Chemical Profile and Antioxidant Activity of Zinnia elegans Jacq. Fractions
Abstract
1. Introduction
2. Results
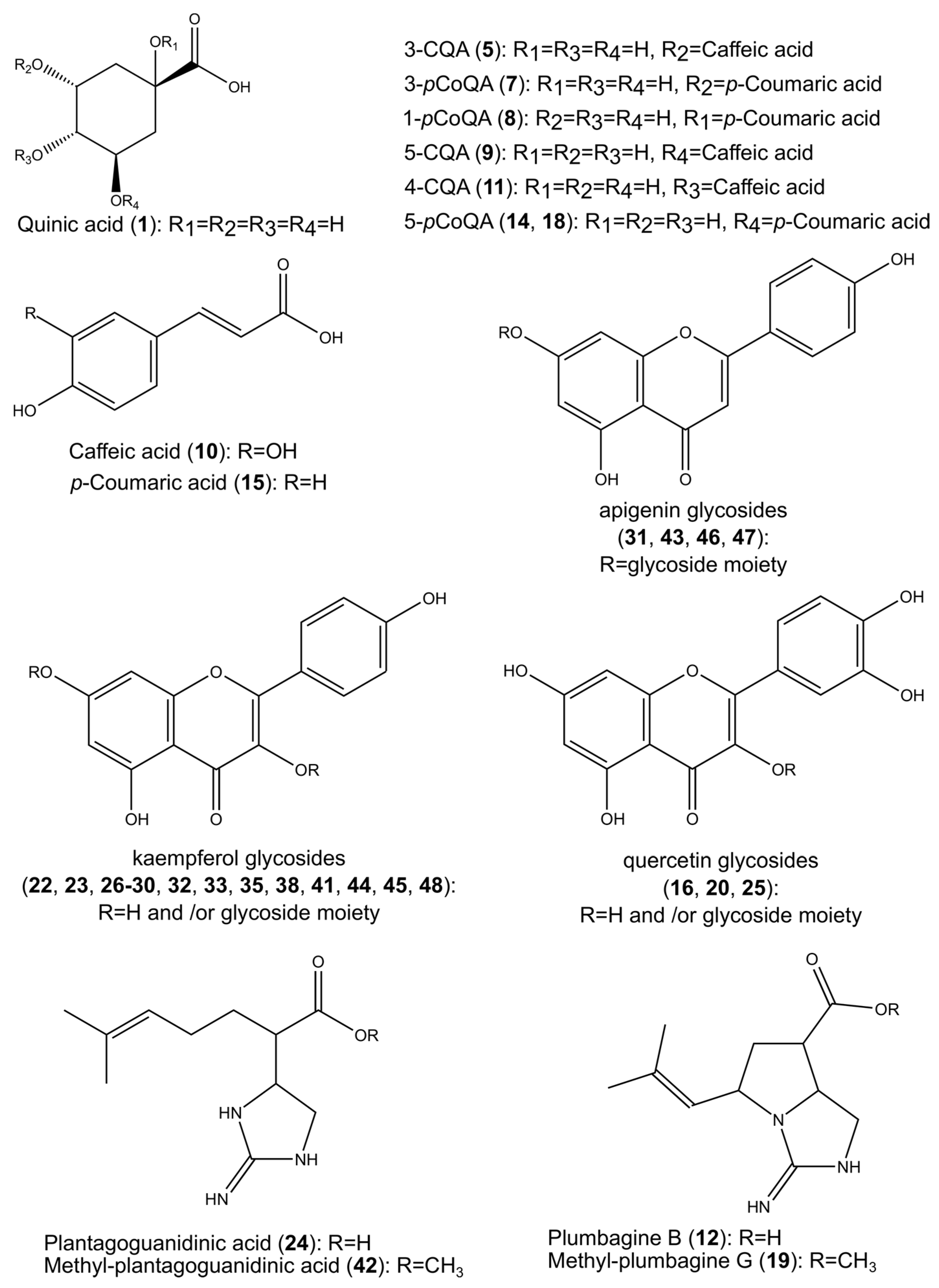
2.1. Identification of the Constituents Found in the Z. elegans Extract
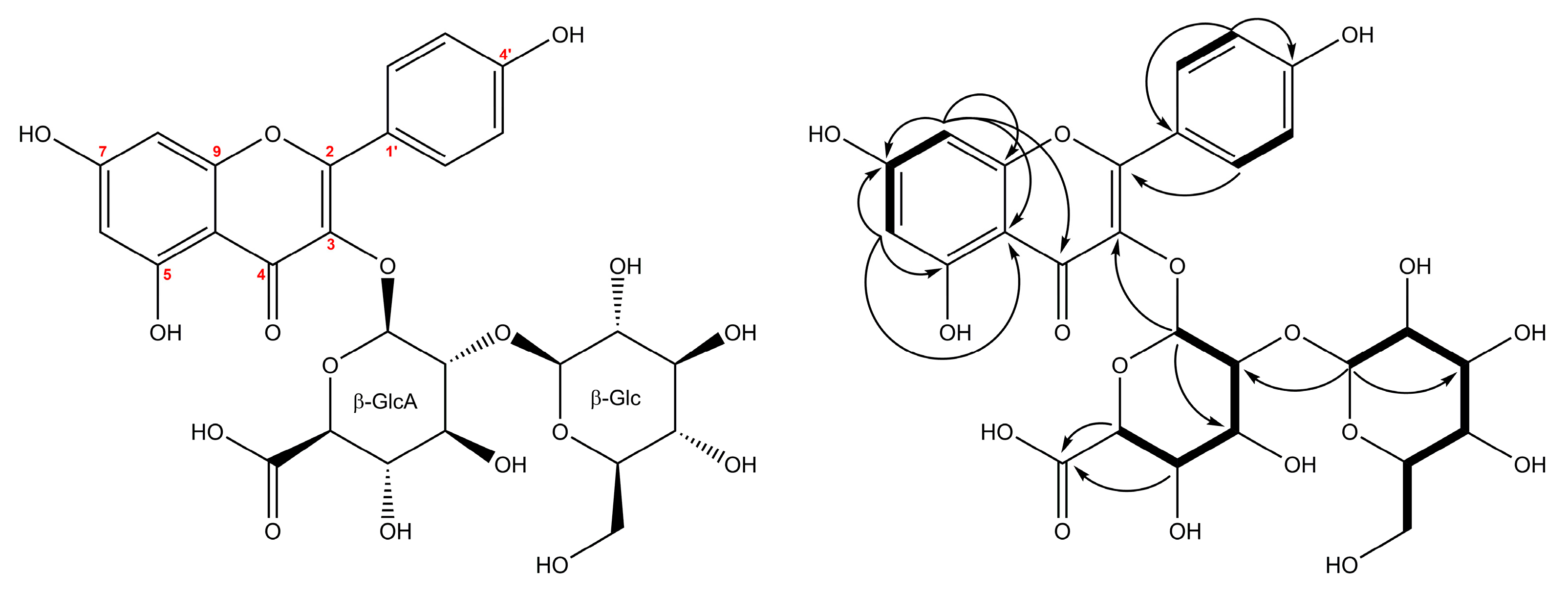
2.2. Structural Characterization of the New Compounds
2.3. Antioxidant Activity
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Plant Material
4.3. Isolation
4.4. Semi-Preparative HPLC
4.5. High-Resolution LC-MS and Qualitative Analysis
4.6. NMR Spectroscopy
Characteristic Data of Isolated Z. elegans Compounds
4.7. Antioxidant Tests
4.7.1. Lipoxygenase Inhibition
4.7.2. Metal Chelation
4.7.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stimart, D.; Boyle, T. Zinnia. In Flower Breeding and Genetics: Issues, Challenges and Opportunities for the 21st Century; Anderson, N.O., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 337–357. [Google Scholar]
- Winter, N. Tough-as-Nails Flowers for the South; University Press of Mississippi: Jackson, MS, USA, 2003; pp. 58–59. [Google Scholar]
- Hill, L.; Hill, N. The Flower Gardener’s Bible: A Complete Guide to Colorful Blooms All Season Long: 400 Favorite Flowers, Time-Tested Techniques, Creative Garden Designs, and a Lifetime of Gardening Wisdom; Storey Publishing: North Adams, MA, USA, 2012; p. 346. [Google Scholar]
