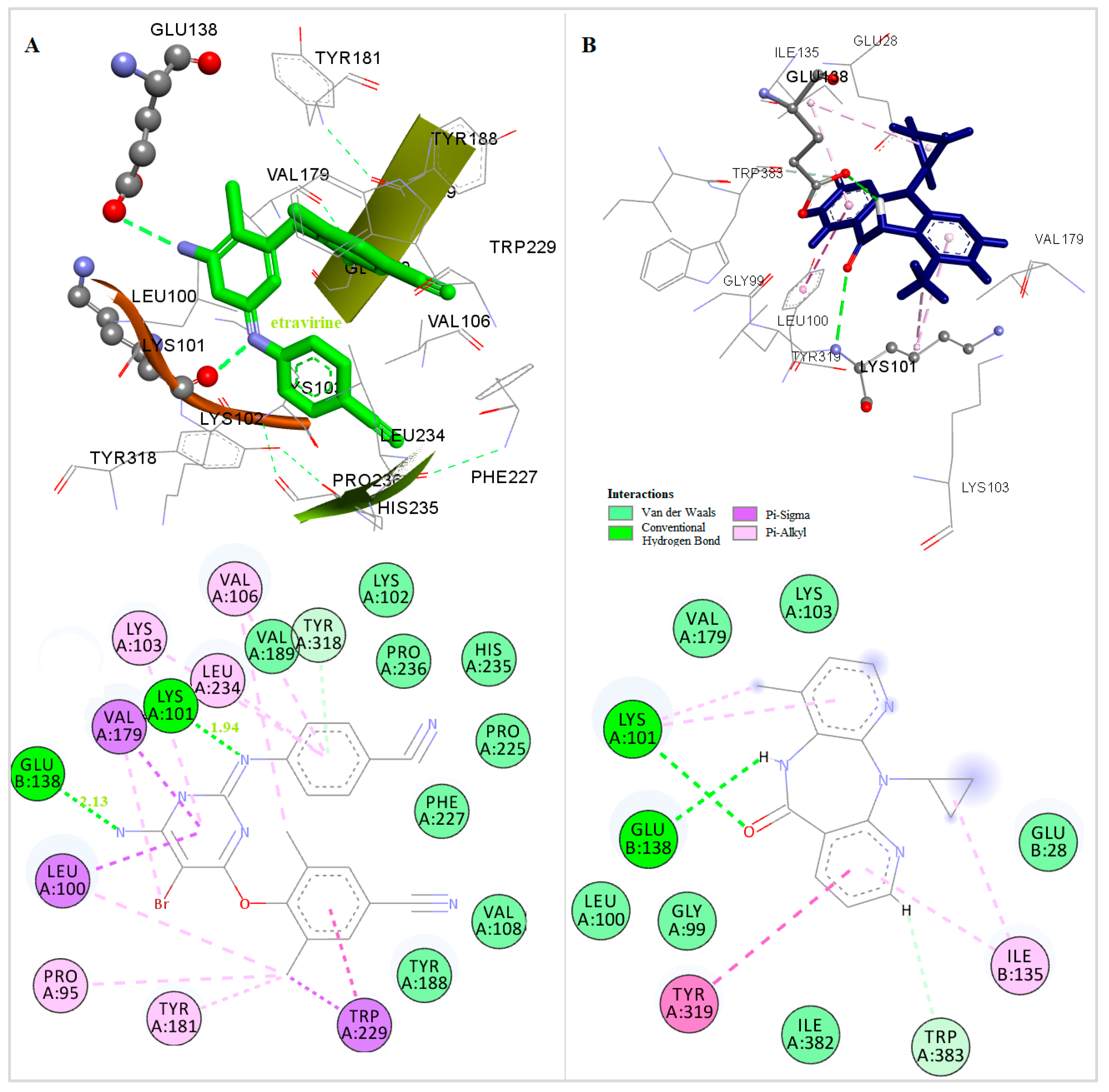
The three-dimensional docked structures of the studied compounds were compared with the X-ray crystallographic structure of TMC 125 (Figure 3). Detailed analysis of the binding mode found in the docked compounds-RT and other NNRTIs-RT crystal complexes showed that the most of the compounds could bind to RT in a butterfly-like conformation. Like TMC 125 (Figure 3), most of the compounds are involved in the formation of a hydrogen bond with Lys101, which was observed in the X-ray crystallographic structure of the TMC 125-RT complex [43].
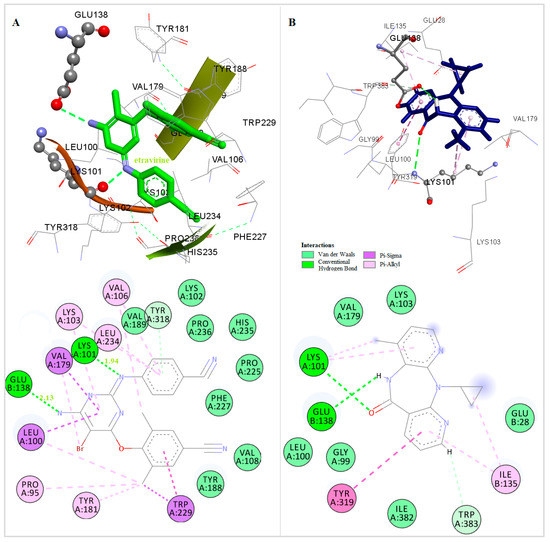
Figure 3.
Docked Conformation of TMC125(etravirine) (A) and Nevirapine (B) in the Allosteric Center of HIV-1 RT.
From the 132 compounds designed, the 32 showed very good binding scores and taking into account also the prediction results by PASS, were chosen for the synthesis (Table 1 and Table 2).

Table 2.
Molecular docking scores and PASS prediction of the selected compounds.
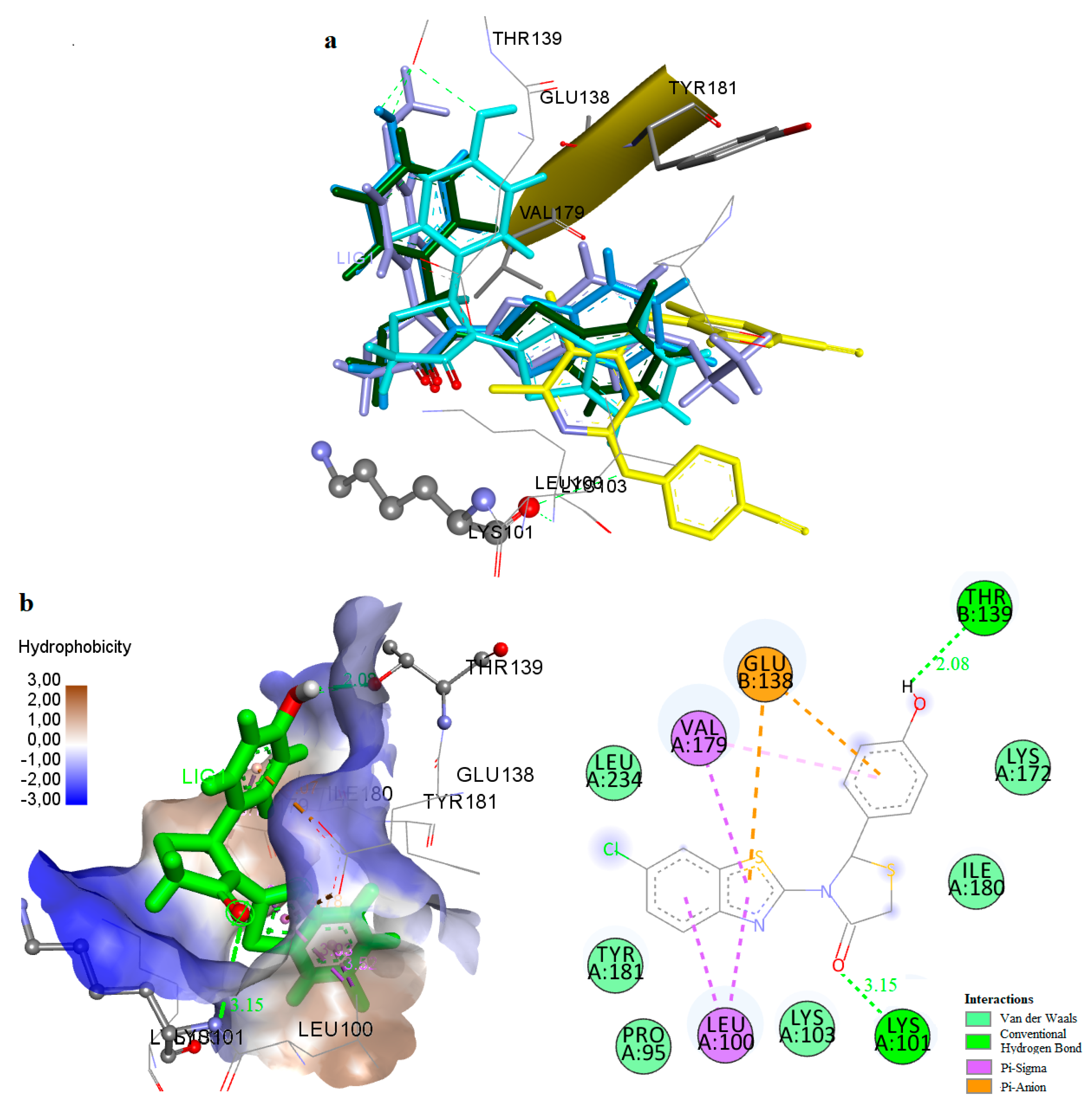
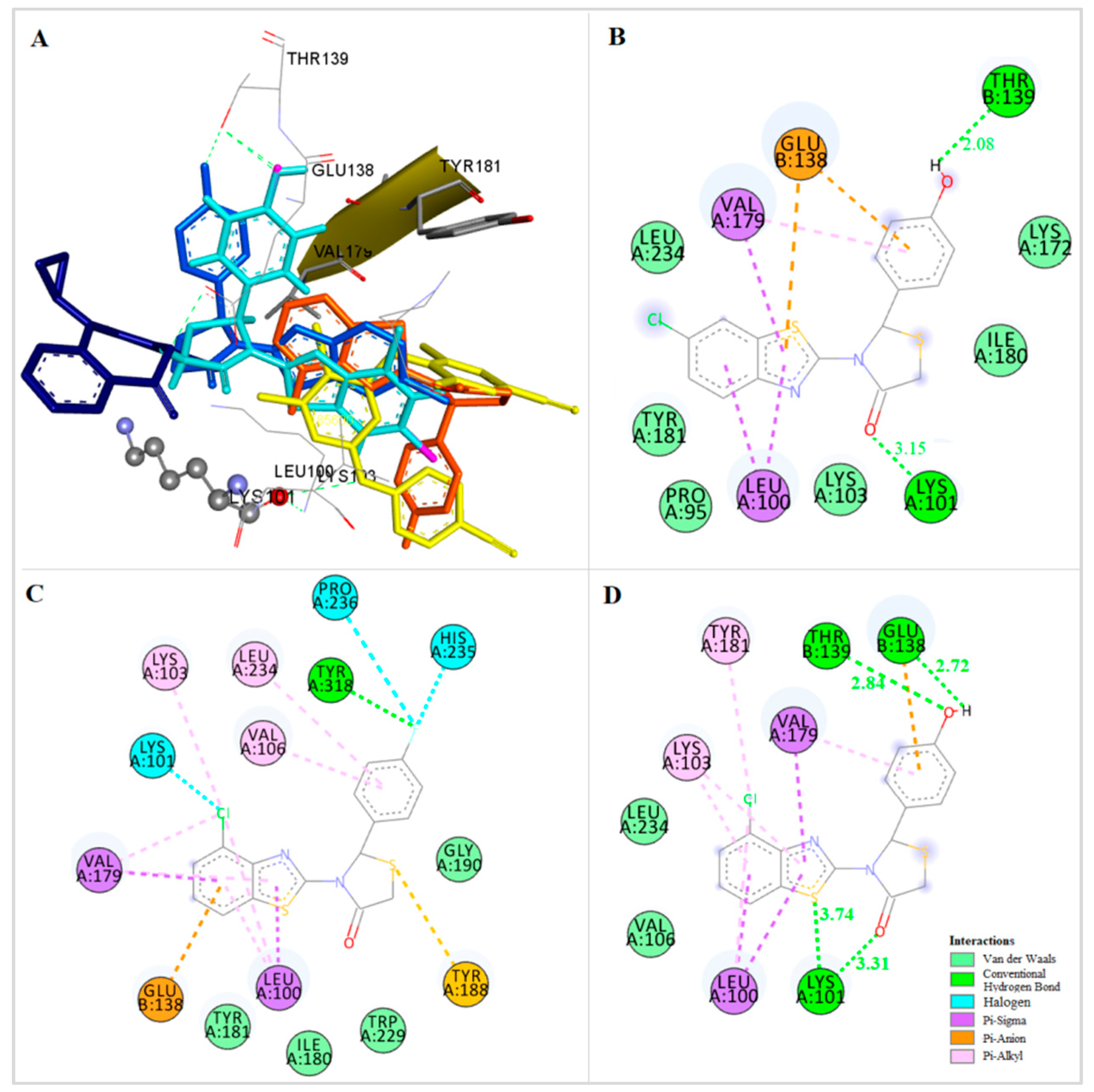
It was observed that these compounds bind to the allosteric center of the enzyme occupying a similar configuration to TMC 125 inhibitor (Figure 4a, best docked conformations of compounds 1, 9, 14, 27). Comparison of the X-ray structure of TMC 125 with compound 9 (−14.63 kcal/mol) (Figure 4b), showed that this compound occupies similar configuration to TMC 125 inhibitor and forms a hydrogen bond with Lys101. Specifically, the CH2-S moiety of 4-thiazolidinone is oriented towards a channel formed between the residues Lys101, Val179 (p66 subunit) and Glu138 (p51 subunit). The nitrogen atom of Lys101 interacts with the oxygen atom of the 4-thiazolidinone forming a hydrogen bond. The substituted benzothiazole ring interacts hydrophobically with the residues Leu100, Val179, Glu138 and Leu234. These hydrophobic interactions contribute significantly to the stabilization of the compound-RT complex. As shown in Figure 4b, the 4-hydroxyphenyl ring is located inside the cavity surrounded by the residues Pro95, Thr139, Glu138, and Ile180. It is important to mention that the presence of the hydrogen bond with Lys101 is characteristic of many NNRTIs inhibitors. This phenomenon is also observed in the crystallographic TMC 125-RT and 9-Cl TIBO-RT complexes [43].
