Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis
Abstract
1. Introduction
2. Results and Discussion
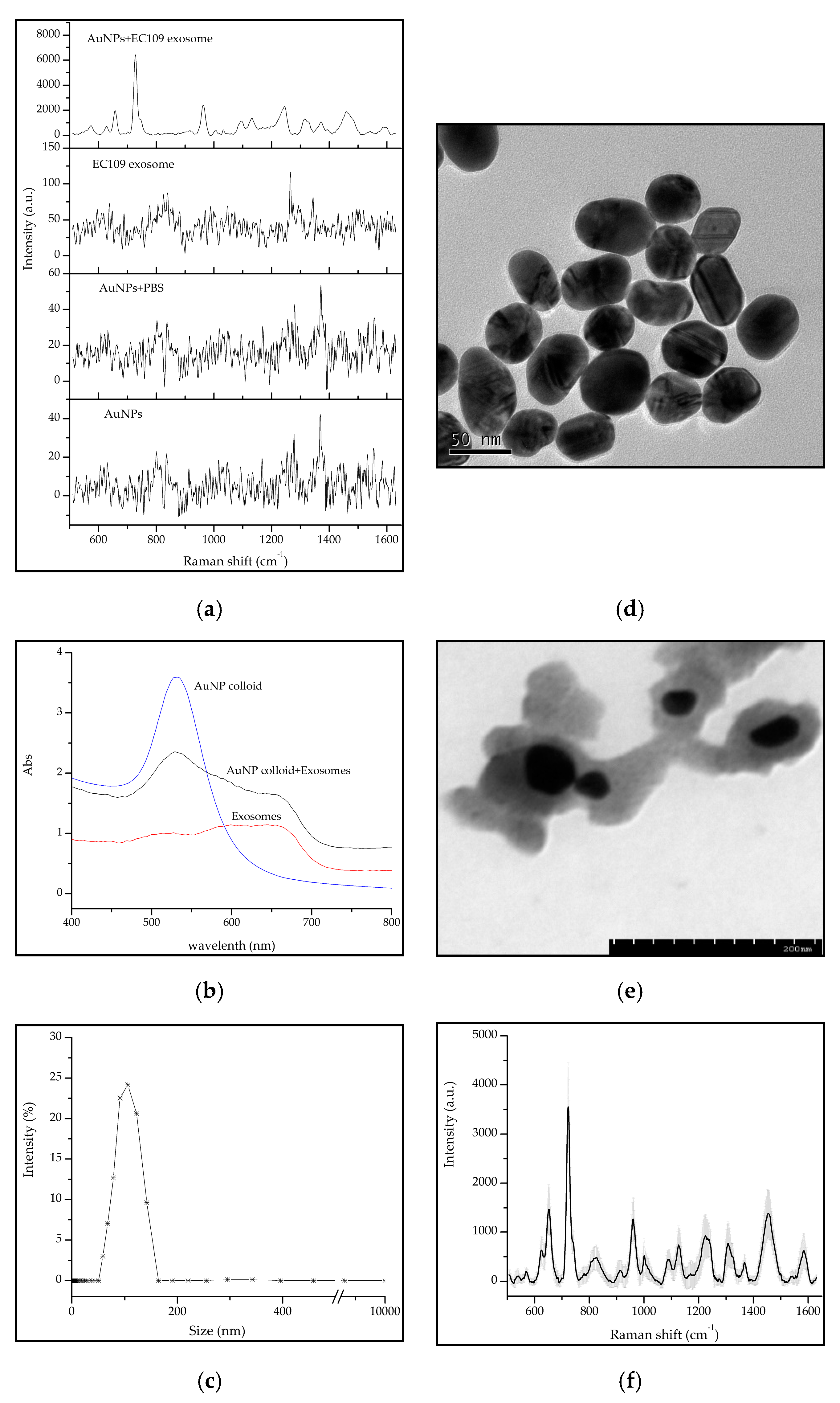
2.1. Exosomes Isolation and Characterization
2.2. The Raman Signals of Exosomes Were Enhanced by Au Nanoparticles
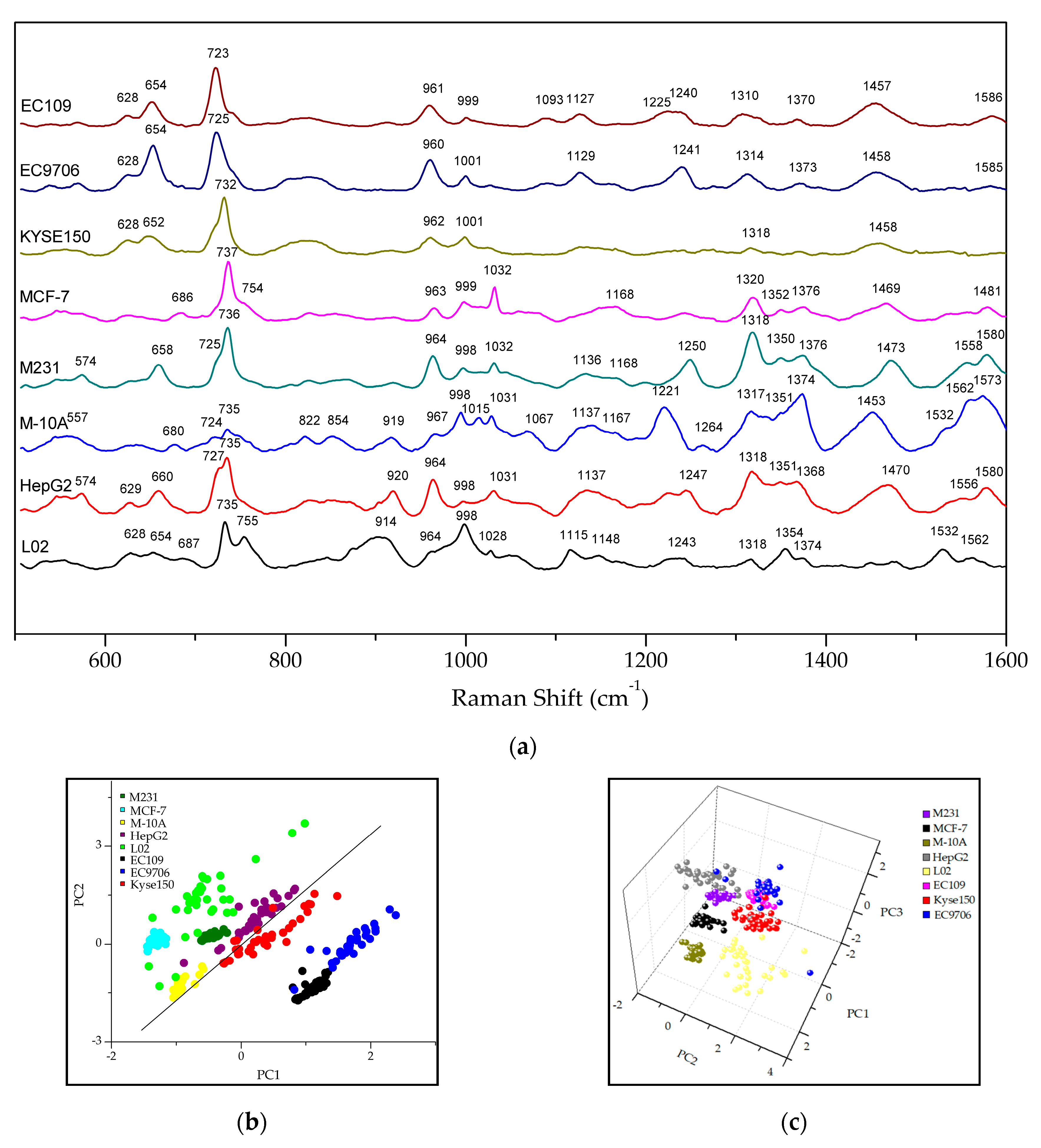
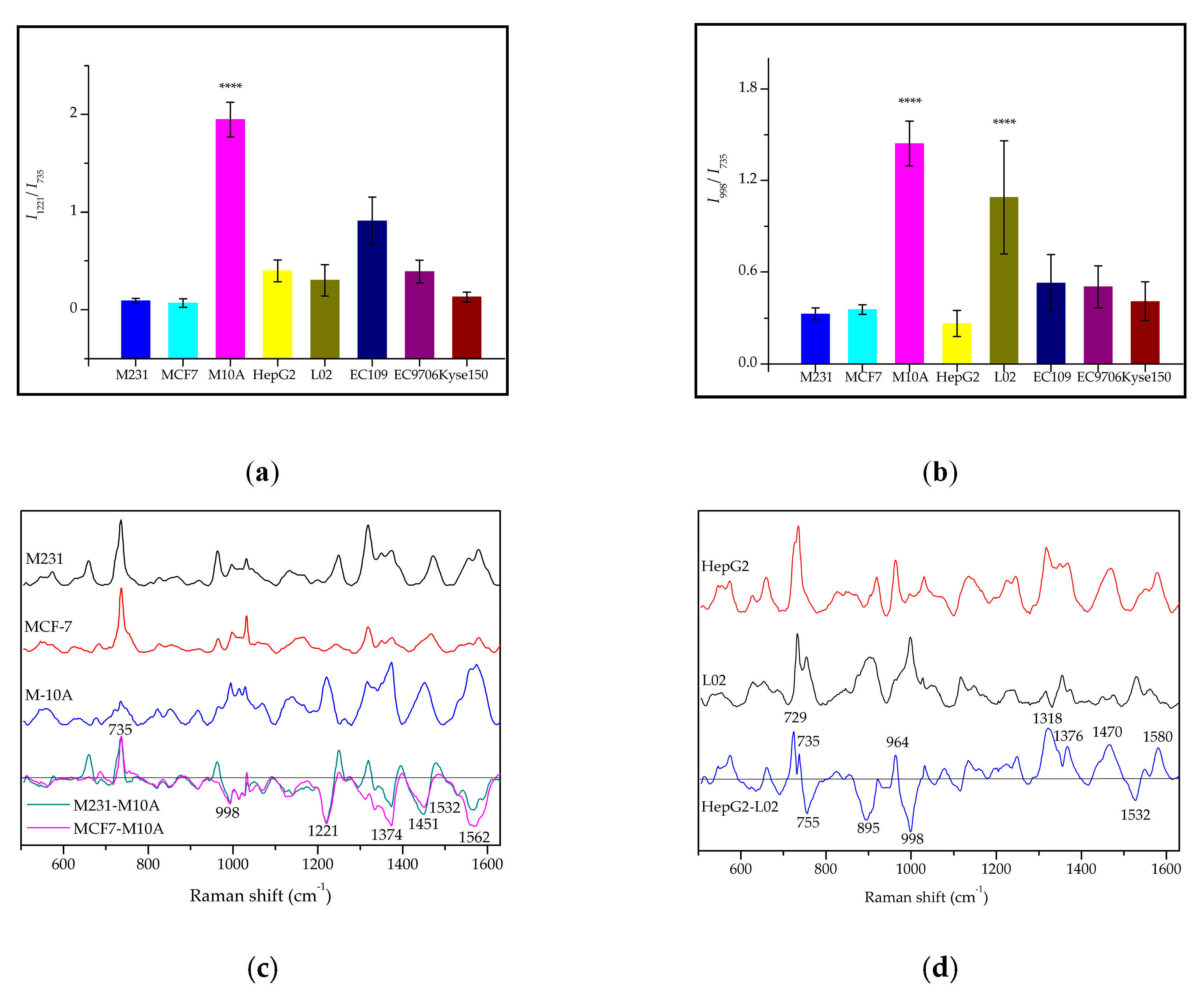
2.3. The Raman Phenotypes of Exosomes Derived from Different Types of Cancer Cells
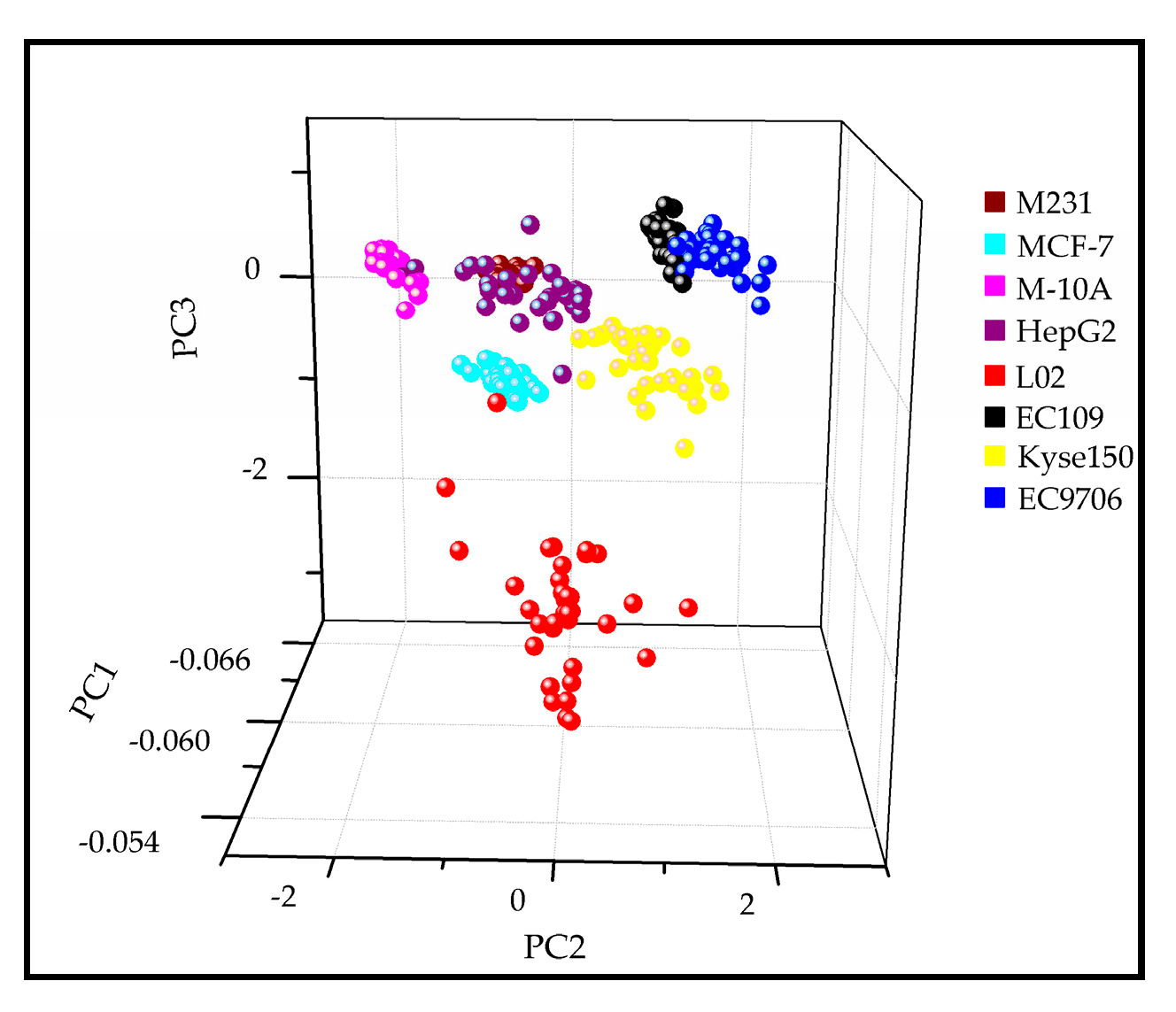
2.4. Discrimination of the Subtypes of Exosomes by PCA-LDA
3. Materials and Methods
3.1. Isolation of Exosomes in Culture Media
3.2. Exosomes Identification
3.3. Synthesis of AuNPs
3.4. SERS Detection
3.5. Data Processing
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2011, 40, D1241–D1244. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteomic 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Abak, A.; Abhari, A.; Rahimzadeh, S. Exosomes in cancer: Small vesicular transporters for cancer progression and metastasis, biomarkers in cancer therapeutics. Peerj 2018, 6, e4763. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic, M.M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Hung, J.Y.; Chang, W.A.; Lin, Y.S.; Pan, Y.C.; Tsai, P.H.; Wu, C.Y.; Kuo, P.L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New technologies for analysis of extracellular vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Stremersch, S.; Marro, M.; Pinchasik, B.; Baatsen, P.; Hendrix, A.; De Smedt, S.C.; Loza-Alvarez, P.; Skirtach, A.G.; Raemdonck, K.; Braeckmans, K. Identification of individual exosome-like vesicles by surface enhanced Raman spectroscopy. Small 2016, 12, 3292–3301. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Hwang, M.; Choi, B.; Jeong, H.; Jung, J.H.; Kim, H.K.; Hong, S.; Park, J.H.; Choi, Y. