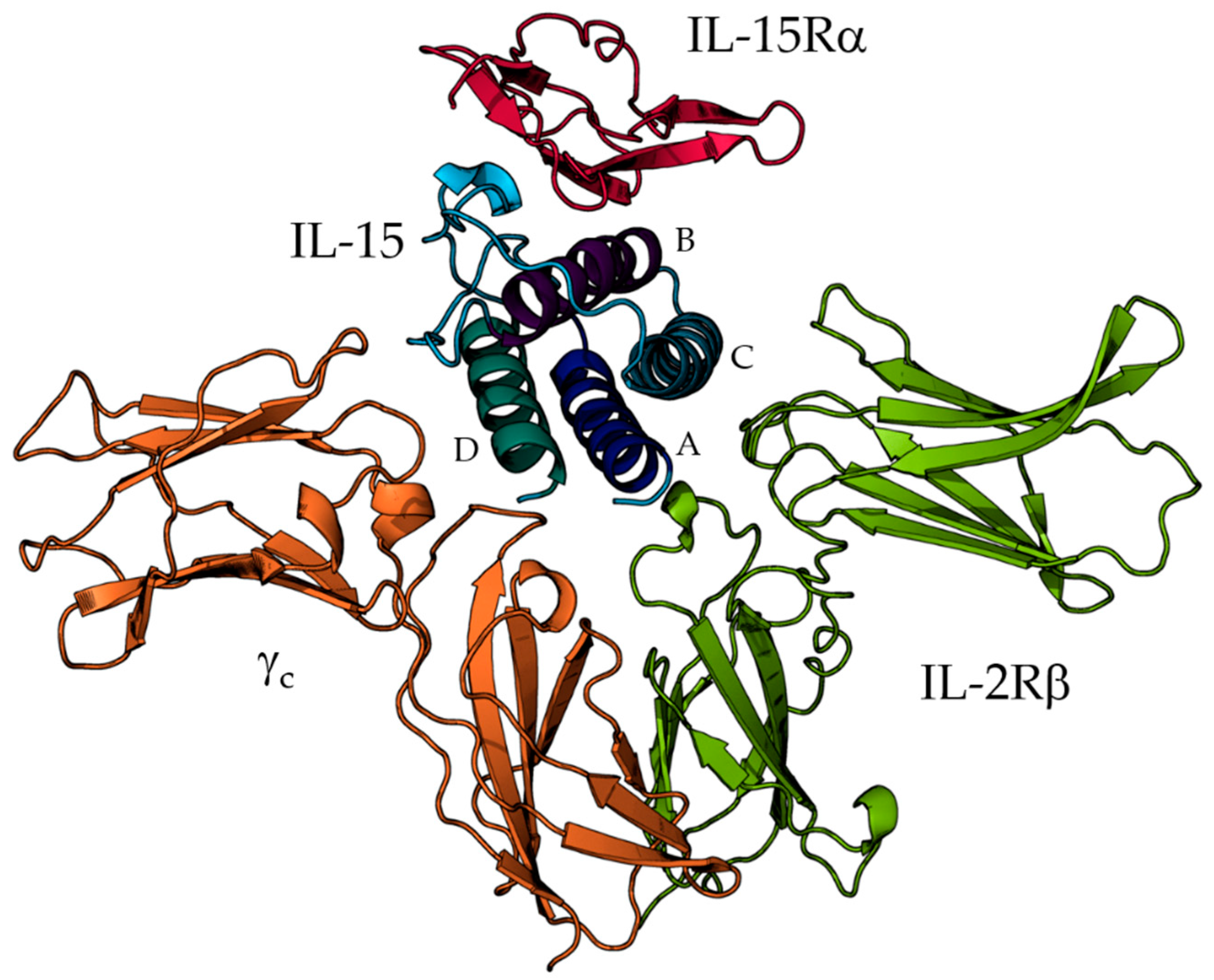
Mechanistic and Structural Insights on the IL-15 System through Molecular Dynamics Simulations
Abstract
1. Introduction
2. Methods
2.1. Structure Preparation
2.2. Molecular Dynamics Simulations
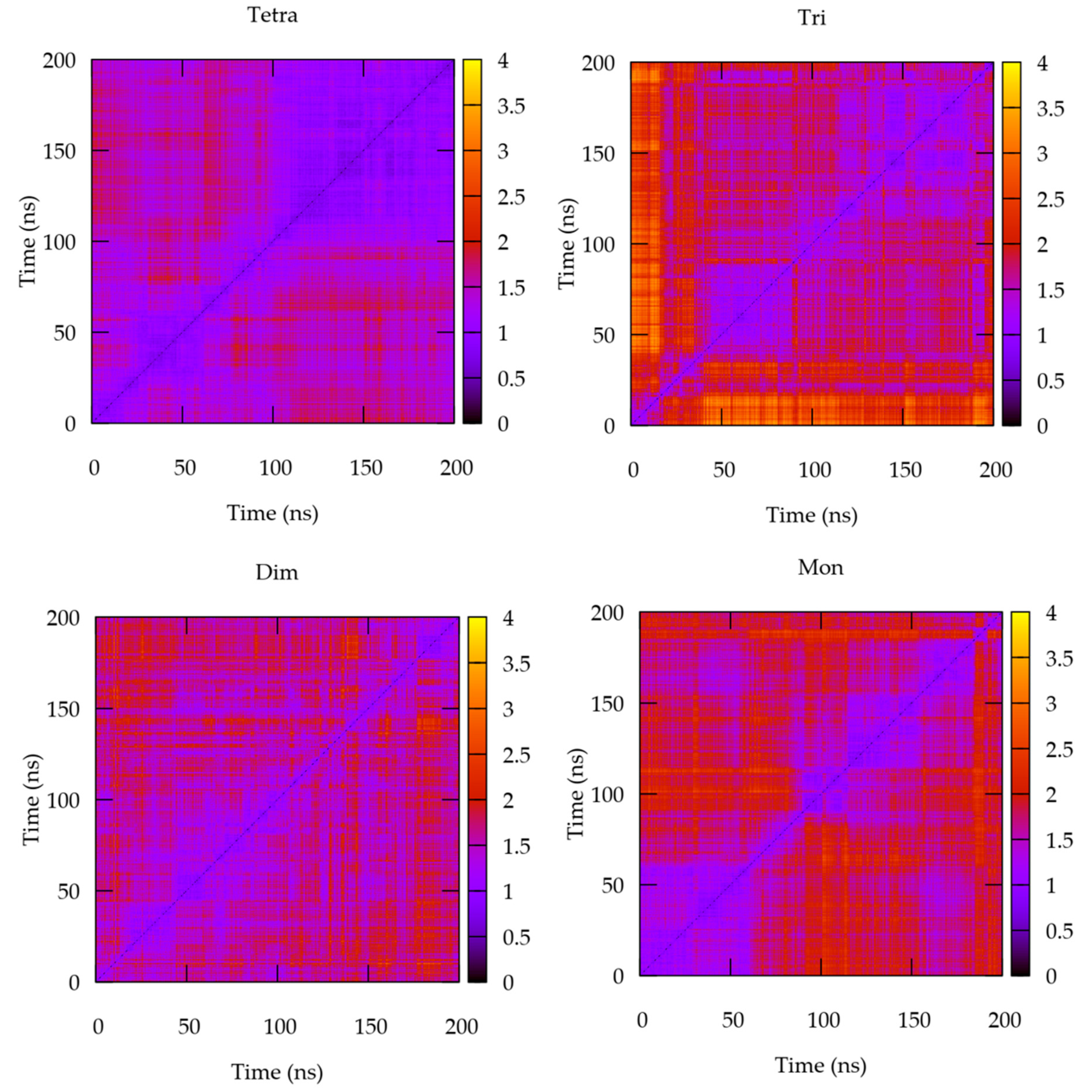
2.3. MD Analyses
3. Results and Discussion
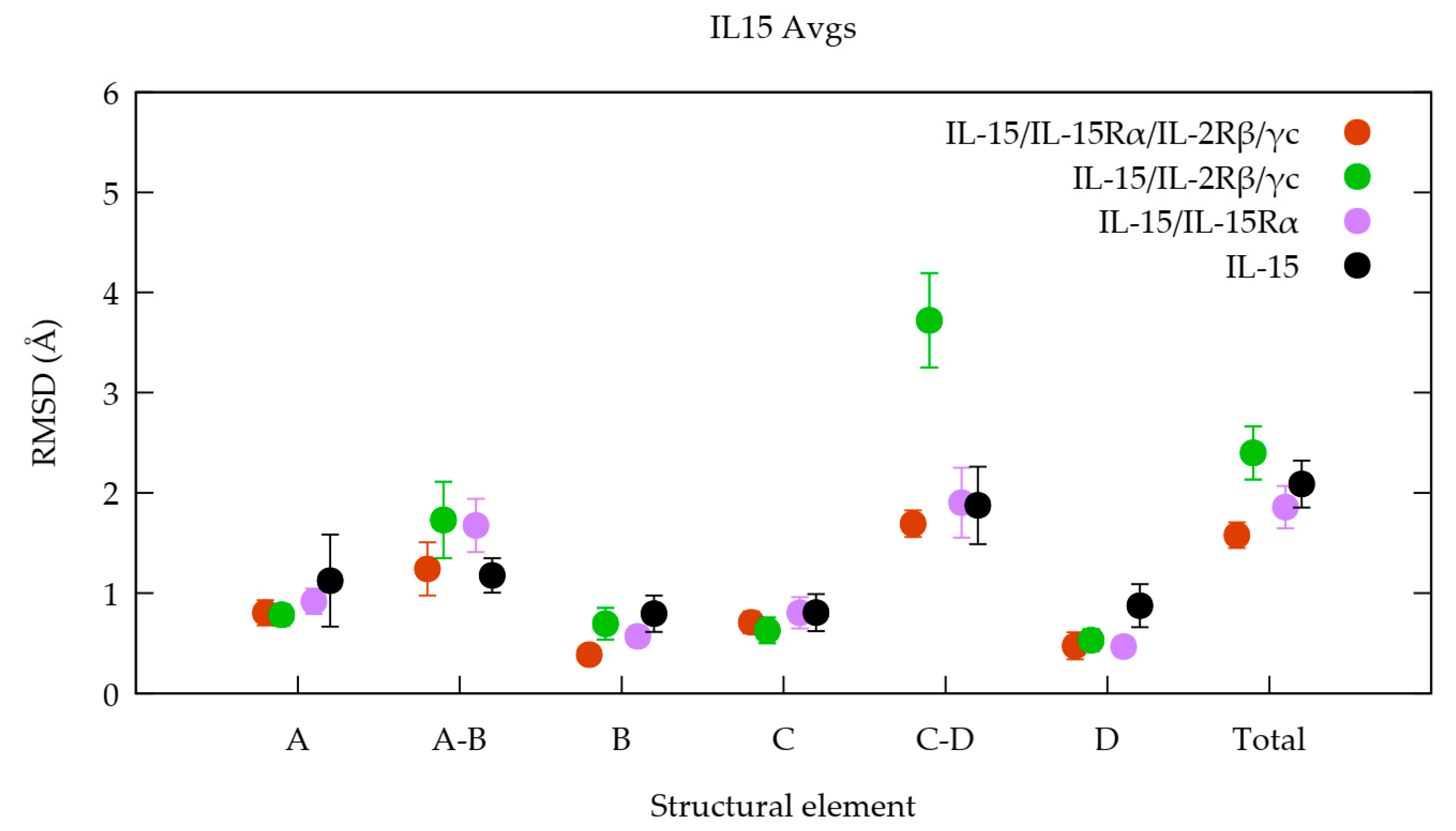
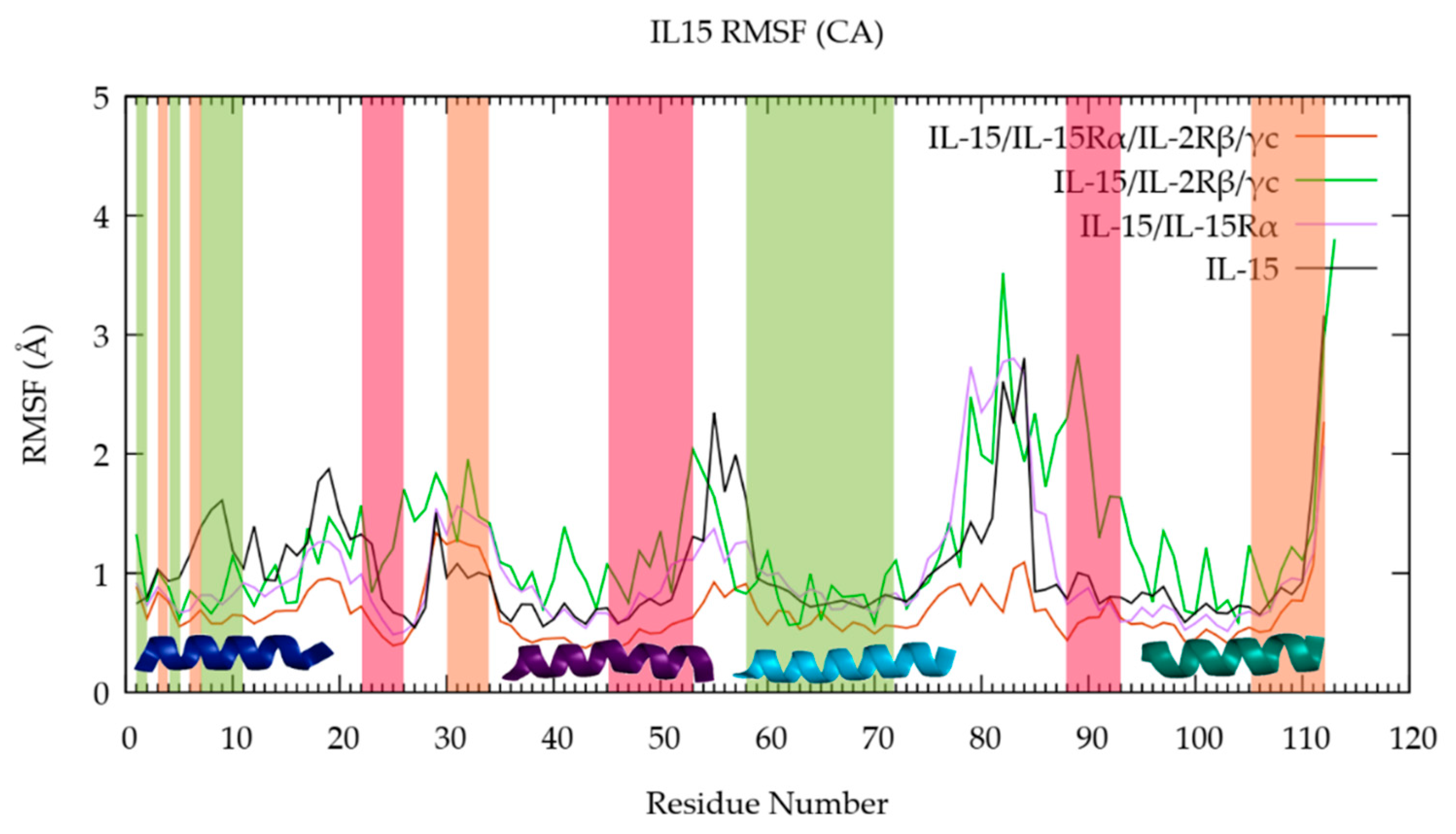
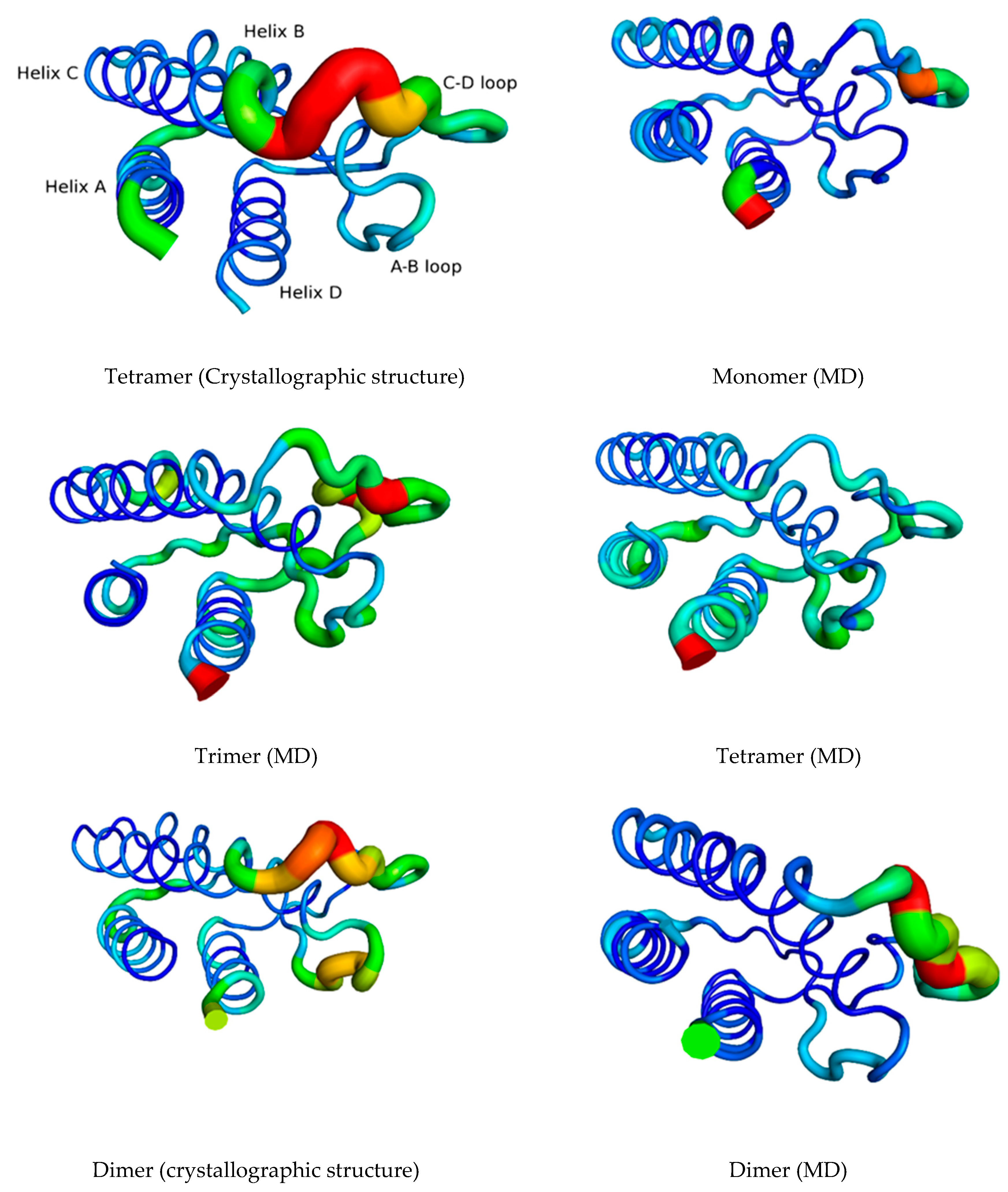
3.1. The Quaternary Structure Impacts the Flexibility of the IL-15 Receptor Interfaces
RMSF
3.2. The Quaternary Structure Impacts the Structural and Energetic Features of the Various Interfaces
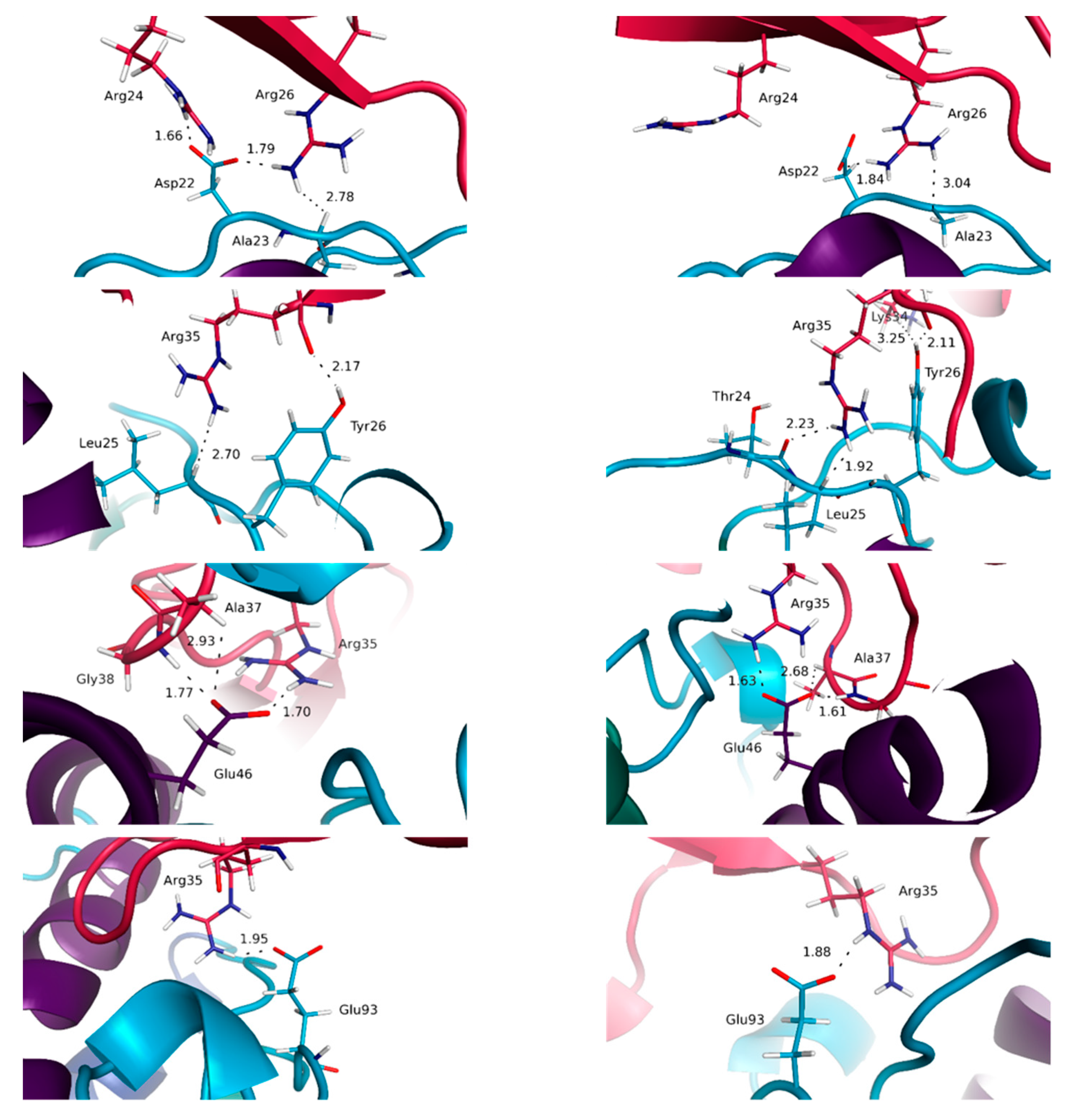
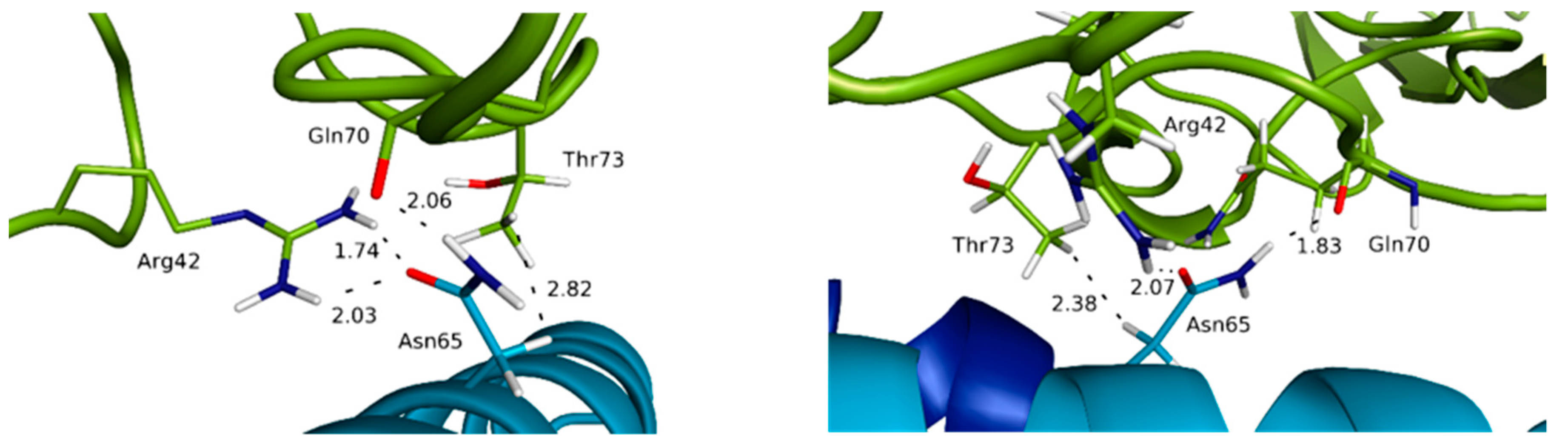
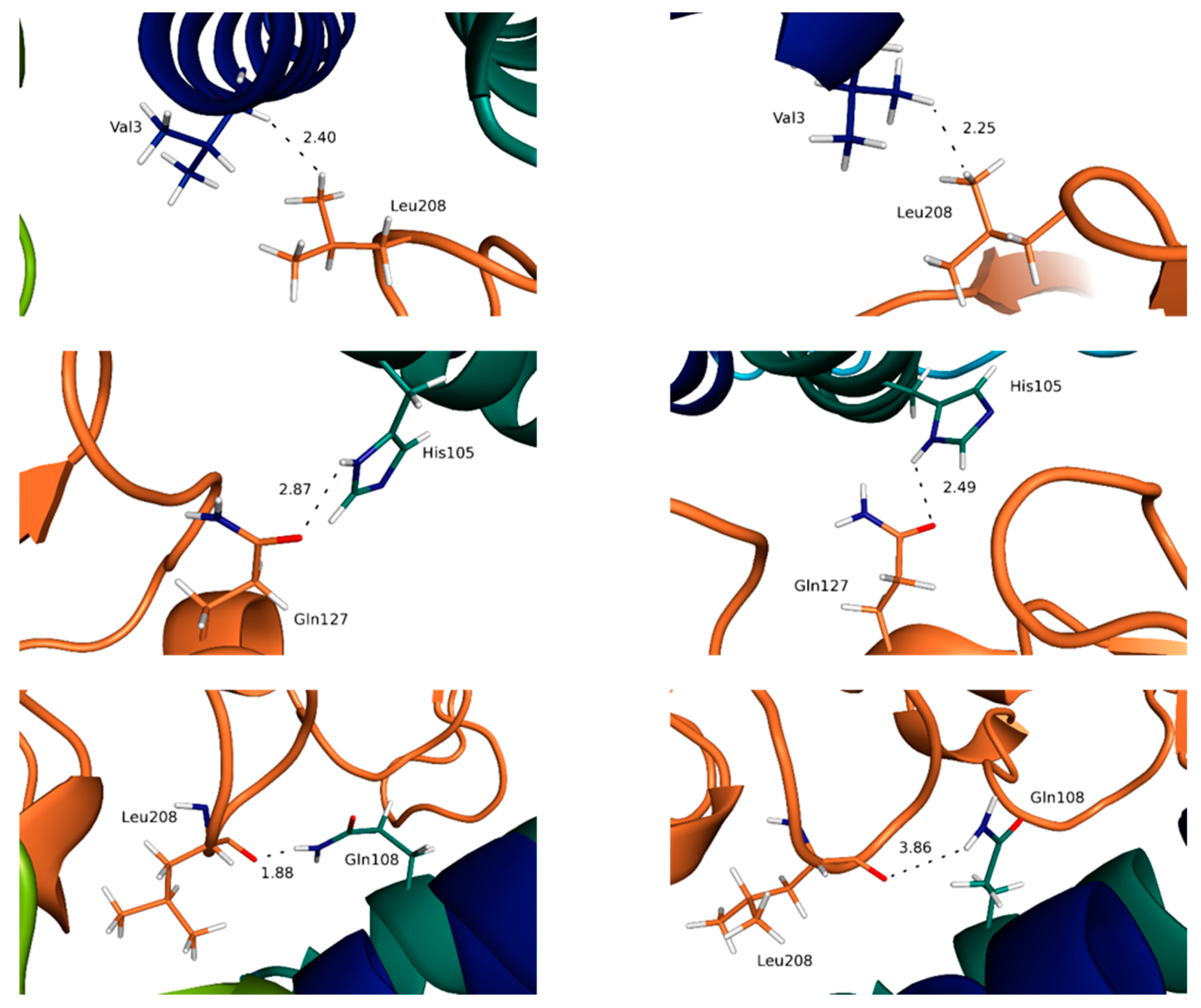
3.3. Highlighting Novel Key Structural Features at IL-15 Interfaces
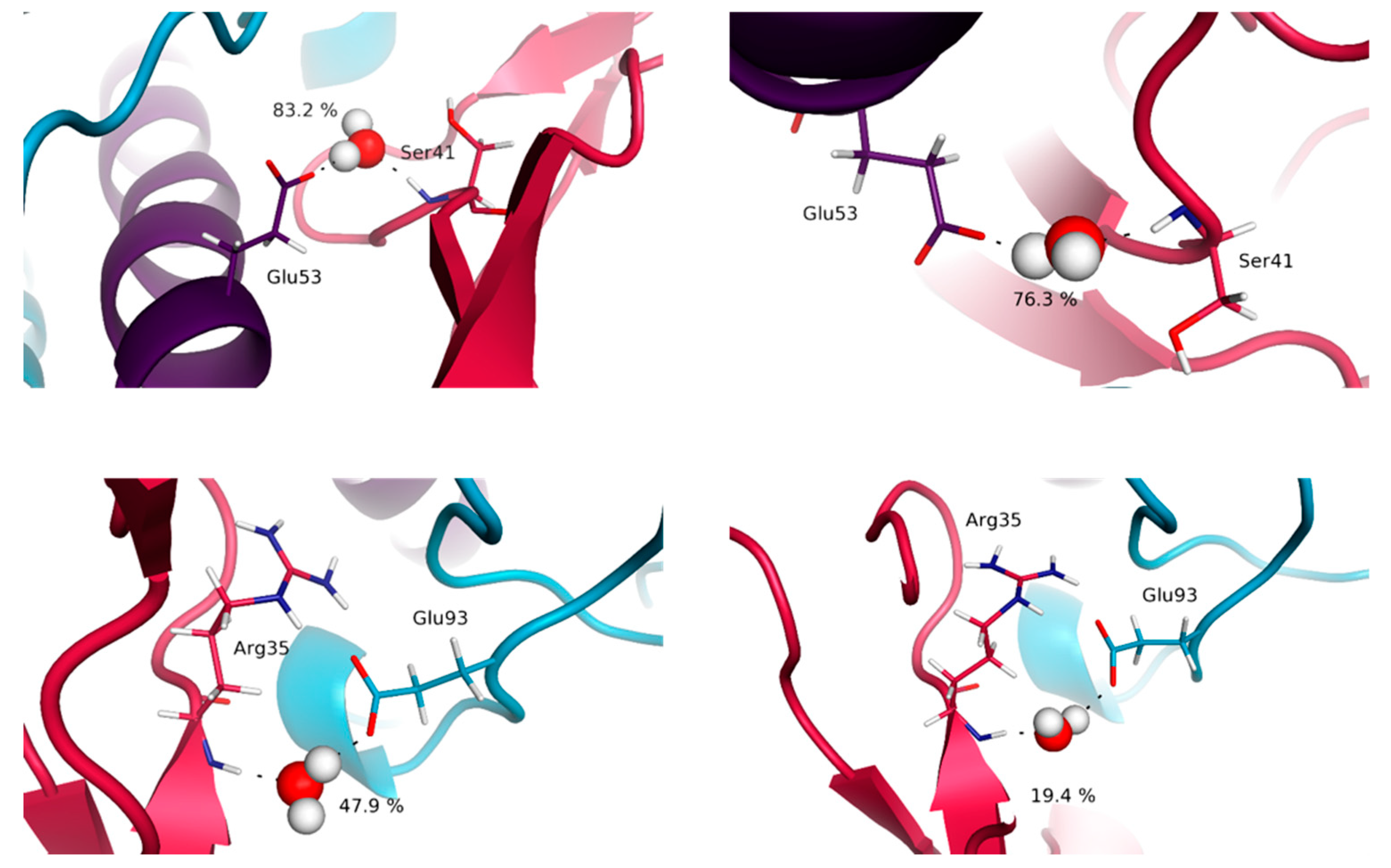
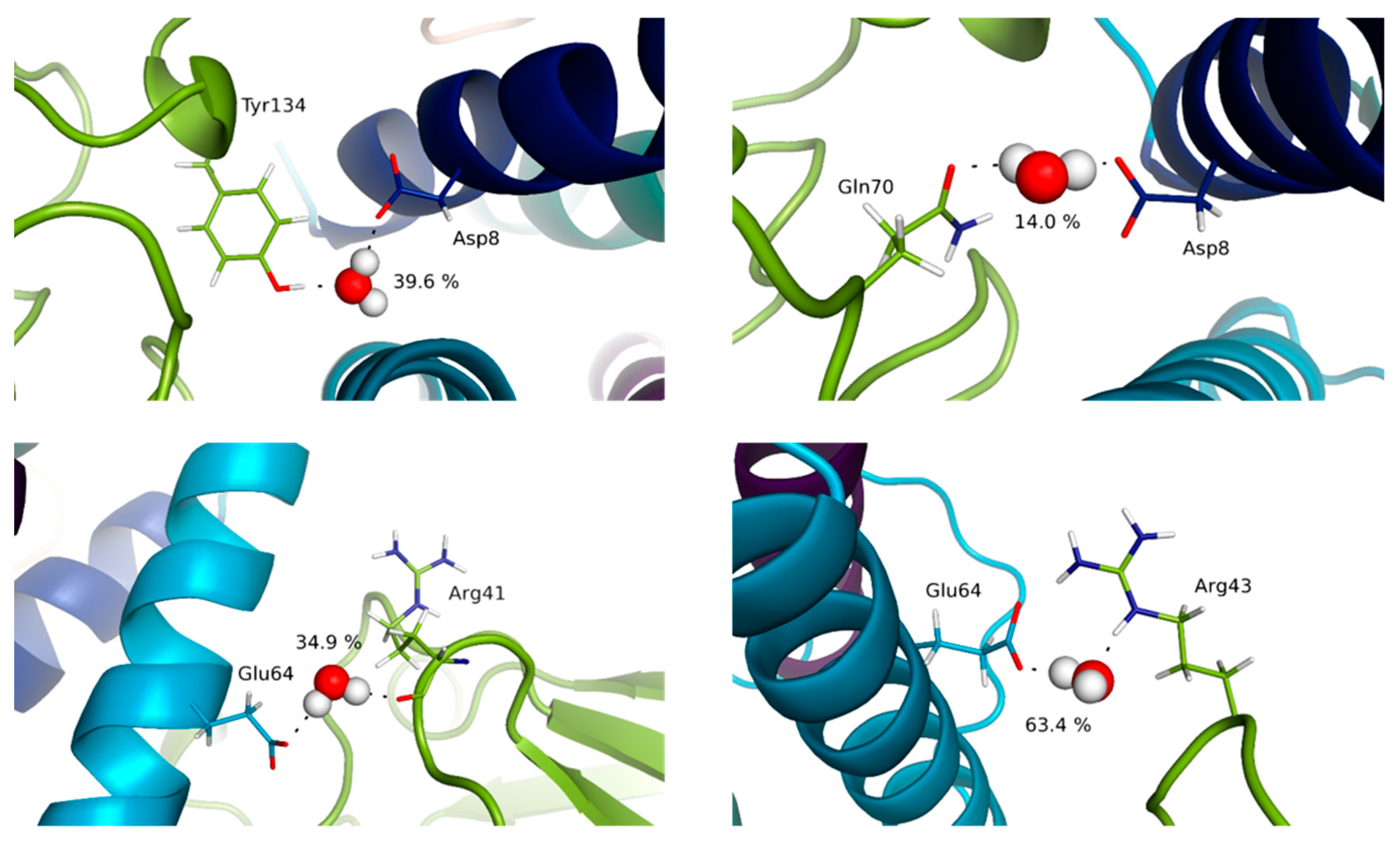