- Hafiza, M.A.; Parveen, B.; Ahmad, R.; Hamid, K. Phytochemical and antifungal screening of Medicago sativa and Zinnia elegans. Online, J. Biol. Sci. 2002, 2, 130–132. [Google Scholar]
- Mohamed, A.H.; Ahmed, F.A.; Ahmed, O.K. Hepatoprotective and antioxidant activity of Zinnia elegans leaves ethanolic extract. Int. J. Sci. Eng. Res. 2015, 6, 154–161. [Google Scholar]
- Gomaa, A.; Samy, M.; Desoukey, S.; Kamel, M. A comprehensive review of phytoconstituents and biological activities of genus Zinnia. J. Adv. Biomed. Pharm. Sci. 2018, 2, 29–37. [Google Scholar] [CrossRef]
- Satorres, S.E.; Chiaramello, A.I.; Tonn, C.E.; Laciar, A.L. Antibacterial activity of organic extracts from Zinnia peruviana (L.) against gram-positive and gram-negative bacteria. Emirates, J. Food Agric. 2012, 24, 344–347. [Google Scholar]
- Gomaa, A.A.-R.; Samy, M.N.; Abdelmohsen, U.R.; Krischke, M.; Mueller, M.J.; Wanas, A.S.; Desoukey, S.Y.; Kamel, M.S. Metabolomic profiling and anti-infective potential of Zinnia elegans and Gazania rigens (Family Asteraceae). Nat. Prod. Res. 2018, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Bashyal, B.P.; McLaughlin, S.P.; Gunatilaka, A.A.L. Zinagrandinolides A−C, cytotoxic δ-elemanolide-type sesquiterpene lactones from Zinnia grandiflora. J. Nat. Prod. 2006, 69, 1820–1822. [Google Scholar] [CrossRef]
- Alexenizer, M.; Dorn, A. Screening of medicinal and ornamental plants for insecticidal and growth regulating activity. J. Pest Sci. 2007, 80, 205–215. [Google Scholar] [CrossRef]