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal. Chem. 2017, 89, 6695–6701. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Nanou, A.; Rikkert, L.; Coumans, F.A.W.; Otto, C.; Terstappen, L.W.M.M.; Offerhaus, H.L. Label-free prostate cancer detection by characterization of extracellular vesicles using Raman spectroscopy. Anal. Chem. 2018, 90, 11290–11296. [Google Scholar] [CrossRef] [PubMed]
- Kwizera, E.A.; O’Connor, R.; Vinduska, V.; Williams, M.; Butch, E.R.; Snyder, S.E.; Chen, X.; Huang, X. Molecular detection and analysis of exosomes using surface-enhanced Raman scattering gold nanorods and a miniaturized device. Theranostics 2018, 8, 2722–2738. [Google Scholar] [CrossRef] [PubMed]
- Carmicheal, J.; Hayashi, C.; Huang, X.; Liu, L.; Lu, Y.; Krasnoslobodtsev, A.; Lushnikov, A.; Kshirsagar, P.G.; Patel, A.; Jain, M.; et al. Label-free characterization of exosome via surface enhanced Raman spectroscopy for the early detection of pancreatic cancer. Nanomedicine 2019, 16, 88–96. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.F.; Ning, C.F.; He, F.; Yin, B.C.; Ye, B.C. Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst 2018, 143, 4915–4922. [Google Scholar] [CrossRef]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Yang, R. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef]
- Kneipp, K.; Kneipp, H.; Kneipp, J. Surface-enhanced Raman scattering in local optical fields of silver and gold nanoaggregates-from single-molecule Raman spectroscopy to ultrasensitive probing in live cells. Acc. Chem. Res. 2006, 39, 443–450. [Google Scholar] [CrossRef]
- Tian, Z.Q.; Ren, B.; Wu, D.Y. Surface-enhanced Raman scattering: From noble to transition metals and from rough surfaces to ordered nanostructures. J. Phys. Chem. B 2002, 106, 9463–9483. [Google Scholar] [CrossRef]