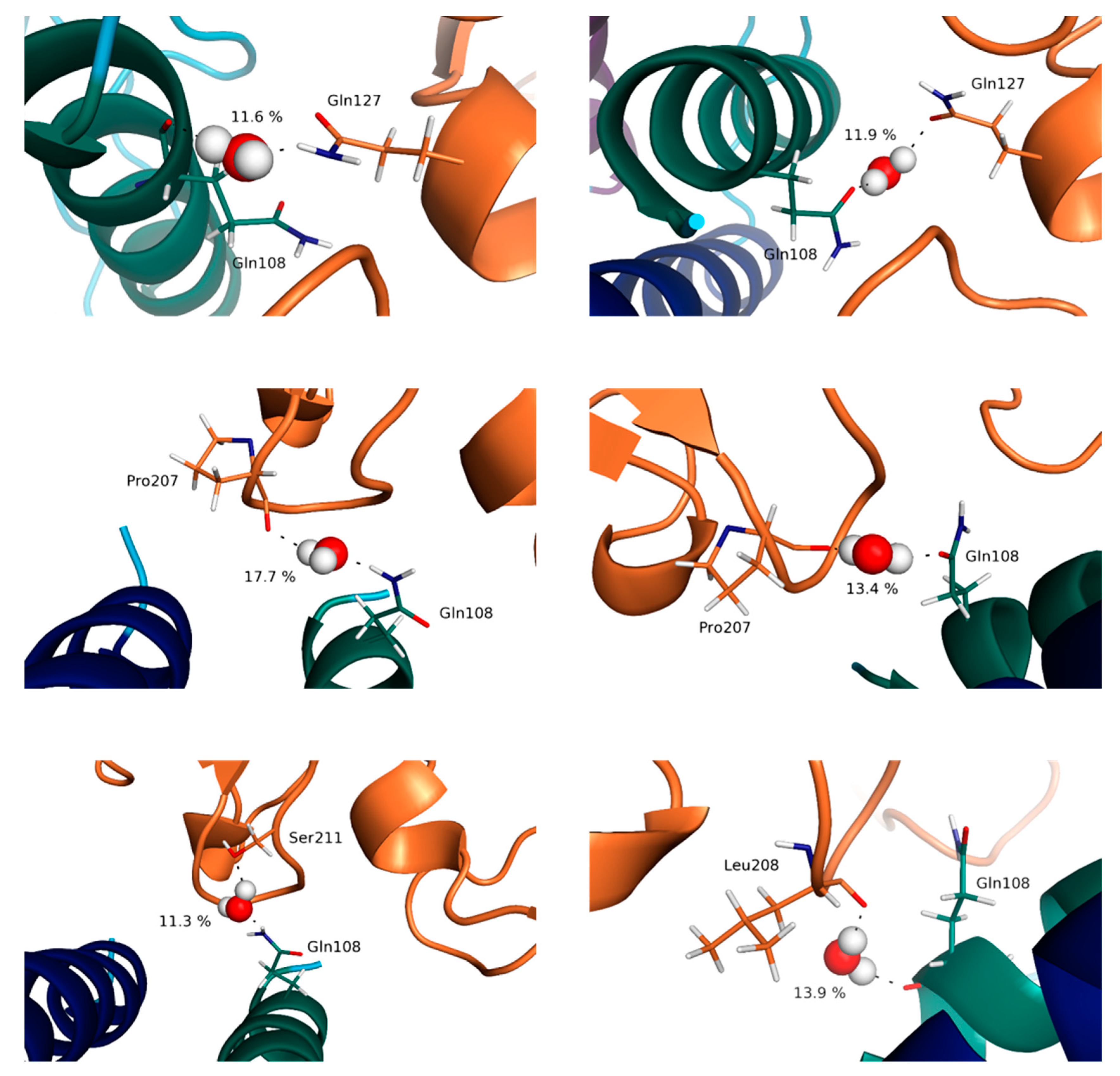
3.4. Water Molecules Stabilize the Interfaces
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burton, J.D.; Bamford, R.N.; Peters, C.; Grant, A.J.; Kurys, G.; Goldman, C.K.; Brennan, J.; Roessler, E.; Waldmann, T.A. A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc. Natl. Acad. Sci. USA 1994, 91, 4935–4939. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Koka, R.; Burkett, P. Diverse Functions of Il-2, Il-15, And Il-7 In Lymphoid Homeostasis. Annu. Rev. Immunol. 2006, 24, 657–679. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Lin, J.-X.; Leonard, W.J. IL-2 Family Cytokines: New Insights into the Complex Roles of IL-2 as a Broad Regulator of T helper Cell Differentiation. Curr. Opin. Immunol. 