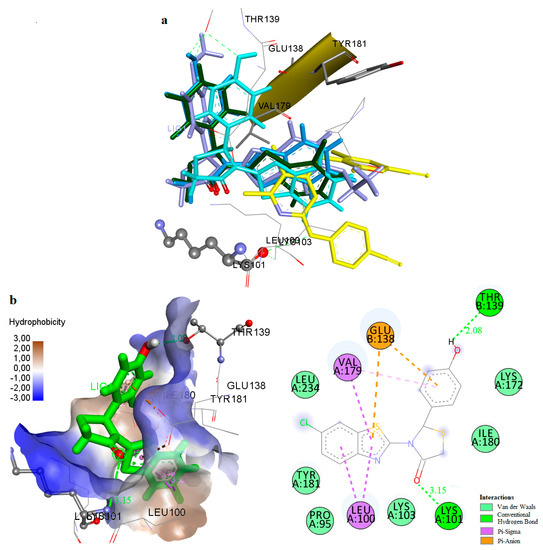
Figure 4.
(a) Docking of TMC125 (yellow) and Compounds 1 (green), 9 (blue), 14 (light blue) and 27 (purple) to the Allosteric Site of HIV-1 RT. (b) Docking of Compound 9 (green) in the Binding Site of HIV-1RT.
Compounds with higher docking scores exhibit a different binding mode from those with lower docking scores. In this type of binding, the substituted phenyl ring is oriented to a “channel” formed by Leu100, Glu138, Ile180 and Val179. This orientation results in the CH2-S moiety of 4-thiazolidinone being removed away from Lys101 and thus, being not able to interact and form a hydrogen bond with it. This fact contributes significantly to the ability of these compounds to form a stable complex with the enzyme and, potentially, to their intended action.
2.4. Prediction of Toxicity
According to Lasar model throughout OpenTox, all the compounds found to be at the category IV with LD50 between 500 mg/kg and 1000 mg/kg and they are saved for use (Table 3).

Table 3.
Predicted toxicity with program PROTOX.
2.5. Chemistry
Synthesis of the compounds was performed by one pot method according to Scheme 1 [44] and for some compounds by microwave assisted one-pot procedures (Scheme 1). According to conventional method, the mixture of the suitable aminobenzothiazole, substituted benzaldehyde and mercaptoacetic acid was refluxed in toluene for 18–32 h. All products were obtained as racemates in low yield. Instead using microwave assisted one-pot synthetic procedure the reaction time was reduced to 30 min. The final products were obtained in good yield. All synthesized compounds were characterized by TLC and spectroscopic methods (IR, 1H NMR, 13C-NMR and MS for some compounds).
Scheme 1.
General synthetic procedure. Reagents and conditions: (a) Conventional method: toluene, reflux for 18–32 h, (b) microwave-assisted technic: 100 °C, power 100 W, 30 min.
In IR spectra stretching absorption bands at 1700 cm−1 (strong) of C=O, 1600 and 1540 cm−1 of -C–C- and 3200 of –OH were detected. In 1H-NMR spectra signal at 8.10–6.89 ppm as well as at 4.35–4.10 ppm are attributed to aromatic protons and proton of the position 2 of thiazolidinone moiety respectively. The rest of the protons appeared at the expected chemical shifts. In 13C-NMR spectra peaks were observed for C=O group at δ 172–170 ppm, for C-2 of benzothiazole ring at δ 161–165 ppm and for C-2 and C-5 of thiazolidinone moiety at 53–60 ppm and at 30–34 ppm respectively (see experimental).
2.6. HIV-1 RT Inhibitory Action
The results of the evaluation of RT inhibitory activity are presented in Table 4. All compounds at the concentration of 4 μM exhibited inhibitory action varying from 21% to 83%.

Table 4.
HIV-1 RT inhibitory action.
Seven compounds were found to have significantly lower IC50 than nevirapine (0.3 μM), the best of which displayed IC50 value lower than 1μM. The order of the compound’s potency can be presented as follows: 10 ≥ 9 > 2 > 14 > 3 > 1 > 8 > nevirapine > 20 > 27 > 13 > 28 > 15 > 16 > 23 > 26 > 17 > 19 > 18 > 22 > 4 > 7 = 12 = 21. Nine compounds, the 5, 6, 11, 24, 25, 29, 30, 31, 32, exhibited IC50 values greater than 4 μM.
The study of structure-activity relationships revealed that the activity of compounds depends not only on the nature of substituent and its position in benzothiazole ring but also on the nature and position of substituents in benzene ring. The presence of 6-Cl substitution in the benzothiazole moiety in combination with the presence of hydroxyl group at position 4′ of the benzene ring (9) appeared to be beneficial for the HIV RT inhibitory action. The same effect was observed in case of 4-Cl substitution in the benzothiazole moiety combined with the presence of 4′-F group in benzene ring (10).
The replacement of 4′-F substitution in compound 10, one of the two most active compounds, by 4′-OH (14) remarkably decreased the activity although compound 14 still remained a very active compound being more active than nevirapine. On the other hand, compound 2 with the presence of 7-Cl substituent in benzothiazole moiety and 2′-F, 6′-Cl (2) substitution in benzene ring was the third most active compound. The replacement of 6′-Cl (compound 2) by 6′-F in benzene ring (1) significantly decreased activity compared to compound 2. While introduction of 6-F in benzothiazole moiety and 4′-F substituent in benzene ring (3) increased the activity compared to compound 1, the replacement of 6-F by 6-Cl led to compound 8 with decreased activity in comparison with compounds 1 and 3. However, despite that compounds 1, 3 and 8 were less active compared to compounds 9, 10, 2 and 14 they still appeared to be more active than the reference compound.
The replacement of 4′-F substituent by nitro-, chloro-, methoxy and hydroxyl group respectively resulted to compounds with decreased activity (4–7). The 6-F benzothiazole substituted derivatives 4–7 showed IC50 values ≥ 4 μM. On the contrary, the 6-Cl benzothiazole substituted derivatives 8 and 9 exhibited good inhibitory action with compound 9 being one of the most active compounds. It seems that in this group of compounds the presence of the hydrogen donor 4′-OH group is beneficial (9). In case of the 4-Cl benzothiazole substituted derivatives (10–14) it appeared that the presence of the hydrogen acceptor 4′-F group was the most beneficial (10) followed by the hydrogen donor 4′-OH (14) and 4′-OMe group (13). The rest of compounds exhibited IC50 values ≥ 4 μM. It is interesting to mention that 4-OCH3 as well as 6-OCH3 benzothiazole substituted derivatives showed activity with IC50 values between 1.5 and 3 μM, showing small differentiation in concern to phenyl substituents. An exception was observed only for the 4′-nitrophenyl derivatives with 6-methoxy group in the benzothiazole moiety (21, IC50 4 μM) being the less active compound of the group. In general, 4′-nitro substitution in benzene ring was detrimental for all groups of compounds with exception of the derivatives with methoxy group in benzothiazole moiety.
The derivatives 26–28 (4′-Cl, 4′-OMe, 4′-OH) with 6-ethoxy group in benzothiazole moiety displayed activity almost similar to that of 6-methoxy substitution (R1) with IC50 1.93–2.48 μM. Regarding the 4′-fluoro- and 4′-nitrophenyl derivatives (24, 25) IC50 values higher than 4 μM were observed.
In conclusion, it is interesting to notice that the presence of chlorine in position 4′ (R2) of benzene ring was negative for all derivatives with halogen substituted benzothiazole moiety independent of its position. The presence of a chloro- substituent at the benzothiazole ring (R1) at positions 4, 6 or 7 significantly increases the inhibitory effect (1, 2, 8, 9, 10, 14). On the other hand, the introduction of -OCF3 substituent into the benzothiazole system further reduces the activity as compared to the chloro-substituted derivatives, with most of the compounds of this group (30–32) displaying IC50 values higher than 4 μM.
2.7. Docking Analysis
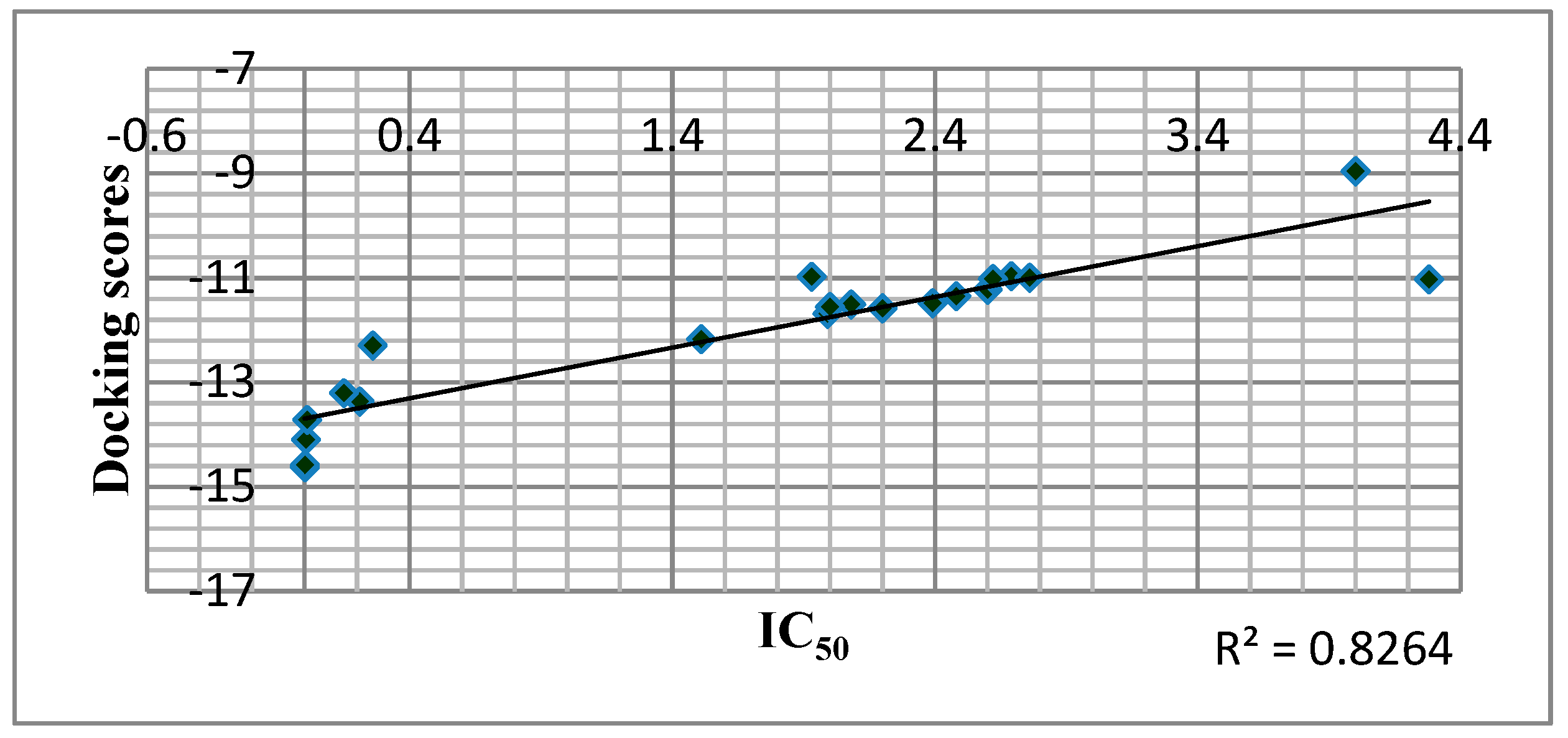
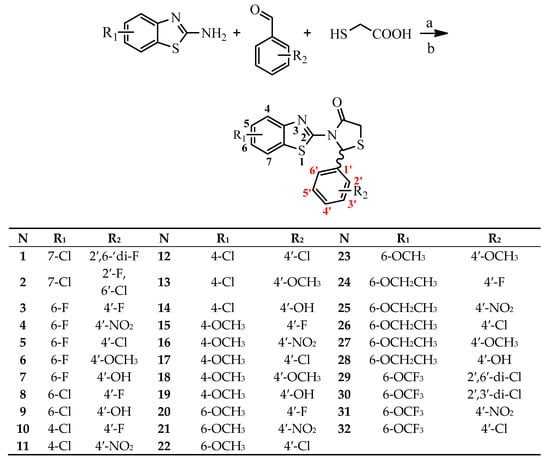
Docking analysis effectively predicted RT inhibitory action in most cases as shown in Figure 5.
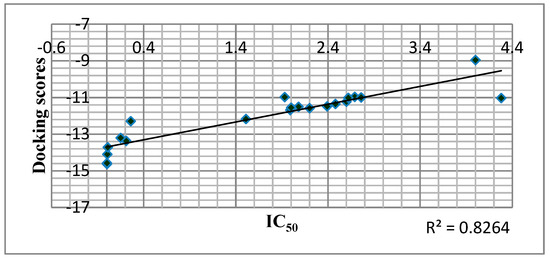
Figure 5.
Correlation of Predicted Free Binding Energy of the Compounds in the Allosteric Center of the Enzyme with IC50 of the Compounds.