- Manciu, F.S.; Ciubuc, J.D.; Parra, K.; Manciu, M.; Bennet, K.E.; Valenzuela, P.; Sundin, E.M.; Durrer, W.G.; Reza, L.; Francia, G. The label-free Raman imaging to monitor breast tumor signatures. Technol. Cancer Res. Treat. 2017, 16, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Kallaway, C.; Almond, L.M.; Barr, H.; Wood, J.; Hutchings, J.; Kendall, C.; Stone, N. Advances in the clinical application of Raman spectroscopy for cancer diagnostics. Photodiagn. Photodyn. Ther. 2013, 10, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Willets, K.A. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal. Bioanal. Chem. 2009, 394, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Tatischeff, I.; Larquet, E.; Falcón-Pérez, J.M.; Turpin, P.Y.; Kruglik, S.G. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.R.; Malkin, A.; Lyng, F.M. Current advances in the application of Raman spectroscopy for molecular diagnosis of cervical cancer. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Kneipp, K.; Haka, A.S.; Kneipp, H.; Badizadegan, K.; Yoshizawa, N.; Boone, C.; Shafer-Peltier, K.E.; Motz, J.T.; Dasari, R.R.; Feld, M.S. Surface-enhanced Raman spectroscopy in single living cells using gold nanoparticles. Appl. Spectrosc. 2002, 56, 150–154. [Google Scholar] [CrossRef]
- Shetty, G.; Kendall, C.; Shepherd, N.; Stone, N.; Barr, H. Raman spectroscopy: Elucidation of biochemical changes in carcinogenesis of oesophagus. Br. J. Cancer 2006, 94, 1460–1464. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Lee, C.; Carney, R.P.; Hazari, S.; Smith, Z.J.; Knudson, A.; Robertson, C.S.; Lam, K.S.; Wachsmann-Hogiu, S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale 2015, 7, 9290–9297. [Google Scholar] [CrossRef]