2011, 23, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Marks-Konczalik, J.; Dubois, S.; Losi, J.M.; Sabzevari, H.; Yamada, N.; Feigenbaum, L.; Waldmann, T.A.; Tagaya, Y. IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proc. Natl. Acad. Sci. USA 2000, 97, 11445–11450. [Google Scholar] [CrossRef] [PubMed]
- Ku, C.C.; Murakami, M.; Sakamato, A.; Kappler, J.; Marrack, P. Control of homeostasis of CD8(+) memory T cells by opposing cytokines. Science 2000, 288, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 2006, 6, 595–601. [Google Scholar] [CrossRef] [PubMed]
- van de Vosse, E.; van Agtmael, M.A. Targets of anticytokine therapy and the risk of infections in humans and mice. Curr. Opin. Rheumatol. 2007, 19, 626–635. [Google Scholar] [CrossRef]
- Johnston, J.A.; Bacon, C.M.; Finbloom, D.S.; Rees, R.C.; Kaplan, D.; Shibuya, K.; Ortaldo, J.R.; Gupta, S.; Chen, Y.Q.; Giri, J.D.; et al. Tyrosine Phosphorylation and Activation of Stat5, Stat3, And Janus Kinases by Interleukin-2 And Interleukin-15. Proc. Natl. Acad. Sci. USA 1995, 92, 8705–8709. [Google Scholar] [CrossRef]