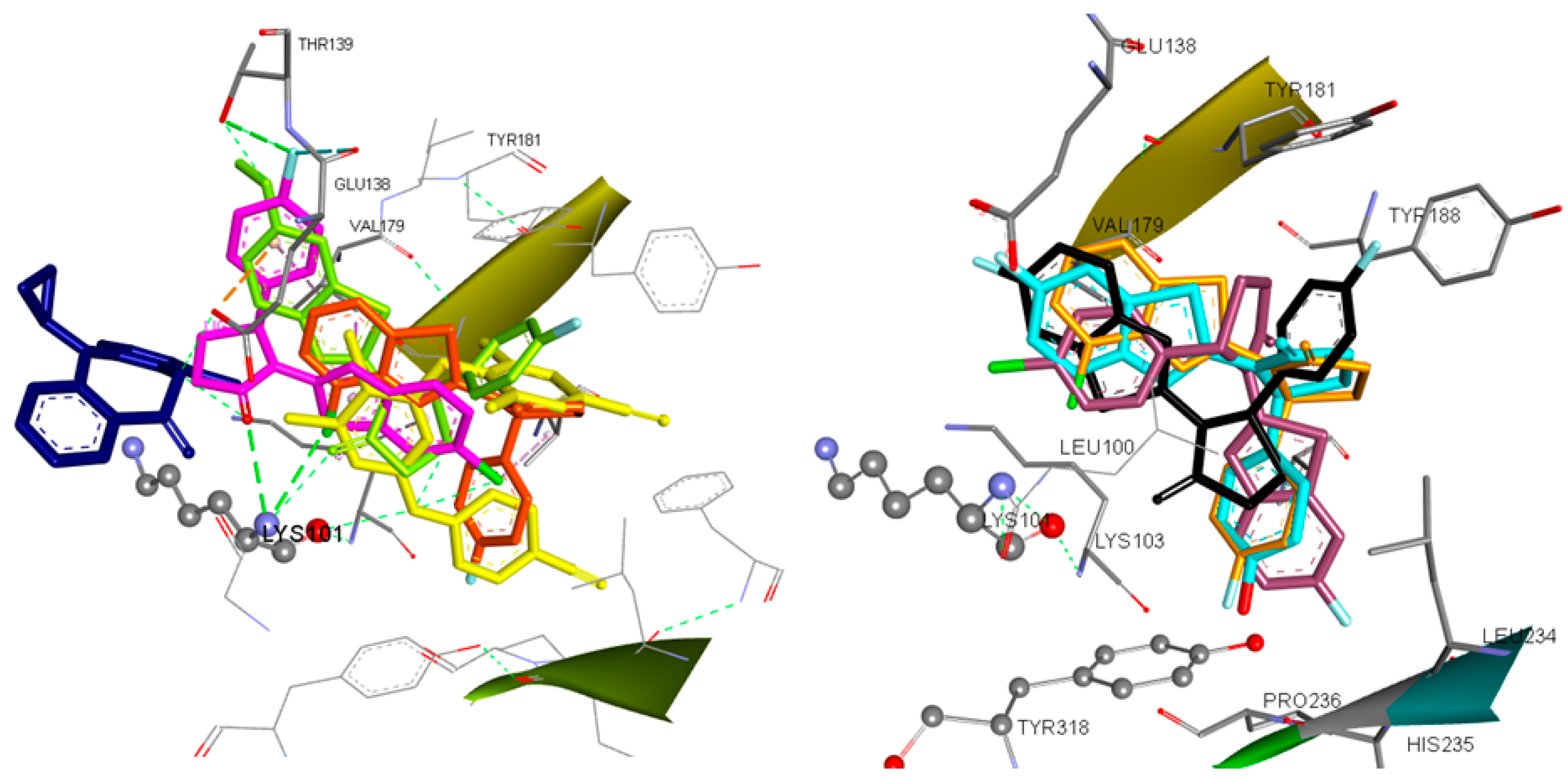
According to docking results, most of the compounds are placed within the NNRTIs binding pocket between the place equipped by the classic butterfly shaped compound, nevirapine, and the binding site of etravirine or within the etravirine binding pocket (Figure 6).
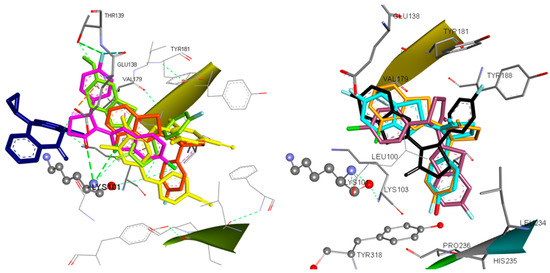
Figure 6.
(Left) Docking of Etravirine (yellow), Nevirapine (blue) and Compounds 8 (magenta), 20 (light green) and 10 (orange) in the Binding Site of HIV-1RT. (Right) Docked Conformations of Compounds 3 (black), 5 (dark purple), 7 (light blue) and 10 (orange) in the Binding Site of HIV-1RT.
Interestingly, the most active compounds structurally belong to three different groups and adopt three different orientations.
An orientation most close to the nevirapine binding place is adopted by compound 2, the 2′-F,6′-Cl derivative of the subgroup of compounds bearing a Cl substituent at the 7-position of the benzothiazolyl moiety (Figure 7A,B). More precisely, the benzothiazole and thiazolidinone moieties occupy positions similar with the “wings” of nevirapine. The benzothiazolyl moiety is placed, in vicinity to the amino acids Trp383 and Ile135 with which it forms pi interactions. The thiazolidinone ring is placed in the area of Lys103, while the phenyl ring is placed near Glu138, Tyr181 and Lys101, forming pi interactions with Glu138. Halogen and hydrogen bonds interactions are also formed between the F atom and the amino acids Glu138 and Lys101, respectively, stabilizing the complex.
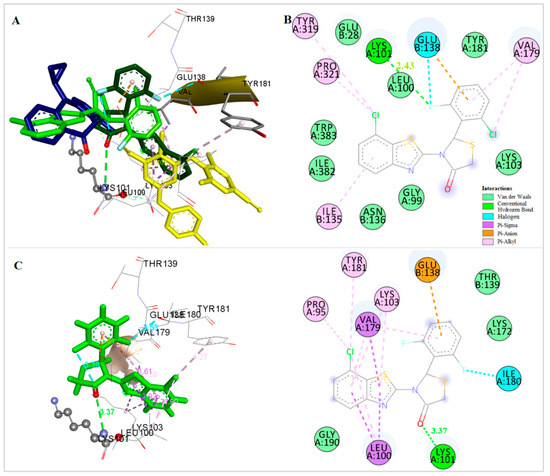
Figure 7.
(A) Docking of Compounds 1 (dark green), 2 (light green), Nevirapine (dark blue) and Etravirine (yellow) at the Allosteric Site of HIV-1 RT. (B) Docking of Compound 2 at the Allosteric Site of HIV-1 RT. (C) Docking of Compound 1 at the Allosteric Site of HIV-1 RT.
A different orientation is adapted by the 2′,6′-di-fluoro derivative of the same subgroup, compound 1 (Figure 7A,C). In this case, the thiazolidinone ring is placed in vicinity to Lys 101, which forms a hydrogen bond with the CO group of this moiety. The benzothiazolyl group is placed in the same area where the central aromatic ring of etravirine is bound, towards Tyr181, Leu100 and Val179 with which it participates in pi interactions. The two fluoro-atoms at the o-positions of the phenyl moiety participate in halogen bond interactions, an intramolecular one with CO of the thiazolidinone ring stabilizing the conformation of the molecule and a second one with Ile180.
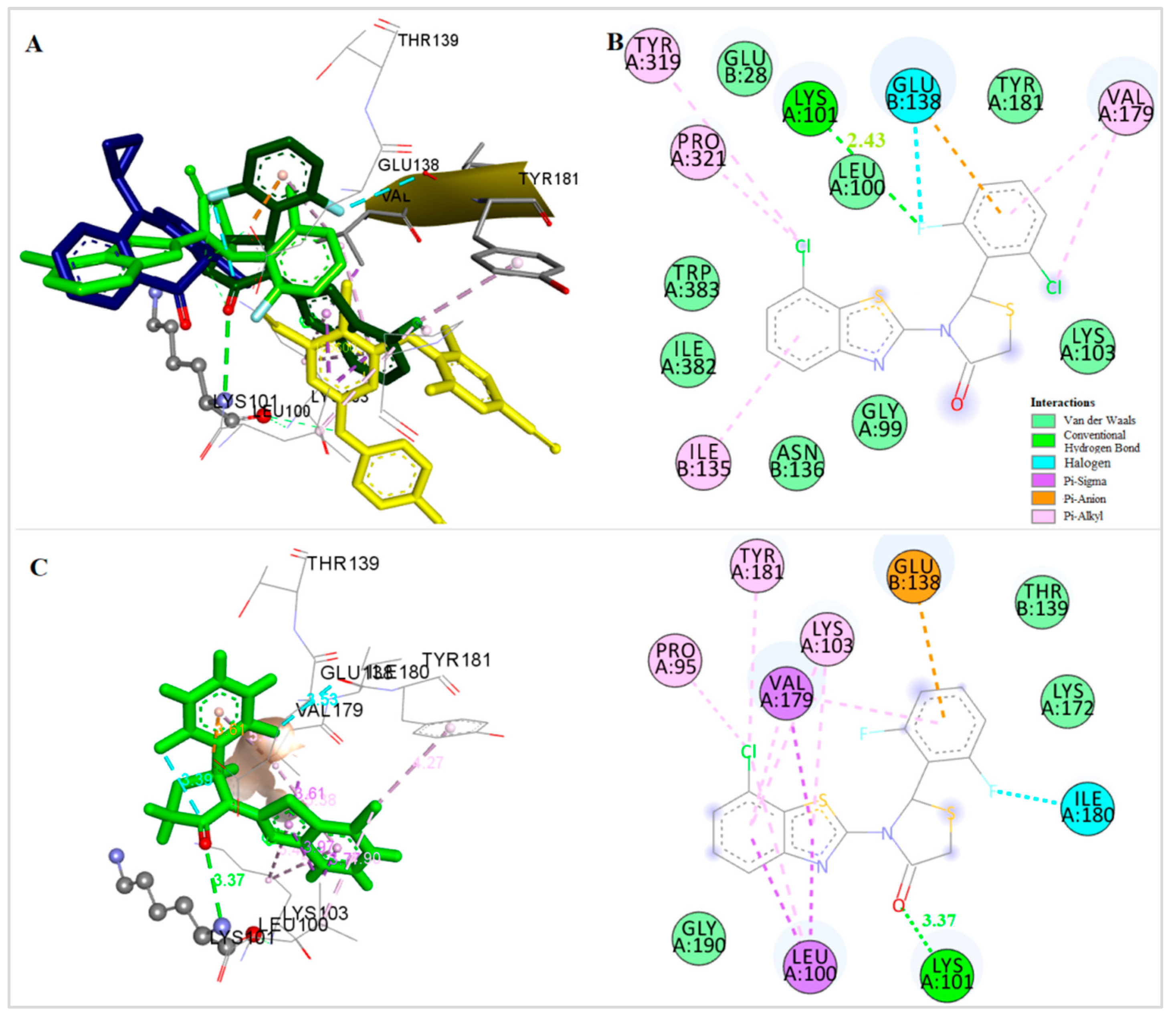
A similar orientation is adopted by several active compounds like compounds 8, 9, the two more active compounds of the subgroup bearing a Cl-atom at position 6 of the benzothiazolyl moiety (R1) (Figure 8A,B,D) and compound 14, the most active compound of the subgroup bearing a Cl-atom at the 4-position of the benzothiazolyl moiety (Figure 8A,D). Among these, compounds 9 and 14 belong in the four more potent out of the thirty-two tested compounds with IC50 values at the nanomolar range. In case of compounds 9 and 14, a hydrogen bond is formed between the CO group of the thiazolidinone ring and Lys101 while a second hydrogen bond is formed between hydrogen acceptor substituents of the 4-position of the phenyl ring and Thr139. An additional hydrogen bond between the H-atom of the 4′-OH substituent and the Glu138 is also formed in compound 14. Pi interactions between the rings of the benzothiazolyl moieties and the amino acids Leu100 and Val179 were observed in all these cases.
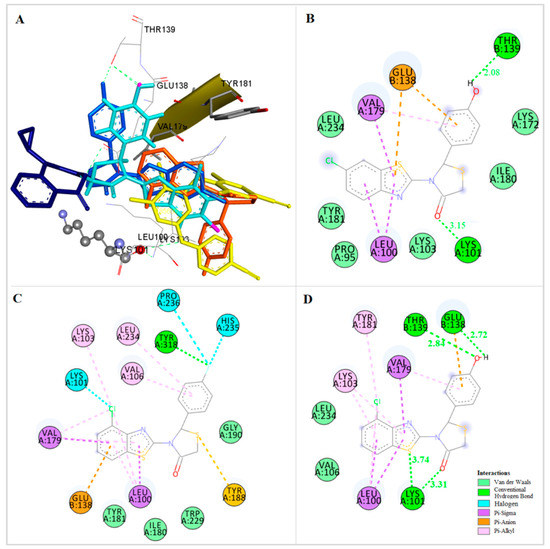
Figure 8.
(A) Docking of compounds 8 (magenta), 9 (light blue), 10 (orange), 14 (cyan), nevirapine (dark blue) and etravirine (yellow) at the allosteric site of HIV-1 RT. (B) Docking of compound 9 at the allosteric site of HIV-1 RT. (C) Docking of compound 10 at the allosteric site of HIV-1 RT. (D) Docking of compound 14 at the allosteric site of HIV-1 RT.