- Galler, K.; Requardt, R.P.; Glaser, U.; Markwart, R.; Bocklitz, T.; Bauer, M.; Popp, J.; Neugebauer, U. Single cell analysis in native tissue: Quantification of the retinoid content of hepatic stellate cells. Sci. Rep. 2016, 6, 24155. [Google Scholar] [CrossRef] [PubMed]
- Tirinato, L.; Gentile, F.; Di Mascolo, D.; Coluccio, M.L.; Das, G.; Liberale, C.; Pullano, S.A.; Perozziello, G.; Francardi, M.; Accardo, A.; et al. SERS analysis on exosomes using super-hydrophobic surfaces. Microelectron. Eng. 2012, 97, 337–340. [Google Scholar] [CrossRef]
- Griffin, J.L.; Shockcor, J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer 2004, 4, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zheng, D.; Zhang, P.; Lin, T.; Wang, H.; Zhu, Y. Surface enhanced Raman scattering investigation of bovine albumin with different size of Au nanoparticles. J. Appl. Biomater. Funct. 2018, 16, 157–162. [Google Scholar] [CrossRef]
Sample Availability: Samples of the cells are available from the authors. |

| Raman Shift/cm−1 | Assignments | |||||||
|---|---|---|---|---|---|---|---|---|
| EC109 | EC9706 | Kyse150 | MCF7 | M231 | M10A | HepG2 | L02 | |
| 540 | 547 | 546 | 546 | 546 | 536 | Cholesterol [12], C-S stretching [23] | ||
| 558 | 556 | 557 | 557 | 556 | Glycogen [26] | |||
| 570 | 570 | 574 | 574 | 574 | C=S tensile vibration, Glycogen [26] | |||
| 628 | 628 | 628 | 627 | 629 | 636 | 629 | 628 | Deformation vibration of adenine ring, phenylalanine C-C torsional vibration [22] |
| 654 | 654 | 652 | 658 | 660 | 654 | Tyrosine vibration, Guanine [27] | ||
| 686 | 680 | 687 | Tyrosine, phenylalanine [26] | |||||
| 723 | 725 | 727 | Adenine respiratory vibration, nucleotides [23], C-N symmetric stretching band (phospholipid) [22] | |||||
| 732 | 735 | 735 | 735 | 735 | 735 | Adenine [22,23] | ||
| 742 | 754 | 749 | 755 | Lactic acid [14,28], DNA, nucleic acids [23], symmetric breathing of tryptophan [22] | ||||