- Xq, W.; Orchardson, M.; A Gracie, J.; Leung, B.P.; Bm, G.; Guan, H.; Niedbala, W.; Paterson, G.K.; McInnes, I.B.; Liew, F.Y. The Sushi domain of soluble IL-15 receptor alpha is essential for binding IL-15 and inhibiting inflammatory and allogenic responses in vitro and in vivo. J. Immunol. 2001, 167, 277–282. [Google Scholar]
- Chirifu, M.; Hayashi, C.; Nakamura, T.; Toma, S.; Shuto, T.; Kai, H.; Yamagata, Y.; Davis, S.J.; Ikemizu, S. Crystal structure of the IL-15-IL-15R alpha complex, a cytokine-receptor unit presented in trans. Nat. Immunol. 2007, 8, 1001–1007. [Google Scholar] [CrossRef]
- Ring, A.M.; Lin, J.X.; Feng, D.; Mitra, S.; Rickert, M.; Bowman, G.R.; Pande, V.S.; Li, P.; Moraga, I.; Spolski, R.; et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol. 2012, 13, 1187. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A. Cytokines in Cancer Immunotherapy. Cold Spring Harb. Perspect. Biol. 2018, 10, a028472. [Google Scholar] [CrossRef] [PubMed]
- Wrangle, J.M.; Patterson, A.; Johnson, C.B.; Neitzke, D.J.; Mehrotra, S.; Denlinger, C.E.; Paulos, C.M.; Li, Z.; Cole, D.J.; Rubinstein, M.P. IL-2 and Beyond in Cancer Immunotherapy. J. Interf. Cytokine Res. 2018, 38, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.M.; StGallay, S.; Kleywegt, G.J.; A St-Gallay, S. Limitations and lessons in the use of X-ray structural information in drug design. Drug Discov. Today 2008, 13, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Skwarczynska, M.; Ottmann, C. Protein–protein interactions as drug targets. Future Med. Chem. 2015, 7, 2195–2219. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J. Inhibition of protein–protein interactions using designed molecules. Chem. Soc. Rev. 2009, 38, 3289. [Google Scholar] [CrossRef] [PubMed]