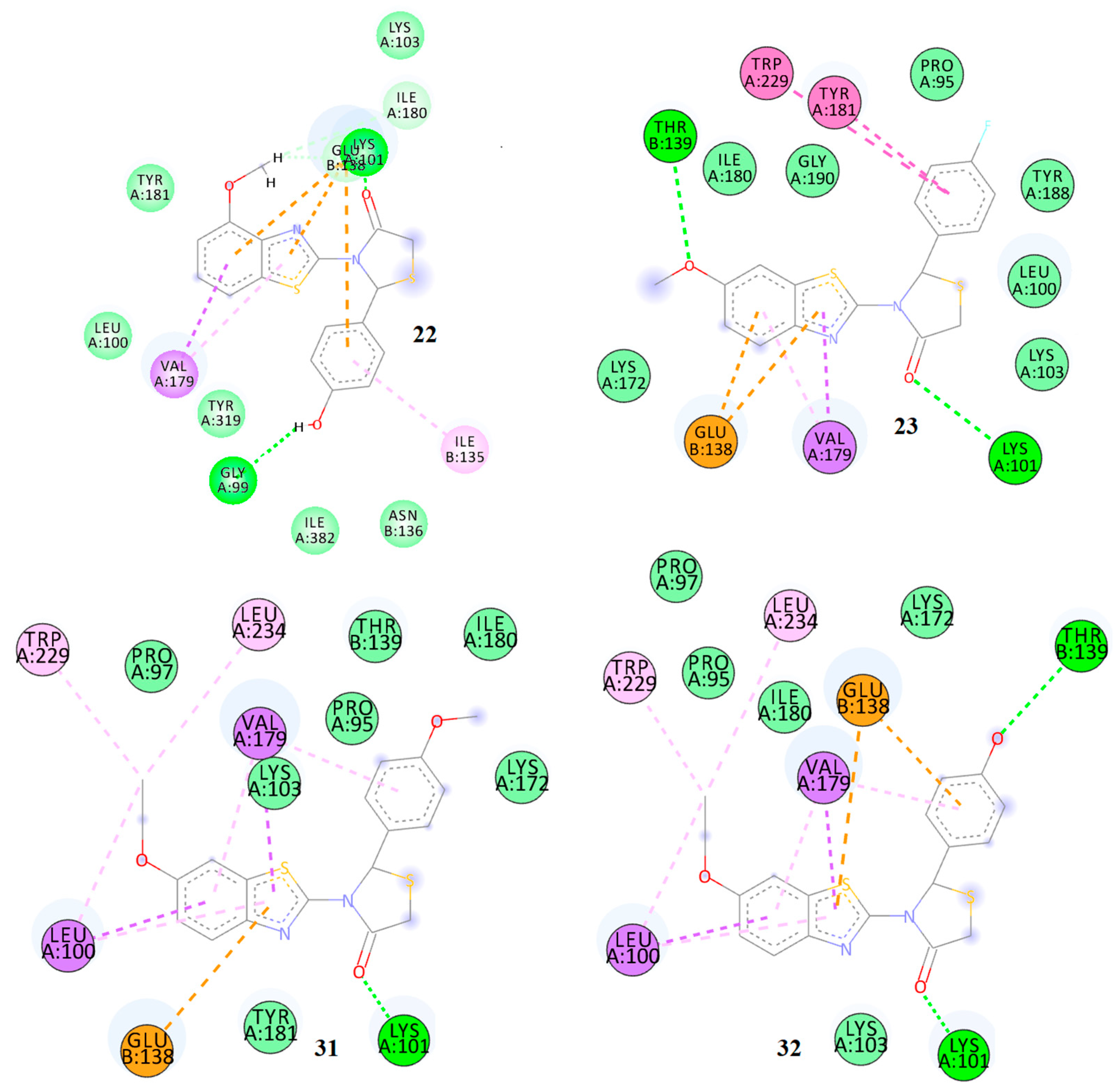
An orientation enabling pi interactions between the aromatic rings of the benzothiazolyl moiety and the amino acids Leu100 and Val179, as well as hydrogen bond formation between Lys101 and the CO group of the thiazolidinone ring were also observed in the active compounds of the 4-OMe, 6-OMe and 6-OEt subgroups such as compounds 18, 20, 27 and 28 (Figure 9).
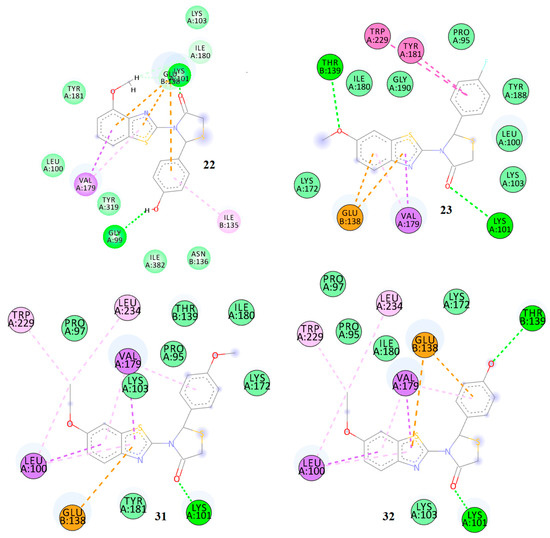
Figure 9.
2D docking diagrams of compounds 19, 20, 27, and 28 at the allosteric site of HIV-1 RT.
On the contrary, the presence of the strongly electro-negative 4′-F (R2) substituent at compound 10 (Figure 8A) leads to a totally different orientation placing the compound with the phenyl moiety in vicinity to Tyr318 with which the F-atom forms a hydrogen bond and close to Pro236 and His235 forming halogen bonds. The differentiation of this compound from the other derivatives and most known NNRTIs where hydrogen bond interactions with Lys101 play a crucial role in complex stabilization is an interesting characteristic. However, the benzothiazolyl moiety is still placed in the area of Lys101, Val179 and Leu100 forming pi interactions with the last two amino acids (Figure 8A,C). In this conformation, the phenyl and the thiazolidinone moieties occupy the places occupied by the edge aromatic rings of etravirine while the benzothiazolyl moiety is placed in the area of the central ring.
Similar orientations, with the phenyl or benzothiazole moiety placed towards Tyr 318 are also observed in other cases (Figure 6).
In general, the presence of a Cl substituent at the 7-, 6- or 4- position of the benzothiazolyl moiety facilitates an orientation which enables complex stabilization via pi-interactions and hydrogen bond formation involving either the CO group of the thiazolidinone ring or hydrogen acceptor/donor substituents of the phenyl ring. In case of subgroups bearing hydrogen acceptor substituents at the benzothiazole moiety, involvement of these substituents in hydrogen bond formation also favors complex stabilization such as in case of compound 3 of the 6F-subgroup, 18 of the 4-OMe subgroup, 23 of the -OEt subgroup and 33 of the -CF3 subgroup.
2.8. Cytotoxicity Assessment
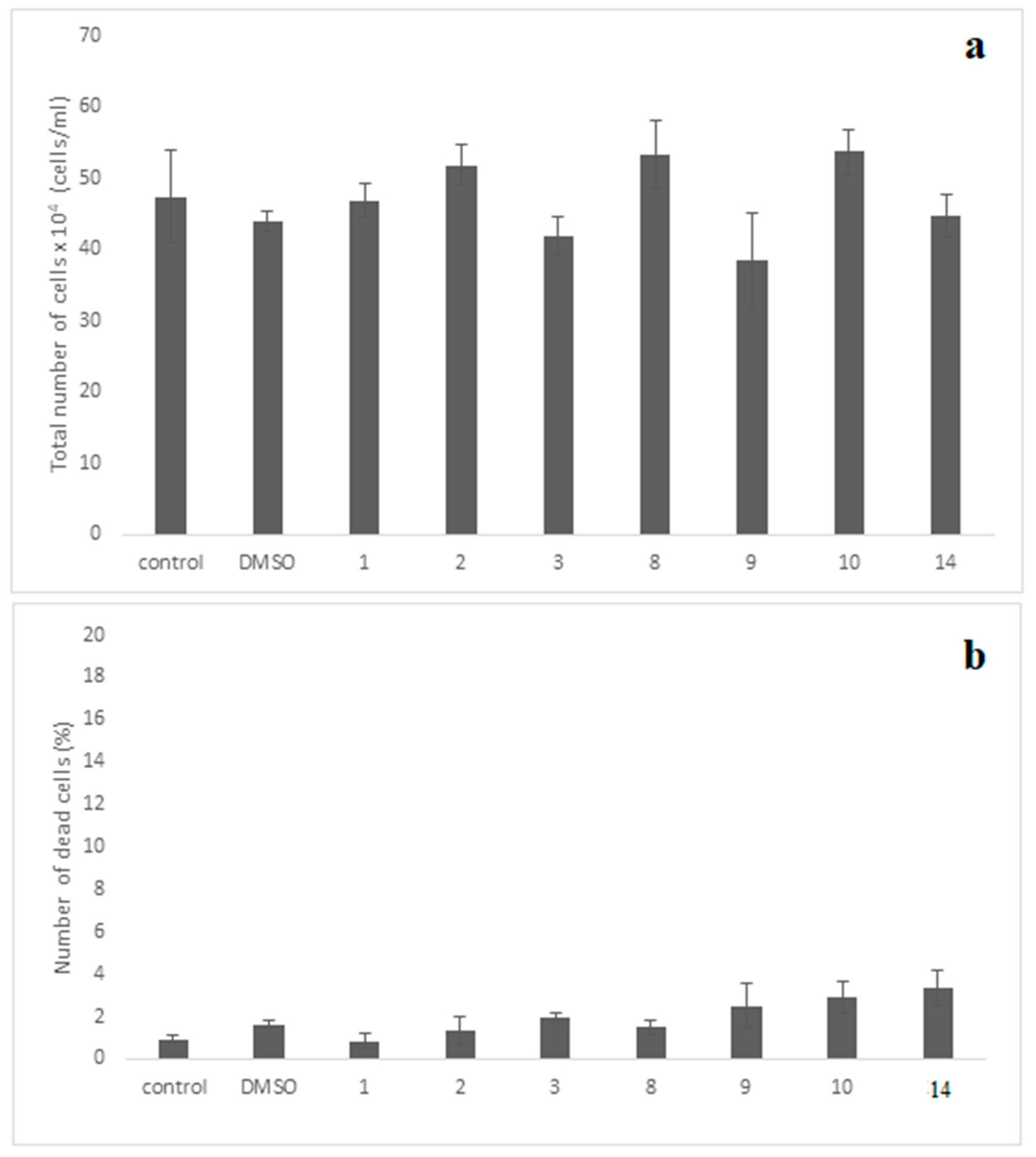
For the evaluation of cellular cytotoxicity of the compounds, the MRC-5 normal cells were incubated for 48 h separately with each compound at the concentration of 10 μΜ. This concentration is much higher than IC50 values of all tested compounds (IC50 0.001 (compounds 9, 10) and 0.01 (compound 14). The evaluation revealed that compounds exhibited no evidence of cytotoxicity against this normal cell line employed, since no statistically significant change was observed with the cell growth to be ≥80% as compared to control-untreated culture (Figure 10a). Importantly, no significant dead cells accumulation was observed in the treated cultures, since their maximum number did not exceed that of 3.3% (Figure 10b).
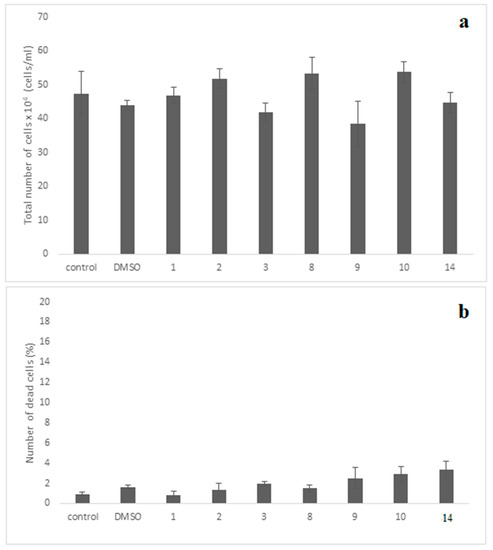
Figure 10.
Assessment cell growth (a) and cell death (b) of MRC-5 cells exposed to various compounds. The MRC-5 cell cultures were incubated separately with each of the compounds at 10−5 M (10μΜ) for 48 h. The cell growth was determined by measuring the cell number in cultures under the microscope using the Neubauer counting chamber, as described in the “Materials and Methods”. Moreover, the dead cells accumulated in these cultures were also evaluated using the Trypan-blue exclusion-dye method, as presented in “Materials and Methods”. The results shown above indicate the mean numbers ± SD of two independent biological experiments. The diagrams shown above and the t-test statistical analysis were carried out using the GraphPad Prism 6.0 program. Notably, no statistical significance between the control-untreated culture with each one of compounds-treated cultures is seen (p values > 0.05).
3. Materials and Methods
3.1. Computer Simulation Methods
For docking calculations, the software AutoDock 4.2 was used [45]. The free energy of binding (ΔG) of HIV-1 Reverse Transcriptase complex with the tested compounds was generated using this molecular docking program.
The crystal structure of HIV-1 Reverse Transcriptase in Complex with TMC125 (PDB code: 3MEC) was downloaded from the Protein Data Bank [46]. For RT preparation, water molecules were deleted, polar hydrogens were added and Kollman United Atom charges were assigned. For the preparation of ligand structures, 2D structure was sketched in chemdraw12.0 software (Chemical Structure Drawing Standard; Perkin Elmer Informatics, Waltham, MA, USA) and converted to three-dimensional structures, mol2 format, for each ligand. Hydrogens were added to the structures and used for docking. The Grid center was calculated: 9.7037 12.7492 16.914 (xyz-coordinates), for HIV-1 Reverse Transcriptase. The grid size was set to 110 × 110 × 110 xyz points with grid spacing of 0.375 Å. All parameters used in docking were default. A primary blind docking was performed in order to discriminate the preferential binding sites of the ligand to the receptor. The ligand TMC125, as it is binded in the crystal structure was extracted and docked back into the analogous binding pocket to determine the ability of Autodock to replicate the position and orientation of the inhibitor in the crystal structure. Control docking showed that Autodock was able to determine the orientation, position and the same interactions of the crystallographic TMC125. The number of docking runs was 100. After docking, the 100 solutions were clustered into groups with RMS lower than 1.0 Ε. The resulting poses and interactions were visualized in Discovery studio 4.1 silent.
3.2. PASS Prediction
For the prediction of activity spectra of the designed compounds the program PASS was used. PASS, as already described in our paper [47] predicts the biological activity spectrum of a compound based on the analysis of structure–activity relationships of more than 1 million of known compounds. These compounds possess over eight thousand biological activities. The average accuracy of prediction is about 96%.
3.3. Prediction of Toxicity
For the prediction of toxicity of the designed compounds the Lazar model over the Open Tox Predict Program was used. OpenTox is a free online program, that provides access to experimental toxicity data, in Silico models (including (Q)SAR), and validation/reporting procedures. Lazar model predicts the toxicity of the compounds based on their structure by searching for similar compounds. Lazar is a k-nearest-neighbor approach to predict chemical endpoints from a training set based on structural fragments. It uses a SMILES file and precomputed fragments with occurrences as well as target class information for each compound as training input. It also features regression; in which case the target activities consist of continuous values. Lazar uses activity-specific similarity (i.e., each fragment contributes with its significance for the target activity) that is the basis for predictions and confidence index for every single prediction [48,49].
3.4. Chemistry
3.4.1. General Procedure for the Synthesis of Thiazolidinones by Conventional Method
The appropriate heteroaromatic amine (1.0 mmol) and substituted benzaldehyde (1.2 mmol) were stirred in dry toluene under reflux condition followed by addition of mercatoacetic acid (2.0 mmol). The reaction mixture was refluxed for 18–32 h and then concentrated to dryness under reduce pressure. The residue was diluted in ethyl acetate and the organic layer was washed with 5% aq citric acid, water and 5% aq sodium hydrogen carbonate. The organic layer was dried over sodium sulfate and concentrated under reduced pressure to give a clear product. All the synthesized compounds were characterized by TLC, 1H-NMR, 13C-NMR and MS.