| 829 | 826 | 828 | 828 | 826 | 822 | 829 | Sugar–phosphate backbone vibration [27], protein [28], C-O-O vibration typical of phospholipids [29], | |
| 855 | 870 | 854 | 852 | 847 | Cholesterol, oxyproline, tryptophan, glycogen [28], C-C stretch proline ring in collagen [22] | |||
| 873 | 874 | Tryptophan, CH2 deformation (e.g., protein) [25,30] | ||||||
| 914 | 917 | 920 | 921 | 919 | 920 | 914 | C=C stretching vibration, proline [24] | |
| 961 | 960 | 962 | 963 | 964 | 967 | 964 | 964 | Adenine, C-N deformed vibration, carbohydrates [24] |
| 999 | 1001 | 1001 | 999 | 998 | 998 | 998 | 998 | symmetric respiratory vibration of phenylalanine [23] |
| 1015 | C–O vibration in DNA/RNA, C–C vibration [24] | |||||||
| 1029 | 1032 | 1032 | 1031 | 1031 | 1028 | CH2CH3 bending (e.g., phospholipid); C-C vibration (e.g., polysaccharide) [25] | ||
| 1055 | Glycogen [28] | |||||||
| 1067 | 1071 | C-C vibrations in lipid and protein [2], C–O vibration in DNA/RNA [24], collagen [26] | ||||||
| 1093 | 1093 | Phosphate: PO2− vibration, C-C vibration, C-O-C vibration, glycoside link [24] | ||||||
| 1115 | C-O ribose (e.g., nucleic acid) [30], O–P–O DNA backbone [27], C-N vibration [24] | |||||||
| 1127 | 1129 | 1130 | C-C vibrations in lipid [12,22], C-N stretching vibration in protein [22], | |||||
| 1137 | 1136 | 1136 | Proline [24] | |||||
| 1150 | 1143 | 1148 | CH vibration in protein [31], ribose-phosphate [24] | |||||
| 1167 | 1168 | 1168 | 1167 | Carotenoids [31], CH deformation in protein [30], Ribose-phosphate [24] | ||||
| 1221 | Amide III [24] | |||||||
| 1225 | 1229 | 1229 | Lipids, protein [28], cytosine [24] | |||||
| 1240 | 1241 | 1246 | 1250 | 1247 | 1243 | Amide III [24], asymmetric phosphate stretching (e.g., nucleic acid) [25] | ||
| 1264 | Amide III (e.g., protein), C=C (e.g., fatty acids) [24,25,28] | |||||||