- Schillinger, O.; Panwalkar, V.; Strodel, B.; Dingley, A.J. Molecular Dynamics Simulations Reveal Key Roles of the Interleukin-6 Alpha Receptor in the Assembly of the Human Interleukin-6 Receptor Complex. J. Phys. Chem. B 2017, 121, 8113–8122. [Google Scholar] [CrossRef]
- Bobby, R.; Robustelli, P.; Kralicek, A.V.; Mobli, M.; King, G.F.; Grötzinger, J.; Dingley, A.J. Functional implications of large backbone amplitude motions of the glycoprotein 130-binding epitope of interleukin-6. FEBS J. 2014, 281, 2471–2483. [Google Scholar] [CrossRef]
- Silva, D.-A.; Yu, S.; Ulge, U.Y.; Spangler, J.B.; Jude, K.M.; Labão-Almeida, C.; Ali, L.R.; Quijano-Rubio, A.; Ruterbusch, M.; Leung, I.; et al. De novo design of potent and selective mimics of IL-2 and IL-15. Nature 2019, 565, 186–191. [Google Scholar] [CrossRef]
- Quéméner, A.; Maillasson, M.; Arzel, L.; Sicard, B.; Vomiandry, R.; Mortier, E.; Dubreuil, D.; Jacques, Y.; Lebreton, J.; Mathé-Allainmat, M. Discovery of a Small-Molecule Inhibitor of Interleukin 15: Pharmacophore-Based Virtual Screening and Hit Optimization. J. Med. Chem. 2017, 60, 6249–6272. [Google Scholar] [CrossRef]
- Żyżyńska-Granica, B.; Trzaskowski, B.; Niewieczerzał, S.; Filipek, S.; Zegrocka-Stendel, O.; Dutkiewicz, M.; Krzeczyński, P.; Kowalewska, M.; Koziak, K. Pharmacophore guided discovery of small-molecule interleukin 15 inhibitors. Eur. J. Med. Chem. 2017, 136, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins Struct. Funct. Bioinformat. 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, J.; Impey, R.W.; Jorgensen, W.L.; Madura, J.D.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- The Amber Home Page. Available online: http://ambermd.org/index.php (accessed on 2 September 2019).