3.4.2. Microwave Irradiation Experiments
All microwave irradiation experiments were carried out in CEM-Discover monomode microwave device, operating at a frequency of 2.45 GHz. Substituted aminobenzothiazole, equimolar amount of substituted benzaldehyde (1.5 mmol) and mercaptoacetic acid in absolut ethanol (3 mL) were placed in a 10 mL reaction vial containing a stirring bar. The vial was sealed with a Teflon septum and placed into the microwave cavity. It was irradiated at 100 °C using 100 W as maximum power for 30 min. at the end of the reaction the mixture was rapidly cooled with gas jet cooling to room temperature. The clean product was obtained after filter under reduced pressure.
3.4.3. Synthesis of 3-(7-Chloro-benzo[d]thiazol-2-yl)-2-(2,6-difluorophenyl)thiazolidin-4-one (1)
Yield: 39%, m.p. 200–202 °C, Rf: 0.66 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1705 (C=O), 1541, 1102. 1H-NMR (500 MHz, CHCl3-d): δ = 3.86 (d, J = 16.38 Hz, 1H, CH2), 4.10 (d, J = 16.63 Hz, 1H, CH2), 6.83 (d, J = 18.59 Hz, 1H, N-CH-S), 7.21 (t, J = 8.56 Hz, 2H, Ar-C18, C20), 7.06–7.15 (m, 1H, Ar-C5), 7.61–7.72 (m, 3H, Ar-C5, C6, C19), 8.05 (d, J = 8.02 Hz, 1H, Ar-C4). 13C-NMR (500 MHz, CHCl3-d): δ = 33.12, 60.42, 113.05, 119.34, 123.02, 124.15, 125.03, 126.11, 126.79, 129.01, 130.00, 135.65 150.42, 162.16, 164.17, 171.12. MS: m/z = 382 (M+, 100%), 353 (33.3%), 372 (36%), 363 (28%), 348 (21%), 242 (72%).
3.4.4. Synthesis of 3-(7-Chloro-benzo[d]thiazol-2-yl)-2-(2-chloro-6-fluorophenyl)thiazolidin-4-one (2)
Yield: 48%, m.p. 159–160 °C, Rf: 0.52 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1708 (C=O), 1541, 1101, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.85 (d, J = 16.38 Hz, 1H, CH2), 4.12 (d, J = 16.63 Hz, 1H, CH2), 6.71 (d, J = 18.59 Hz, 1H, N-CH-S), 7.07 (t, J = 8.56 Hz, 1H, Ar-C19), 7.30–7.55 (m, 4H, Ar-C5, C-6, C18, C19), 8.06 (d, J = 8.02 Hz, 1H, Ar-C4). 13C-NMR (500 MHz, CHCl3-d): δ = 32.88, 63.15, 111.07, 112.25, 119.11, 123.02, 126.53, 128.02, 128.25, 150.44, 162.13, 164.12, 171.02. MS: m/z = 400 (M+, 39%), 363 (100%), 321 (32%), 289 (88%), 254 (13%), 227 (10%), 215 (22%), 169 (23%), 153 (66%), 107 (18%).
3.4.5. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (3)
Yield: 21%, m.p. 192–193 °C, Rf: 0.71 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1705 (C=O), 1571, 1231, 1105. 1H-NMR (500 MHz, CHCl3-d): δ = 3.85 (d, J = 16.38 Hz, 1H, CH2), 4.11 (d, J = 16.63 Hz, 1H, CH2), 6.77 (d, J = 18.59 Hz, 1H, N-CH-S), 7.02 (t, J = 8.56 Hz, 1H, Ar-C5), 7.06–7.15 (m, 1H, Ar-C5), 7.27–7.36 (m, 4H, Ar-C17, C18, C20, C21), 7.48 (d, J = 8.02 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.79, 63.09, 109.98, 113.94, 115.84, 120.83, 127.49, 127.55, 130.01, 133.32, 146.77, 156.13, 161.98, 163.52, 170.87 (1C, C=O). MS: m/z = 349 (M+, 7%), 288 (3%), 272 (25%), 245 (14%), 244 (100%), 240 (11%), 200 (7%), 163 (5%), 130 (6%).
3.4.6. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (4)
Yield: 55%, m.p. 204–205 °C, Rf: 0.44 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1706 (C=O), 1572, 1235. 1H-NMR (500 MHz, CHCl3-d): δ = 4.04 (d, J = 16.63 Hz, 1H, CH2), 4.29 (d, J = 16.63 Hz, 1H, CH2), 6.97 (s, 1H, N-CH-S), 7.22 (t, J = 8.43 Hz, 1H, Ar-C5), 7.62 (br s, 1H, Ar-C4), 7.68 (d, J = 7.34 Hz, 2H, Ar-C17, C21), 7.92 (br d, J = 8.80 Hz, 1H, Ar-C7), 8.17 (d, J = 8.31 Hz, 2H, Ar-C18, C20). 13C-NMR (500 MHz, CHCl3-d): δ = 32.77, 63.08, 109.95, 113.97, 117.94, 120.83, 123.32, 127.49(2C), 133.32, 145.78, 146.77, 156.12, 163.56, 170.96. MS: m/z = 376 (M+, 100%), 342 (25%), 272 (41%), 150 (38%), 115 (17%), 73 (11%).
3.4.7. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (5)
Yield: 74%, m.p. 175–176 °C, Rf: 0.67 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1708 (C=O), 1554, 1231, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.86 (d, J = 16.38 Hz, 1H, CH2), 4.10 (br d, J = 16.63 Hz, 1H, CH2), 6.73 (s, 1H, N-CH-S), 7.06–7.19 (m, 1H, Ar-C5), 7.22–7.40 (m, 4H, Ar-C17, C18, C20, C21), 7.48 (br d, J = 7.83 Hz, 1H, Ar-C4), 7.56–7.79 (m, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.79, 63.09, 109.98, 113.94, 117.94, 127.94, 130.32, 131.31, 132.35, 137.38, 146.77, 156.13, 163.52, 170.87. MS: m/z = 365 (M+, 100%), 343 (47%), 320 (7%), 291 (70%), 279 (34%), 259 (32%), 225 (28%).
3.4.8. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (6)
Yield: 29%, m.p. 198–200 °C, Rf: 0.63 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1706 (C=O), 1572, 1230, 1213. 1H-NMR (500 MHz, CHCl3-d): δ = 3.76 (s, 3H, O-CH3), 3.91 (d, J = 16.38 Hz, 1H, CH2), 4.10 (d, J = 16.63 Hz, 1H, CH2), 6.73 (s, 1H, N-CH-S), 6.84 (d, J = 8.80 Hz, 2H, Ar-C18, C20), 7.12 (t, J = 8.56 Hz, 1H, Ar-C5), 7.23–7.35 (m, 1H, Ar-C4), 7.46–7.55 (m, 2H, Ar-C17, C21), 7.58–7.79 (m, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.78, 58.01, 63.08, 109.98, 113.89, 114.13, 117.94, 129.49, 131.98, 132.35, 137.38, 146.73, 156.1, 158.01, 163.51, 171.03. MS: m/z = 361 (M+, 15%), 288 (9%), 272 (25%), 244 (100%), 243 (10%), 153 (13%), 135 (7%).
3.4.9. Synthesis of 3-(6-Fluoro-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (7)
Yield: 21%, m.p. 230–231 °C, Rf: 0.37 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 3258 (-OH), 2361, 1706 (C=O), 1573, 1230. 1H-NMR (500 MHz, CHCl3-d): 4.04 (d, J = 16.63 Hz, 1H, CH2), 4.29 (d, J = 16.63 Hz, 1H, CH2), 5.35 (br s, 1H, -OH), 6.97 (s, N-CH-S), 7.22 (t, J = 8.43 Hz, 1H, Ar-C5), 7.62 (br s, 1H, Ar-C4), 7.68 (d, J = 7.34 Hz, 2H, Ar-C18, C20), 7.92 (br d, J = 8.80 Hz, 1H, Ar-C7), 8.16 (d, J = 8.31 Hz, 2H, Ar-C17, C21). 13C-NMR (500 MHz, CHCl3-d): δ = 32.81, 63.11, 109.94, 113.91, 115.12, 117.94, 130.00, 131.24, 131.53, 146.68, 156.09, 158.04, 163.52, 171.05. MS: m/z = 347 (M+, 42%), 309 (92%), 279 (40%), 263 (70%), 243 (72%).
3.4.10. Synthesis of 3-(6-Chloro-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (8)
Yield: 51%, m.p. 196–197 °C, Rf: 0.76 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2363, 1704 (C=O), 1568, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.85 (d, J = 16.38 Hz, 1H, CH2), 4.11 (d, J = 16.63 Hz, 1H, CH2), 6.75 (s, 1H, N-CH-S), 7.10–7.18 (m, 2H, Ar-C18, C20), 7.27–7.36 (m, 2H, Ar-C17, C21), 7.56 (t, J = 8.56 Hz, 1H, Ar-C5), 7.61 (d, J = 8.02 Hz, 1H, Ar-C4), 8.05 (s, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.86, 63.10, 115.20, 120.82, 122.73, 126.76, 127.73, 133.30, 136.08, 146.90, 156.05, 163.49, 171.11. MS: m/z = 364 (M+, 100%), 343 (10%), 299 (10%), 290 (100%), 279 (9%), 184 (11%), 115 (80%), 88 (9%), 73 (94%).
3.4.11. Synthesis of 3-(6-Chloro-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (9)
Yield: 24%, m.p. 236–237 °C, Rf: 0.40 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2359 (-OH), 2361, 1701 (C=O), 1573. 1H-NMR (500 MHz, CHCl3-d): δ = 3.86 (d, J = 16.38 Hz, 1H, CH2), 4.10 (d, J = 16.63 Hz, 1H, CH2), 5.35 (br s, 1H, -OH), 6.73 (s, 1H, N-CH-S), 7.26–7.43 (m, 5H, Ar-C5, C17, C18, C20, C21), 7.76 (br d, J = 8.80 Hz, 1H, Ar-C4), 8.13 (s, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 33.12, 63.71, 115.75, 118.23, 121.55, 125.14, 129.02, 130.01, 131.52, 132.87, 151.68, 156.71, 163.77, 171.12. MS: m/z = 363 (M+, 100%), 288 (88%), 272 (9%), 225 (33%), 184 (48%), 146 (7%), 136 (21%), 115 (18%), 73 (17%).
3.4.12. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (10)
Yield: 33%, m.p. 215–216 °C, Rf: 0.74 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1702 (C=O), 1573, 1101. 1H-NMR (500 MHz, CHCl3-d): δ = 3.93 (d, J = 16.63 Hz, 1H, CH2), 4.21 (d, J = 16.63 Hz, 1H, CH2), 6.81 (s, 1H, N-CH-S), 7.02 (br t, J = 8.56 Hz, 2H, Ar-C18, C20), 7.15–7.24 (m, 1H, Ar-C6), 7.41 (d, J = 7.34 Hz, 1H, Ar-C5), 7.50 (br dd, J = 8.31, 5.38 Hz, 2H, Ar-C17, C21), 7.68 (d, J = 7.83 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.81, 63.87, 115.02, 119.33, 120.77, 121.13, 125.76, 130.71, 132.45, 134.38, 149.11, 161.33, 164.02, 171.05. MS: m/z = 364 (M+, 72%), 343 (35%), 232 (31%), 290 (100%), 292 (79%), 279 (33%), 242 (64%), 225 (65%), 184 (77%), 174 (44%).
3.4.13. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (11)
Yield: 41%, m.p. 203–204 °C, Rf: 0.45 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1701 (C=O), 1573. 1H-NMR (500 MHz, CHCl3-d): δ = 3.93 (d, J = 16.63 Hz, 1H, CH2), 4.21 (d, J = 16.63 Hz, 1H, CH2), 6.85 (s, 1H, N-CH-S), 7.21–7.30 (d, J = 7.34 Hz, 2H, Ar-C17, C21), 7.40 (br d, J = 8.80 Hz, 1H, Ar-C6), 7.80 (br d, J = 8.80 Hz, 1H, Ar-C5), 8.20 (d, J = 8.31 Hz, 3H, Ar-C7, C18, C20). 13C-NMR (500 MHz, CHCl3-d): δ = 32.86, 63.35, 119.32, 121.14, 121.58, 123.69, 125.81, 129.66, 132.11, 145.38, 146.37, 149.72, 164.01, 171.02. MS: m/z = 373 (M+, 33%), 349(14%), 343 (46%), 332 (40%), 304 (29%), 292 (92%), 290 (100%), 279 (18%), 259 (7%).
3.4.14. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (12)
Yield: 56%, m.p. 184–185 °C, Rf: 0.73 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1704 (C=O), 1568, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.94 (d, J = 16.63 Hz, 1H, CH2), 4.21 (d, J = 16.63 Hz, 1H, CH2), 6.79 (s, 1H, N-CH-S), 7.21–7.31 (m, 3H, Ar-C6, C17, C21), 7.39–7.43 (m, 3H, Ar-C5, C18, C20), 7.68 (d, J = 7.83 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.83, 63.12, 119.89, 121.23, 121.59, 125.83, 128.77, 130.77, 132.31, 132.59, 145.38, 137.36, 149.71, 163.95, 171.03. MS: m/z = 383 (M+, 13%), 381 (43%), 379 (61%), 346 (11%), 341 (11%), 337 (18%), 269 (46%), 227 (32%).
3.4.15. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (13)
Yield: 67%, m.p. 223–224 °C, Rf: 0.54 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1700 (C=O), 1571, 1232. 1H-NMR (500 MHz, CHCl3-d): δ = 3.75 (s, 3H, O-CH3), 3.92 (d, J = 16.38 Hz, 1H, CH2), 4.20 (d, J = 16.63 Hz, 1H, CH2), 6.81 (s, 1H, N-CH-S), 6.84(d, J = 8.80 Hz, 2H, Ar-C18, C20), 7.19 (t, J = 8.56 Hz, 1H, Ar-C6), 7.25 (s, 1H, Ar-C5), 7.41–7.45 (m, 2H, Ar-C17, C21), 7.68 (d, J = 7.83 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.85, 55.81, 63.32, 114.21, 119.31, 121.18, 121.57, 125.86, 129.79, 131.28, 132.38, 149.49, 159.00, 163.52, 171.04. MS: m/z = 377 (M+, 100%), 362 (15%), 353 (31%), 326 (27%), 290 (40%), 288 (82%), 136 (36%), 115 (17%) 73 (14%).