| 1310 | 1314 | 1318 | 1320 | 1318 | 1317 | 1318 | 1318 | Amide III, CH deformation, CH3CH2 wagging (e.g., nucleic acids, collagen) [25], guanine [22,24], |
| 1352 | 1350 | 1351 | 1351 | 1354 | Guanine (nucleic acid) [12], CH2, CH3 wagging in protein [24] | |||
| 1368 | CH3 vibration (e.g., phospholipid) [12] | |||||||
| 1370 | 1373 | 1373 | 1376 | 1376 | 1374 | 1376 | Carbohydrate [12], adenine, guanine, thymine [24] | |
| 1457 | 1458 | 1458 | 1453 | 1451 | CH2CH3 asymmetric and symmetric deformations in proteins, phospholipid and DNA [22,26] | |||
| 1469 | 1473 | 1470 | 1475 | Adenine, C-N stretching, CH deformation (e.g., lipid, protein) [32] | ||||
| 1532 | 1532 | Vibration of (-C=C-) conjugated [25] | ||||||
| 1558 | 1562 | 1556 | 1562 | Tryptophan [12] | ||||
| 1586 | 1585 | 1581 | 1580 | 1573 | 1580 | Guanine [22], adenine, purine, phenylalanine, tyrosine [24] | ||
| Sample | Prediction Group | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| EC109 | EC9706 | Kyse150 | M231 | MCF7 | M-10A | HepG2 | L02 | ||
| EC109 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 |
| EC9706 | 1 | 32 | 0 | 0 | 0 | 0 | 0 | 0 | 33 |
| Kyse150 | 0 | 0 | 36 | 0 | 0 | 0 | 0 | 0 | 36 |
| M231 | 0 | 0 | 0 | 34 | 0 | 0 | 1 | 0 | 35 |
| MCF7 | 0 | 0 | 0 | 0 | 35 | 0 | 0 | 0 | 35 |
| M-10A | 0 | 0 | 0 | 0 | 0 | 35 | 0 | 0 | 35 |
| HepG2 | 0 | 0 | 0 | 2 | 0 | 1 | 32 | 0 | 35 |
| L02 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 32 | 35 |
| Sample | Prediction Group | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| M231 | MCF7 | M10A | EC109 | EC9706 | Kyse150 | HepG2 | L02 | ||
| M231 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 |
| MCF7 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 35 |
| M10A | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | 35 |
| EC109 | 0 | 0 | 0 | 29 | 2 | 4 | 0 | 0 | 35 |