- Stella, L.; Melchionna, S. Equilibration and sampling in molecular dynamics simulations of biomolecules. J. Chem. Phys. 1998, 109, 10115–10117. [Google Scholar] [CrossRef]
- Anderson, D.M.; Kumaki, S.; Ahdieh, M.; Bertles, J.; Tometsko, M.; Loomis, A.; Giri, J.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; et al. Functional Characterization of the Human Interleukin-15 Receptor αChain and Close Linkage of IL15RA and IL2RA Genes. J. Biol. Chem. 1995, 270, 29862–29869. [Google Scholar]
- Sakamoto, S.; Caaveiro, J.M.; Sano, E.; Tanaka, Y.; Kudou, M.; Tsumoto, K. Contributions of Interfacial Residues of Human Interleukin15 to the Specificity and Affinity for Its Private α-Receptor. J. Mol. Biol. 2009, 389, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Mortier, E.; Quéméner, A.; Vusio, P.; Lorenzen, I.; Boublik, Y.; Grötzinger, J.; Plet, A.; Jacques, Y. Soluble Interleukin-15 Receptor α (IL-15Rα)-sushi as a Selective and Potent Agonist of IL-15 Action through IL-15Rβ/γ: Hyperagonist Il-15·Il-15rα Fusion Proteins. J. Biol. Chem. 2006, 281, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Bernard, J.; Harb, C.; Mortier, E.; Meloen, R.H.; Vermot-Desroches, C.; Wijdeness, J.; Van Dijken, P.; Slootstra, J.W.; Plet, A.; Jacques, Y.; et al. Identification of an Interleukin-15α Receptor-binding Site on Human Interleukin-15. J. Biol. Chem. 2004, 279, 24313–24322. [Google Scholar] [CrossRef] [PubMed]
- Quéméner, A.; Bernard, J.; Mortier, E.; Plet, A.; Jacques, Y.; Tran, V. Docking of human interleukin-15 to its specific receptor α chain: Correlation between molecular modeling and mutagenesis experimental data. Proteins Struct. Funct. Bioinformat. 2006, 65, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Chernov-Rogan, T.; Whitehorn, E.; Tate, E.; Bell, M.P.; Barrett, R.W.; Balasubramanian, S.; Davis, A.M.; Zurawski, G. Ligand binding kinetics of IL-2 and IL-15 to heteromers formed by extracellular domains of the three IL-2 receptor subunits. Int. Immunol. 1995, 7, 1839–1849. [Google Scholar]
- Flemming, C.L.; Russell, S.J.; Collins, M.K.L. Mutation of Asp20of human interleukin-2 reveals a dual role of the p55 α chain of the interleukin-2 receptor. Eur. J. Immunol. 1993, 23, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Shanafelt, A.B.; Lin, Y.; Shanafelt, M.-C.; Forte, C.P.; Dubois-Stringfellow, N.; Carter, C.; Gibbons, J.A.; Cheng, S.-L.; Delaria, K.A.; Fleischer, R.; et al. A T-cell-selective interleukin 2 mutein exhibits potent antitumor activity and is well tolerated in vivo. Nat. Biotechnol. 2000, 18, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Bonnert, T.P. Structure-Function Studies of Interleukin 15using Site-specific Mutagenesis, Polyethylene Glycol Conjugation, and Homology Modeling. J. Biol. Chem. 1997, 272, 2312–2318. [Google Scholar]
- Collins, L.; Tsien, W.H.; Seals, C.; Hakimi, J.; Weber, D.; Bailon, P.; Hoskings, J.; Greene, W.C.; Toome, V.; Ju, G. Identification of specific residues of human interleukin 2 that affect binding to the 70-kDa subunit (p70) of the interleukin 2 receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 7709. [Google Scholar] [CrossRef]
- Wang, X.Q.; Rickert, M.; Garcia, K.C. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gamma(c) receptors. Science 2005, 310, 1159–1163. [Google Scholar] [CrossRef]
| Number of Contacts | ΔGbind | ||||||
|---|---|---|---|---|---|---|---|
| Interface | Model | Dimer | Trimer | Tetramer | Dimer | Trimer | Tetramer |
| IL-15/IL-15Rα | 43 (4) | - | 41 (3) | −80.3 (6.6) | −83.8 (8.2) | ||
| IL-15/IL-2Rβ | 28 (4) | 26 (4) | −29.1 (5.6) | −27.2 (6.6) | |||
| IL-15/γc | 18 (5) | 18 (7) | −16.2 (7.7) | −17.3 (10.5) | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, R.P.; Laurent, A.D.; Quéméner, A.; Mortier, E.; Le Questel, J.-Y. Mechanistic and Structural Insights on the IL-15 System through Molecular Dynamics Simulations. Molecules 2019, 24, 3261. https://doi.org/10.3390/molecules24183261
Sousa RP, Laurent AD, Quéméner A, Mortier E, Le Questel J-Y. Mechanistic and Structural Insights on the IL-15 System through Molecular Dynamics Simulations. Molecules. 2019; 24(18):3261. https://doi.org/10.3390/molecules24183261
Chicago/Turabian StyleSousa, Rui P., Adèle D. Laurent, Agnès Quéméner, Erwan Mortier, and Jean-Yves Le Questel. 2019. "Mechanistic and Structural Insights on the IL-15 System through Molecular Dynamics Simulations" Molecules 24, no. 18: 3261. https://doi.org/10.3390/molecules24183261
APA StyleSousa, R. P., Laurent, A. D., Quéméner, A., Mortier, E., & Le Questel, J.-Y. (2019). Mechanistic and Structural Insights on the IL-15 System through Molecular Dynamics Simulations. Molecules, 24(18), 3261. https://doi.org/10.3390/molecules24183261