3.4.16. Synthesis of 3-(4-Chloro-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (14)
Yield: 17%, m.p. 258–259 °C, Rf: 0.39 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 3205 (-OH), 2361, 1701 (C=O), 1569. 1H-NMR (500 MHz, CHCl3-d): δ = 3.93 (d, J = 16.63 Hz, 1H, CH2), 4.21 (d, J = 16.63 Hz, 1H, CH2), 5.32 (br s, 1H, -OH), 6.81 (s, 1H, N-CH-S), 7.02 (br t, J = 8.56 Hz, 2H, Ar-C18, C20), 7.15–7.24 (m, 1H, Ar-C6), 7.41 (d, J = 7.34 Hz, 1H, Ar-C5), 7.50 (br dd, J = 8.31, 5.38 Hz, 2H, Ar-C17, C21), 7.68 (d, J = 7.83 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 33.02, 63.31, 115.18, 119.23, 121.55, 121.89, 125.15, 130.03, 131.52, 132.17, 149.68, 156.73, 163.79, 171.08. MS: m/z = 363 (M+, 75%), 346 (28%), 310 (22%), 290 (21%), 272 (100%), 184 (19%), 136 (22%), 115 (51%), 88 (27%), 73 (15%).
3.4.17. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-flurophenyl)thiazolidin-4-one (15)
Yield: 51%, m.p. 251–252 °C, Rf: 0.73 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1968 (C=O), 1587, 1272, 1100. 1H-NMR (500 MHz, DMSO-d6): δ = 3.68–3.86 (m, 3H, O-CH3), 3.91–4.09 (m, 1H, CH2), 4.25 (br d, J = 9.78 Hz, 1H, CH2), 6.88 (s, 1H, N-CH-S), 7.06 (br d, J = 10.27 Hz, 1H, Ar-C5), 7.26 (br dd, J = 10.03, 8.07 Hz, 1H, Ar-C6), 7.55 (br d, J = 10.27 Hz, 1H, Ar-C7), 7.61–7.76 (m, 2H, Ar-C17, C21), 8.08–8.26 (m, 2H, Ar-C18, C20). 13C-NMR (500 MHz, DMSO-d6): δ = 32.86, 55.82, 63.18, 108.26, 114.11, 115.43, 121.18, 130.33, 131.99, 135.59, 142.31, 150.23, 161.51, 163.91, 171.08. MS: m/z = 361 (M+, 100%), 343 (42%), 321 (36%), 290 (72%), 288 (31%), 279 (39%), 225 (24%).
3.4.18. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (16)
Yield: 63%, m.p. 298–299 °C, Rf: 0.48 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1705 (C=O), 1585, 1273. 1H-NMR (500 MHz, DMSO-d6): δ = 3.77 (s, 3H, O-CH3), 4.03 (d, J = 17.12 Hz, 1H, CH2), 4.20 (d, J = 17.12 Hz, 1H, CH2), 6.93 (s, 1H, N-CH-S), 7.05 (s, 1H, Ar-C5), 7.26 (s, 1H, Ar-C6), 7.56 (d, J = 7.83 Hz, 2H, Ar-C17, C21), 7.67 (d, J = 8.80 Hz, 1H, Ar-C7), 8.17 (d, J = 8.32 Hz, 2H, Ar-C18, C20). 13C-NMR (500 MHz, DMSO-d6): δ = 32.85, 55.85, 63.21, 108.25, 114.12, 121.18, 123.84, 129.66, 131.97, 142.31, 145.26, 146.32, 150.21, 163.95, 171.02. MS: m/z = 388 (M+, 100%), 288 (73%), 272 (3%), 225 (42%), 184 (76%), 136 (42%), 115 (31%), 73 (28%).
3.4.19. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (17)
Yield: 43%, m.p. 218–219 °C, Rf: 0.53 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1708 (C=O), 1589, 1276, 1154. 1H-NMR (500 MHz, DMSO-d6): δ = 3.80 (s, 3H, O-CH3), 3.99 (br d, J = 17.12 Hz, 1H, CH2), 4.23 (br d, J = 16.63 Hz, 1H, CH2), 6.93 (s, 1H, N-CH-S), 6.91–6.97 (m, 1H, Ar-C5), 7.26 (br t, J = 8.07 Hz, 1H, Ar-C4), 7.39 (br d, J = 9.29 Hz, 4H, Ar-C17, C18, C20, C21), 7.54 (br d, J = 7.83 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, DMSO-d6): δ = 32.82, 55.88, 63.27, 108.25, 114.11, 121.32, 128.73, 130.12, 131.93, 132.75, 137.36, 142.32, 150.22, 164.03, 171.00. MS: m/z = 377 (M+, 41%), 272 (23%), 244 (100%), 242 (63%), 165 (17%), 136 (23%).
3.4.20. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (18)
Yield: 52%, m.p. 196–197 °C, Rf: 0.63 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1701 (C=O), 1556, 1273. 1H-NMR (500 MHz, CHCl3-d): δ = 33.76 (s, 3H, O-CH3benz), 3.94 (d, J = 16.63 Hz, 1H, CH2), 3.95 (s, 3H, O-CH3), 4.13 (d, J = 16.63 Hz, 1H, CH2), 6.81–6.85 (m, 2H, Ar-C18, C20), 7.93 (s, 1H, N-CH-S), 7.25 (br d, J = 9.29 Hz, 3H, Ar-C5, C6, C7), 7.39 (br d, J = 7.83 Hz, 2H, Ar-C17, C21). 13C-NMR (500 MHz, CHCl3-d): δ = 32.86, 55.83, 63.35, 108.25, 114.11, 114.76, 121.82, 129.75, 131.55, 131.96, 142.34, 150.23, 160.82, 164.06, 171.11. MS: m/z = 373 (M+, 76%), 272 (33%), 243 (56%), 244 (100%), 242 (8%), 165 (5%), 164 (7%), 135 (11%).
3.4.21. Synthesis of 3-(4-Methoxy-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (19)
Yield: 29%, m.p. 261–262 °C, Rf: 0.72 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 3205 (-OH), 2361, 1707 (C=O), 1582, 1271. 1H-NMR (500 MHz, DMSO-d6): δ = 3.77 (s, 3H, O-CH3), 4.03 (d, J = 17.12 Hz, 1H, CH2), 4.20 (d, J = 17.12 Hz, 1H, CH2), 5.32 (br s, 1H, -OH), 6.93 (d, J = 7.83 Hz, 2H, Ar-C18, C20), 7.05 (s, 1H, N-CH-S), 7.26 (br t, J = 8.56 Hz, 1H, Ar-C5), 7.56 (d, J = 7.83 Hz, 1H, Ar-C6), 7.67 (d, J = 8.80 Hz, 1H, Ar-C7), 8.17 (d, J = 8.32 Hz, 2H, Ar-C17, C21). 13C-NMR (500 MHz, CHCl3-d): δ = 32.88, 55.82, 63.33, 108.21, 114.13, 115.81, 121.73, 130.13, 131.52, 131.96, 142.31, 150.22, 156.95, 164.13, 171.04. MS: m/z = 359 (M+, 82%), 344 (30%), 328 (64%), 189 (27%), 244 (100%), 165 (54%), 135 (21%).
3.4.22. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (20)
Yield: 52%, m.p. 242–243 °C, Rf: 0.62 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1701 (C=O), 1531, 1225. 1H-NMR (500 MHz, DMSO-d6): δ = 3.76 (s, 3H, O-CH3), 3.99 (br d, J = 16.63 Hz, 1H, CH2), 4.26 (br d, J = 16.63 Hz, 1H, CH2), 6.84 (s, 1H, N-CH-S), 6.97 (dd, J = 8.80, 2.45 Hz, 1H, Ar-C5), 7.13 (br t, J = 8.80 Hz, 2H, Ar-C18, C20), 7.44 (br dd, J = 8.31, 5.38 Hz, 2H, Ar-C17, C21), 7.49–7.71 (m, 2H, Ar-C4, C7). 13C-NMR (500 MHz, DMSO-d6): δ = 32.96, 55.82, 63.33, 108.21, 114.13, 115.81, 121.73, 130.13, 131.52, 131.96, 142.31, 150.22, 156.95, 164.13, 171.04. MS: m/z = 361 (M+, 14%), 288 (8%), 272 (25%), 260 (5%), 258 (8%), 244 (100%), 243 (8%), 153 (13%), 165 (4%), 135 (7%), 109 (8%).
3.4.23. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (21)
Yield: 41%, m.p. 220–221 °C, Rf: 0.49 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1701 (C=O), 1532, 1226. 1H-NMR (500 MHz, DMSO-d6): δ = 3.76 (s, 3H, O-CH3), 4.03 (br d, J = 16.63 Hz, 1H, CH2), 4.27 (br d, J = 16.63Hz, 1H, CH2), 6.89–7.06 (m, 2H, Ar-C5, N-CH-S), 7.51 (br d, J = 9.29 Hz, 1H, Ar-C5), 7.60 (br s, 1H, Ar-C7), 7.67 (br d, J = 7.34 Hz, 2H, Ar-C17, C21), 8.17 (br d, J = 8.31 Hz, 2H, Ar-C18, C20). 13C-NMR (500 MHz, DMSO-d6): δ = 32.87, 55.86, 63.32, 104.12, 114.33, 117.25, 124.38, 125.96, 131.95, 145.13, 145.51, 146.79, 156.81, 164.03, 170.92. MS: m/z = 388 (M+, 62%), 288 (100%), 225 (29%), 184 (61%), 136 (27%), 115 (23%), 107 (27%), 73 (18%).
3.4.24. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (22)
Yield: 52%, m.p. 179–180 °C, Rf: 0.53 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1706 (C=O), 1536, 1219, 1108. 1H-NMR (500 MHz, DMSO-d6): δ = 3.77 (s, 3H, O-CH3), 3.99 (d, J = 17.12 Hz, 1H, CH2), 4.26 (d, J = 16.63 Hz, 1H, CH2), 6.84 (s, 1H, N-CH-S), 6.97 (dd, J = 8.80, 2.45 Hz, 1H, Ar-C5), 7.32–7.47 (m, 4H, Ar-C17, C18, C20, C21), 7.47–7.65 (m, 2H, Ar-C4, C7). 13C-NMR (500 MHz, DMSO-d6): δ = 32.76, 55.85, 63.31, 104.99, 114.61, 118.23, 127.81(2C), 129.16, 131.93, 132.15, 137.33, 145.52, 156.74, 164.03, 170.85. MS: m/z = 377 (M+, 100%), 288 (82%), 272 (33%), 225 (21%), 136 (35%), 115 (20%), 73 (18%).
3.4.25. Synthesis of 3-(6-Methoxy-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (23)
Yield: 33%, m.p. 225–226 °C, Rf: 0.62 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1700 (C=O), 1540, 1225. 1H-NMR (500 MHz, CHCl3-d): δ = 3.76 (s, 3H, O-CH3benz), 3.94 (d, J = 16.63 Hz, 1H, CH2), 3.98 (s, 3H, O-CH3), 4.13 (d, J = 16.63 Hz, 1H, CH2), 6.84 (s, 1H, N-CH-S), 6.93 (br d, J = 8.31 Hz, 2H, Ar-C18, C20), 7.51 (br d, J = 9.29 Hz, 1H, Ar-C5), 7.25 (br d, J = 9.29 Hz, 2H, Ar-C4, C7, C21), 7.39 (br d, J = 7.83 Hz, 1H, Ar-C17). 13C-NMR (500 MHz, CHCl3-d): δ = 32.81, 55.84, 63.33, 104.92, 114.81, 115.01, 118.22, 127.79, 131.91, 132.28, 145.55, 156.72, 159.85, 163.92, 170.93. MS: m/z = 373 (M+, 78%), 299 (10%), 344 (10%), 272 (33%), 244 (100%), 242 (7%), 165 (5%), 164 (7%), 150 (9%), 135 (3%).
3.4.26. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-fluorophenyl)thiazolidin-4-one (24)
Yield: 63%, m.p. 144–145 °C, Rf: 0.44 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2316, 1701 (C=O), 1531, 1225. 1H-NMR (500 MHz, CHCl3-d): δ = 1.25 (t, J = 6.85 Hz, 3H, -OCH2CH3), 3.82–3.85 (m, 3H, CH2,-OCH2CH3), 4.10 (d, J = 16.63 Hz, 1H, CH2), 6.79 (s, 1H, N-CH-S), 6.97 (dd, J = 8.80, 2.45 Hz, 2H, Ar-C18, C20), 7.25 (s, 1H, Ar-C5), 7.30–7.33 (m, 2H, Ar-C17, C21), 7.59 (d, J = 8.80 Hz, 2H, Ar-C4, C7). 13C-NMR (500 MHz, CHCl3-d): δ = 14.13, 32.35, 55.89, 63.12, 104.52, 115.13, 115.52, 115.61, 130.02, 131.51, 134.03, 144.00, 153.62, 161.13, 164.05, 171.21. MS: m/z = 375 (M+, 100%), 288 (28%), 272 (36%), 225 (18%), 197 (19%), 136 (20%), 115 (16%), 73 (16%).
3.4.27. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (25)
Yield: 72%, m.p. 155–156 °C, Rf: 0.58 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2316, 1701 (C=O), 1527, 1225. 1H-NMR (500 MHz, CHCl3-d): δ = 1.42 (t, J = 6.85 Hz, 3H, -OCH2CH3), 3.89 (d, J = 16.63 Hz, 1H, CH2), 4.10 (d, J = 16.63 Hz, 1H, CH2), 4.25–4.32 (q, J = 6.68 Hz, 2H, -OCH2CH3), 6.80 (s, 1H, N-CH-S), 6.97 (dd, J = 8.80, 2.45 Hz, 1H, Ar-C5), 7.19–7.33 (m, 1H, Ar-C7), 7.42–7.59 (m, 3H, Ar-C4, C17, C21), 8.19 (d, J = 8.80 Hz, 2H, Ar-C18, C20). 13C-NMR (500 MHz, CHCl3-d): δ = 14.12, 32.31, 55.97, 63.17, 104.52, 115.25, 117.31, 122.15, 129.75, 131.21, 142.02, 143.85, 145.06, 153.72, 164.02, 170.75. MS: m/z = 402 (M+, 100%), 382 (49%), 373 (42%), 357 (21%), 348 (18%), 115 (12%), 73 (7%).