| EC9706 | 0 | 0 | 0 | 5 | 26 | 0 | 2 | 0 | 33 |
| Kyse150 | 1 | 0 | 0 | 2 | 4 | 29 | 0 | 0 | 36 |
| HepG2 | 1 | 0 | 0 | 0 | 0 | 2 | 32 | 0 | 35 |
| L02 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 31 | 35 |
| Sample | Prediction Group | Total | Sensitivity (%) | Specificity (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M231 | MCF7 | M10A | HepG2 | L02 | EC109 | Kyse150 | EC9706 | ||||
| M231 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 100 | 98 |
| MCF7 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 100 | 99.6 |
| M10A | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | 35 | 100 | 100 |
| HepG2 | 5 | 0 | 0 | 29 | 0 | 0 | 1 | 0 | 35 | 82.9 | 100 |
| L02 | 0 | 1 | 0 | 0 | 34 | 0 | 0 | 0 | 35 | 97.1 | 100 |
| EC109 | 0 | 0 | 0 | 0 | 0 | 35 | 0 | 0 | 35 | 100 | 99.2 |
| Kyse150 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 0 | 36 | 100 | 99.6 |
| EC9706 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 31 | 33 | 93.9 | 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Wang, L.; Fang, Y.; Zheng, D.; Lin, T.; Wang, H. Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis. Molecules 2019, 24, 2947. https://doi.org/10.3390/molecules24162947
Zhang P, Wang L, Fang Y, Zheng D, Lin T, Wang H. Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis. Molecules. 2019; 24(16):2947. https://doi.org/10.3390/molecules24162947
Chicago/Turabian StyleZhang, Ping, Limin Wang, Yaping Fang, Dawei Zheng, Taifeng Lin, and Huiqin Wang. 2019. "Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis" Molecules 24, no. 16: 2947. https://doi.org/10.3390/molecules24162947
APA StyleZhang, P., Wang, L., Fang, Y., Zheng, D., Lin, T., & Wang, H. (2019). Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis. Molecules, 24(16), 2947. https://doi.org/10.3390/molecules24162947