3.4.28. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4-one (26)
Yield: 82%, m.p. 180–181°C, Rf: 0.66 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1696 (C=O), 1546, 1227, 698. 1H-NMR (500 MHz, CHCl3-d): δ = 1.43 (t, J = 6.85 Hz, 3H, -OCH2CH3), 3.83 (d, J = 16.63 Hz, 1H, CH2), 4.04–4.10 (m, 3H, CH2, -OCH2CH3), 6.74 (s, 1H, N-CH-S), 6.98 (dd, J = 8.80, 2.45 Hz, 1H, Ar-C5), 7.25–7.30 (m, 5H, Ar-c4, C17, C18, C20, C21), 7.59 (d, J = 8.80 Hz, 1H, Ar-C7). 13C-NMR (500 MHz, CHCl3-d): δ = 14.12, 32.31, 55.92, 63.17, 104.83, 114.77, 117.81, 128.72, 130.05, 131.24, 132.74, 137.52, 144.86, 145.06, 153.59, 164.07, 170.69. MS: m/z = 391 (M+, 100%), 363 (68%), 272 (88%), 244 (67%), 215 (26%), 153 (79%).
3.4.29. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-methoxyphenyl)thiazolidin-4-one (27)
Yield: 55%, m.p. 210–211°C, Rf: 0.63 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1700 (C=O), 1531, 1227. 1H-NMR (500 MHz, CHCl3-d): δ = 1.42 (t, J = 6.85 Hz, 3H, -OCH2CH3), 3.89 (d, J = 16.63 Hz, 1H, CH2), 4.03–4.10 (m, 4H, CH2, -OCH3), 4.25–4.32 (q, J = 6.68 Hz, 2H, -OCH2CH3), 6.80 (s, 1H, N-CH-S), 6.97 (dd, J = 8.80, 2.45 Hz, 2H, Ar-C18, C20), 7.19–7.33 (m, 1H, Ar-C5), 7.42–7.59 (m, 2H, Ar-C4, C7), 8.19 (d, J = 8.80 Hz, 2H, Ar-C17, C21). 13C-NMR (500 MHz, CHCl3-d): δ = 14.15, 32.36, 55.81, 55.96, 63.17, 105.26, 114.21, 114.75, 117.82, 129.85, 131.58, 144.82, 153.56, 159.35, 164.15, 170.57. MS: m/z = 387 (M+, 21%), 344 (16%), 313 (15%), 299 (8%), 272 (37%), 244 (100%), 242 (15%), 165 (35%), 164 (9%), 150 (23%), 135 (8%).
3.4.30. Synthesis of 3-(6-Ethoxy-benzo[d]thiazol-2-yl)-2-(4-hydroxyphenyl)thiazolidin-4-one (28)
Yield: 81%, m.p. 191–192°C, Rf: 0.37 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 3200 (-OH), 2361, 1706 (C=O), 1530, 1225. 1H-NMR (500 MHz, DMSO-d6): δ = 1.33 (br t, J = 6.60 Hz, 3 H), 3.37–3.50 (m, 4 H), 3.51–3.68 (m, 1 H), 4.04 (q, J = 6.68 Hz, 2 H), 5.24 (s, 1 H), 6.71 (m, J = 7.83 Hz, 2 H), 6.99 (br d, J = 8.80 Hz, 1 H), 7.20 (m, J = 7.83 Hz, 2 H), 7.52 (br s, 1 H), 7.60 (br d, J = 8.31 Hz, 1 H), 9.55 (br s, 1 H). 13C-NMR (500 MHz, DMSO-d6): δ = 14.11, 32.31, 55.93, 63.19, 104.97, 114.67, 115.32, 117.82, 130.05, 130.87, 132.56, 144.81, 154.59, 156.73, 164.07, 170.63. MS: m/z = 373 (M+, 9%), 288 (17%), 272 (29%), 244 (100%), 243 (8%), 197 (11%), 109 (4%).
3.4.31. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(2,6-dichlorophenyl) thiazolidin-4-one (29)
Yield: 73%, m.p. 164–165°C, Rf: 0.53 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1700 (C=O), 1532, 1209, 1134, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.99 (d, J = 16.63 Hz, 1H, CH2), 4.26 (d, J = 16.63 Hz, 1H, CH2), 6.84 (s, 1H, N-CH-S), 6.97 (dd, J = 8.80, 2.45 Hz, 1H, Ar-C5), 7.42–7.46 (m, 3H, Ar-C18, C19, C20), 7.49–7.71 (m, 2H, Ar-C4, C7).13C-NMR (500 MHz, CHCl3-d): δ = 32.86, 63.33, 104.97, 114.22, 118.71, 126.82, 128.53, 129.27, 130.05, 135.11, 139.11, 145.91, 156.74, 164.13, 171.17. MS: m/z = 446 (M+, 47%), 340 (21%), 336 (16%), 321 (44%), 293 (100%), 292 (11%), 204 (46%), 165 (21%).
3.4.32. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(2,3-dichlorophenyl) thiazolidin-4-one (30)
Yield: 84%, m.p. 142–143 °C, Rf: 0.81 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 2361, 1700 (C=O), 1532, 1202, 1131, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.94 (d, J = 16.63 Hz, 1H, CH2), 4.11 (d, J = 16.63 Hz, 1H, CH2), 6.83 (s, 1H, N-CH-S), 6.91–7.01 (m, 2H, Ar-C5, C17), 7.10–7.18 (m, 1H, Ar-C18), 7.49–7.53 (m, 2H, Ar-C4, C7), 7.60 (dd, J = 8.80, 2.45 Hz, 1H, Ar-C19). 13C-NMR (500 MHz, CHCl3-d): δ = 32.85, 63.34, 103.85, 104.92, 114.66, 118.97, 127.95, 128.05, 128.77, 129.23, 129.56, 131.81, 132.05, 145.52, 156.73, 164.13, 170.93. MS: m/z = 466 (M+, 3%), 355 (6%), 272 (18%), 244 (36%), 228 (15%), 226 (22%), 165 (100%).
3.4.33. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(4-nitrophenyl)thiazolidin-4-one (31)
Yield: 33%, m.p. 158–159 °C, Rf: 0.62 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1706(C=O), 1539, 1209, 1135. 1H-NMR (500 MHz, CHCl3-d): δ = 3.85 (d, J = 16.63 Hz, 1H, CH2), 4.14 (d, J = 16.63 Hz, 1H, CH2), 6.76 (s, 1H, N-CH-S), 7.02 (d, J = 7.83 Hz, 1H, Ar-C5), 7.41–7.45 (m, 4H, Ar-C4, C7, C17, C21), 8.05 (d, J = 7.83 Hz, 2H, Ar-C18, C20). 13C-NMR (500 MHz, CHCl3-d): δ = 32.76, 63.09, 104.92, 114.02, 118.27 123.88, 129.12, 129.93, 131.95, 145.62, 145.94, 149.79, 156.71, 164.19, 170.92. MS: m/z = 442 (M+, 56%), 395 (51%), 386 (19%), 272 (100%), 244 (22%), 165 (7%).
3.4.34. Synthesis of 3-(6-Trifluoromethoxy-benzo[d]thiazol-2-yl)-2-(4-chlorophenyl)thiazolidin-4- one (32)
Yield: 35%, m.p. 243–244 °C, Rf: 0.77 (petroleum ether/ethyl acetate 8:2). IR (Nujol), cm−1: 1698 (C=O), 1528, 1209, 1198, 1131, 721. 1H-NMR (500 MHz, CHCl3-d): δ = 3.85 (d, J = 16.63 Hz, 1H, CH2), 4.10 (d, J = 16.63 Hz, 1H, CH2), 6.74 (s, 1H, N-CH-S), 7.24–7.31(m, 5H, Ar-C5, C17, C18, C20, C21), 7.66–7.71 (m, 2H, Ar-C4, C7). 13C-NMR (500 MHz, CHCl3-d): δ = 32.75, 63.05, 104.93, 114.01, 120.11, 128.27, 129.65, 129.81, 131.74, 132.92, 137.33, 145.95, 156.63, 161.62, 164.27, 170.94. MS: m/z = 431 (M+, 46%), 395 (27%), 378 (34%), 377 (100%), 373 (18%), 363 (55%).
3.5. Evaluation of RT Inhibitory Action
For the evaluation of the HIV-1 Reverse Transcriptase activity, the colorimetric photometric immunoassay kit provided by Roche was used. A two hours incubation at 37 °C was performed, as previously described in our paper [44] following a pre-incubation step in which reverse transcriptase was incubated in the presence of the potential inhibitor for 45 min at room temperature before the addition of the substrate. The total-activity was measured in the absence of inhibitors (compounds) with the addition of an equal volume of the solvent of the compounds to the reaction mixture. For the determination of enzyme activity, incubation in the absence of enzyme was used as blank. Different inhibitor concentrations were added for the calculation of IC50 values.
3.6. Toxicity
Cell culture and cytotoxicity assessment.
The normal MRC-5 cell line is stored and used in our laboratory in a routine manner. MRC-5 cells were grown in culture (37 °C, humidified atmosphere containing 5% v/v CO2) in DMEM medium supplemented with 10% v/v FBS, 100 μg/mL penicillin and 100 μg/mL streptomycin. The compounds under evaluation were dissolved in DMSO and stored in 4 °C (DMSO concentration ≤ 0.2% v/v). The cytotoxicity of the 1, 2, 3, 8, 9, 10, 14 compounds were evaluated in MRC-5 cell cultures. In particular cells were seeded in a 24- well plate at an initial concentration of 1 × 105 cells/mL and allowed to stand overnight before the addition of the compounds at high concentration of 10−5 M. Note that DMSO concentration in culture was 0.2% v/v, where this concentration exhibits no cytotoxicity at all [50]. To estimate the cytotoxicity of each compound, cells were allowed to grow for additional 48h before being harvested by trypsinization and counted with the aid of the optical microscope (Neubauer counting chamber). Cell growth in each treated culture is expressed as a percentage compared to that seen for the untreated control cells. Moreover, cell death was also determined using the Trypan-blue method, as previously described [51,52]. Statistical t-test analysis was performed via the use of GraphPad Prism 6.0 program.
4. Conclusions
Thirty two compounds out of a hundred thirty two compounds designed were selected based on PASS prediction results and molecular docking for the synthesis and evaluation of HIV-1 RT inhibitory activity. Twenty-four out of the thirty-two tested compounds exhibited inhibitory action equal or lower than 4 μΜ. Seven of them showed better activity than nevirapine under experimental condition, while three of the compounds exhibited IC50 values lower than 5 nM. Two compounds 9 and 10 exhibited very good inhibitory activity with IC50 1 nM.
According to docking results, most of the compounds are placed within the NNRTIs binding pocket between the place equipped by the classic butterfly shaped compound, nevirapine and the binding site of etravirine or within the etravirine binding pocket.
It is interesting to mention that, the most active compounds structurally belong to three different groups and adopt three different orientations.
In general, according to docking studies and in vitro results, it seems that the presence of a Cl substituent at the 7-, 6- or 4-position of the benzothiazolyl moiety facilitates an orientation which enables complex stabilization via pi-interactions and hydrogen bond formation involving either the CO group of the thiazolidinone ring or hydrogen acceptor/donor substituents of the phenyl ring. In case of subgroups bearing hydrogen acceptor substituents at the benzothiazole moiety, their involvement in hydrogen bond formation also is favorable for complex stabilization.
The absence of the traditional hydrogen bond interactions with Lys101, in case of one of the most active compounds, 10, is an interesting characteristic in concern of a probable use of such structures in future drugs, since mutations in Lys101 are common in resistant mutants.
Finally, it should be mentioned that no statistically significant changes were observed in cell growth in normal cell line treated with the tested compounds compared to control.
Author Contributions
A.P.—synthesis and characterization, molecular modeling; P.E.—docking, original draft preparation, data curation; A.G.—PASS, data curation, review and editing, supervision, M.G.A. and I.V.—evaluation of cytotoxicity.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ravichandran, S.; Veerasamy, F.; Raman, S.; Krishnan, P.N.; Agrawal, R.K. An overview on HIV-1 reverse transcriptase inhibitors. Dig. J. Nanomater. Biostructures 2008, 3, 171–187. [Google Scholar]
- Asahchop, E.L.; Wainberg, M.A.; Sloan, R.D.; Tremblay, C.L. Antiviral drug resistance and the need for development of new HIV-1 reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 2012, 56, 5000–5008. [Google Scholar] [CrossRef] [PubMed]
- Reeves, J.D.; Doms, R.W. Human immunodeficiency virus type 2. J. Gen. Virol. 2002, 83, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Andries, K.; Azijn, H.; Thielemans, T.; Ludovici, D.; Kukla, M.; Heeres, J.; Janssen, P.; De Corte, B.; Vingerhoets, J.; Pauwels, R.; et al. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2004, 48, 4680–4686. [Google Scholar] [CrossRef]
- Kaushik, N.K.; Kaushik, N.; Attri, P.; Kumar, N.; Kim, C.H.; Verma, A.K.; Choi, E.H. Biomedical importance of indoles. Molecules 2013, 18, 6620–6662. [Google Scholar] [CrossRef]
- Kadow, J.F.; Regueiro-Ren, A.; Xue, Q.M. Indole, Azaindole and Related Heterocyclic Sulfonylureido Piperazine Derivatives. US Patent 6,900,206, 9 June 2003. [Google Scholar]
- Pauwels, R.; Andries, K.; Debyser, Z.; Van Daele, P.; Schols, D.; Stoffels, P.; De Vreese, K.; Woestenborghs, R.; Vandamme, A.M.; Janssen, C.G. Potent and highly selective human immunodeficiency virus type 1 (HIV-1) inhibition by a series of alpha-anilinophenylacetamide derivatives targeted at HIV-1 reverse transcriptase. Proc. Nat. Acad. Sci. USA 1993, 90, 1711–1717. [Google Scholar] [CrossRef]
- Pauwels, R.; Andries, K.; Desmyter, J.; Schols, D.; Kukla, M.J.; Breslin, H.J.; Raeymaeckers, A.; VanGelder, J.; Woestenborghs, R.; Heykants, J.; et al. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBOderivatives. Nature 1990, 343, 470–474. [Google Scholar] [CrossRef]
- Debyser, Z.; Pauwels, R.; Andries, K.; Desmyter, J.; Kukla, M.; Janssen, P.A.; De Clercq, E. An antiviral target on reverse transcriptase of human immunodeficiency virus type 1 revealed bytetrahydroimidazo-[4,5,1-jk][1,4]benzodiazepin-2 (1H)-one and -thione derivatives. Proc. Nat. Acad. Sci. USA 1991, 88, 1451–1455. [Google Scholar] [CrossRef]
- Le Van, K.; Cauvin, C.; Walque, S.; Georges, B.; Boland, S.; Martinelli, V.; Demonté, D.; Durant, F.; Hevesi, L.; Van Lint, C. New pyridinone derivatives as potent HIV-1 nonnucleoside reverse transcriptase inhibitors. J. Med. Chem 2009, 52, 3636–3643. [Google Scholar] [CrossRef][Green Version]
- Tanaka, H.; Takashima, H.; Ubasawa, M.; Sekiya, K.; Inouye, N.; Baba, M.; Shigeta, S.; Walker, R.T.; De Clercq, E.; Miyasaka, T.J. Synthesis and antiviral activity of 6-benzyl analogs of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT)as potent and selective anti-HIV-1 agents. Med. Chem. 1995, 38, 2860–2865. [Google Scholar] [CrossRef]
- Penta, A.; Ganguly, S.; Murugesan, S. Design and synthesis of tetrahydrophthalimide derivatives as inhibitors of HIV-1 reverse transcriptase. Org. Med. Chem. Lett. 2013, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Rawal, R.K.; Tripathi, R.; Kulkarni, S.; Paranjape, R.; Katti, S.B.; Pannecouque, C.; De Clercq, E. 2-(2,6-Dihalo-phenyl)-3-heteroaryl-2-ylmethyl-1, 3-thiazolidin-4-ones: Anti-HIVagents. Chem. Biol. Drug Des. 2008, 72, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Khalid, Z.; Aslam, S.; Ahmad, M.; Munawar, A.; Montero, K.; Detorio, M.; Parvez, M.; Schinazi, R.F. Anti-HIV activity of new pyrazolobenzothiazine 5,5-dioxide-based acetohydrazides. Med. Chem. Res. 2015, 24, 3671–3680. [Google Scholar] [CrossRef]
- Novikov, M.S.; Valuev-Elliston, V.T.; Babkov, D.A.; Paramonova, M.P.; Ivanov, A.V.; Gavryushov, S.A.; Khandazhinskaya, A.L.; Kochetkov, S.N.; Pannecouque, C.; Andrei, G.; et al. N1,N3-disubstituted uracils as nonnucleosides inhibitors of HIV-1 reverse transcriptase. Bioorg. Med. Chem. 2013, 21, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Chen, F.-E.; De Clercq, E.; Balzarini, J.; Pannecouque, C. Synthesis and in vitroanti-HIV evaluation of a new series of 6-arylmethyl-substituted S-DABOs as potentialnon-nucleoside HIV-1 reverse transcriptase inhibitors. Eur. J. Med. Chem. 2009, 41, 1016–1023. [Google Scholar] [CrossRef]
- Pitta, E.; Tsolaki, E.; Geronikaki, A.; Petrović, J.; Glamočlija, J.; Soković, M.; Crespan, E.; Maga, G.; Bhunia, S.S.; Saxena, A.K. 4-Thiazolidinone derivatives as potent antimicrobial agents: Microwave-assisted synthesis, biological evaluation and docking studies. MedChemComm. 2015, 6, 319–326. [Google Scholar] [CrossRef]
- Zahid, M.; Yasin, K.A.; Akhtar, T.; Rama, N.H.; Hameed, S.; Al-Masoudi, N.A.; Loddo, R.; Colla, P. Synthesis and in vitro antiproliferative activity of new adamantylthiazolyl-1,3,4-oxadiazoles. ARKIVOC 2009, xi, 85–93. [Google Scholar]
- La Regina, G.; Coluccia, A.; Piscitelli, F.; Bergamini, A.; Sinistro, A.; Cavazza, A.; Maga, J.; Samuele, A.; Zanoli, S.; Novellino, E.; et al. Indolyl aryl sulfones as HIV-1 non-nucleoside reverse transcriptase inhibitors: Role of two halogen atoms at the indole ring in developing new analogues with improved antiviral activity. J. Med. Chem. 2007, 50, 5034–5038. [Google Scholar] [CrossRef]
- Akkouh, O.; Ng, T.B.; Singh, S.S.; Yin, C.; Dan, X.; Chan, Y.S.; Pan, W.; Cheung, R.C.F. Lectins with Anti-HIV activity: A review. Molecules 2015, 20, 648–668. [Google Scholar] [CrossRef]
- Famiglini, V.; Coluccia, A.; Brancale, A.; Pelliccia, S.; La Regina, G.; Silvestri, R. Arylsulfone-based HIV-I non-nucleoside reverse transcriptase inhibitors. Future. Med. Chem. 2013, 5, 2141–2156. [Google Scholar] [CrossRef]
- De Clercq, E. Dancing with chemical formulae of antivirals: A personal account. Biochem. Pharm. 2013, 86, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Veljkovic, N.; Glisic, S.; Prljic, J.; Perovic, V.; Veljkovic, V. Simple and general criterionfor “in silico” screening of candidate HIV drugs. Curr. Pharm. Biotechnol. 2013, 14, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhan, P.; Liu, H.; Pannecouque, C.; Balzarini, J.; De Clercq, E. Synthesis and biological evaluation of pyrazine derivatives as novel HIV-1 NNRTIs. Bioorg. Med. Chem. 2013, 21, 2128–2134. [Google Scholar] [CrossRef] [PubMed]
- La Regina, G.; Coluccia, A.; Brancale, A.; Piscitelli, F.; Gatti, V.; Maga, G.; Sa-muele, A.; Pannecouque, C.; Schols, D.; Balzarini, J.; et al. Indolylarylsulfones as HIV-1 non-nucleoside reverse transcriptase inhibitors: New cyclic substituents at indole-2-carboxamide. J. Med. Chem. 2011, 54, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- La Regina, G.; Coluccia, A.; Brancale, A.; Piscitelli, F.; Famiglini, V.; Cosconati, S.; Maga, G.; Samuele, A.; Gonzales, E.; Clotet, B.; et al. New nitrogen containing substituents at the indole-2-carboxamide yield high potent and broad spectrum indolylarylsulfone HIV-1 non-nucleoside reverse transcriptase inhibitors. J. Med. Chem. 2012, 55, 6634–6638. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Cheng, X.; Sun, L.; Song, S.; Álvarez, M.; Luczkowiak, J.; Pannecouque, C.; De Clercq, E.; Menéndez-Arias, L.; Zhan, P.; et al. Design, synthesis and biological evaluation of 3-hydroxyquinazoline-2,4(1H,3H)-diones as dual inhibitors of HIV-1 reverse transcriptase-associated RNase H and integrase. Bioorg. Med. Chem. 2019, 27, 3836–3845. [Google Scholar] [CrossRef]
- Tian, Y.; Zhan, P.; Rai, D.; Zhang, J.; Clercq, E.D.; Liu, X. Recent advances in the research of 2,3-diaryl-1,3-thiazolidin-4-one derivatives as potent HIV-1 non-nucleoside reverse transcriptase inhibitors. Curr. Med. Chem. 2012, 19, 2026–2037. [Google Scholar] [CrossRef]
- Suryawanshi, R.; Jadhav, S.; Makwana, N.; Desai, D.; Chaturbhuj, D.; Sonawani, A.; Idicula-Thomas, S.; Murugesan, V.; Katti, S.; Tripathy, S.; et al. Evaluation of 4-thiazolidinone derivatives as potential reverse transcriptase inhibitors against HIV-1 drug resistant strains. Bioorg. Chem. 2017, 71, 211–218. [Google Scholar] [CrossRef]
- Rao, A.; Balzarini, J.; Carbone, A.; Chimirri, A.; De Clercq, E.; Monforte, A.; Monforte, P.; Pannecouque, C.; Zappala, M. 2-(2,6-Dihalophenyl)-3-(pyrimidin-2-yl)-1,3-thiazolidin-4-ones as non-nucleoside HIV-1 reverse transcriptase inhibitors. Antivir. Res. 2004, 63, 79–84. [Google Scholar] [CrossRef]
- Chen, H.; Guo, Z.; Yin, Q.; Duan, X.; Gu, Y.; Li, X. Design, synthesis and HIV-RT inhibitory activity of novel thiazolidin-4-one derivatives. Front. Chem. Sci. Eng. 2011, 5, 231–237. [Google Scholar] [CrossRef]
- Rawal, K.; Prabhakar, Y.; Katti, B.V. Molecular surface features in modeling the HIV-1 RT inhibitory activity of 2-(2,6-disubstituted phenyl)-3-(substituted pyrimidin-2-yl)-thiazolidin-4-ones. QSAR Comb. Chem. Sci. 2007, 26, 398. [Google Scholar] [CrossRef]
- Kumar, S.; Purohit, D.; Pandey, P. Molecular docking and its application towards modern drug discovery. World J. Pharm. Pharm. Sci. 2019, 6, 691–696. [Google Scholar]
- Eweas, A.F.; Maghrabi, I.A.; Namarneh, A.I. Advances in molecular modeling and docking as a tool for modern drug discovery. Der Pharma Chem 2014, 6, 211–228. [Google Scholar]
- Zhou, Z.; Lin, X.; Madura, J.D. HIV-1 RT Nonnucleoside inhibitors and their interaction with RT for antiviral drug development. Infect. Disord. Drug Targets 2006, 6, 391–413. [Google Scholar] [CrossRef]
- De Clercq, E. Where rilpivirine meets with tenofovir, the start of a new anti-HIV drug combination era. Biochem. Pharm. 2012, 84, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskiy, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar]
- Geronikaki, A.; Dearden, J.C.; Filimonov, D.; Galaeva, I.; Garibova, T.L.; Gloriozova, T.; Kraineva, V.; Lagunin, A.; Macaev, F.Z.; Molodavkin, G.; et al. Design of new cognition enhancers: From computer prediction to synthesis and biological evaluation. J. Med. Chem. 2004, 47, 2870–2876. [Google Scholar] [CrossRef] [PubMed]
- Geronikaki, A.; Babaev, E.; Dearden, J.; Dehaen, W.; Filimonov, D.; Galaeva, I.; Kraineva, V.; Lagunin, A.; Macaev, F.; Molodavkin, G.; et al. Design, synthesis, computational and biological evaluation of new anxiolytics. Bioorg. Med. Chem. 2004, 12, 6559–6568. [Google Scholar] [CrossRef] [PubMed]
- Famiglini, V.; Silvestri, R. Focus on chilarity of HIV-1 non-nucleoside reverse transcriptase inhibitors. Molecules 2016, 21, 221. [Google Scholar] [CrossRef]
- Balzarini, J.; Orzeszko, B.; Maurin, J.; Orzerszko, A. Synthesis and anti-HIV studies of 2-adamantyl-substituted thiazolidin-4-ones. Eur. J. Med. Chem. 2007, 42, 993–1003. [Google Scholar] [CrossRef]
- Chimirri, A.; Grasso, S.; Molica, C.; Monforte, A.-M.; Monforte, P.; Zappalà, M.; De Clercq, E. Structural features and anti-human immunodeficiency virus (HIV) activity of the isomers of 1-(2′,6′-difluorophenyl)-1H,3H-thiazolo[3,4-a]benzimidazole, a potent non-nucleoside HIV-1 reverse transcriptase inhibitor. Antivir. Chem. Chemother. 1997, 8, 363–370. [Google Scholar] [CrossRef]
- Prajapati, D.; Ramajayam, R.; Ram Yadar, M.; Giridhar, R. The search for potent, small molecule NNRTIs: A review. Biorg. Med. Chem. 2009, 17, 5744–5762. [Google Scholar] [CrossRef] [PubMed]
- Pitta, E.; Geronikaki, A.; Surmava, S.; Eleftheriou, P.; Mehta, V.P.; Van der Eycken, E.V. Synthesis and HIV-1 RT inhibitory action of novel (4/6-substituted benzo[d]thiazol-2-yl)thiazolidin-4-ones. Divergence from the non-competitive inhibition mechanism. J. Enzyme Inhib. Med. Chem. 2013, 8, 113–122. [Google Scholar] [CrossRef]
- Kouatly, O.; Eleftheriou, P.; Petrou, A.; Hadjipavlou-Litina, D.; Geronikaki, A. Docking assisted design of novel 4-adamantanyl-2-thiazolylimino-5-arylidene-4-thiazolidinones as potent NSAIDs. SAR QSAR Environ. Res. 2018, 29, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.rcsb.org/structure/3MEC (accessed on 9 November 2016).
- Nadaraia, N.S.; Amiranashvili, L.S.; Merlani, M.; Kakhabrishvili, M.L.; Barbakadze, N.N.; Geronikaki, A.; Petrou, A.; Poroikov, V.; Ciric, A.; Glamoclija, J.; et al. Novel antimicrobial agents’ discovery among the steroid derivatives. Steroids 2019, 144, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Helma, C. Lazy structure-activity relationships (lazar) for the prediction of rodent carcinogenicity and Salmonella mutagenicity. Mol. Divers. 2006, 10, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Maunz, A.; Helma, C. Prediction of chemical toxicity with local support vector regression and activity-specific kernels. Sar. Qsar Environ. Res. 2008, 19, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Vizirianakis, I.S.; Tsiftsoglou, A.S. Blockade of murine erythroleukemia cell differentiation by hypomethylating agents causes accumulation of discrete small poly(A)- RNAs hybridized to 3′-end flanking sequences of beta(major) globin gene. Biochim. Biophys. Acta 2005, 1743, 101–114. [Google Scholar] [CrossRef][Green Version]
- Akrivou, M.G.; Demertzidou, V.P.; Theodoroula, N.F.; Chatzopoulou, F.M.; Kyritsis, K.A.; Grigoriadis, N.; Zografos, A.L.; Vizirianakis, I.S. Uncovering the pharmacological response of novel sesquiterpene derivatives that differentially alter gene expression and modulate the cell cycle in cancer cells. Int. J. Oncol. 2018, 53, 2167–2179. [Google Scholar] [CrossRef]
- Tseligka, E.D.; Rova, A.; Amanatiadou, E.P.; Calabrese, G.; Tsibouklis, J.; Fatouros, D.G.; Vizirianakis, I.S. Pharmacological Development of Target-Specific Delocalized Lipophilic Cation-Functionalized Carboranes for Cancer Therapy. Pharm. Res. 2016, 33, 1945–1958. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).