- Burlec, A.F.; Arsene, C.; Cioanca, O.; Mircea, C.; Tuchilus, C.; Corciova, A.; Iancu, C.; Hancianu, M. Chemical and biological investigations of different Zinnia elegans extracts. In Proceedings of the Romanian National Congress of Pharmacy, 17th edition: 21st Century Pharmacy-Between Intelligent Specialization and Social Responsibility, Bucharest, Romania, 26–29 of September 2018; Draganescu, D., Arsene, A., Eds.; Filodiritto Publisher: Bologna, Italy, 2018; pp. 54–57. [Google Scholar]
- Rates, S.M.K. Plants as source of drugs. Toxicon 2001, 39, 603–613. [Google Scholar] [CrossRef]
- Bieski, I.G.C.; Rios Santos, F.; de Oliveira, R.M.; Espinosa, M.M.; Macedo, M.; Albuquerque, U.P.; de Oliveira Martins, D.T. Ethnopharmacology of medicinal plants of the Pantanal Region (Mato Grosso, Brazil). Evidence-Based Complement. Altern. Med. 2012, 2012, 1–36. [Google Scholar] [CrossRef]
- Bursal, E.; Aras, A.; Kılıç, Ö. Evaluation of antioxidant capacity of endemic plant Marrubium astracanicum subsp. macrodon: Identification of its phenolic contents by using HPLC-MS/MS. Nat. Prod. Res. 2019, 33, 1975–1979. [Google Scholar] [CrossRef]
- Garcia, C.J.; García-Villalba, R.; Garrido, Y.; Gil, M.I.; Tomás-Barberán, F.A. Untargeted metabolomics approach using UPLC-ESI-QTOF-MS to explore the metabolome of fresh-cut iceberg lettuce. Metabolomics 2016, 12, 138. [Google Scholar] [CrossRef]
- Clifford, M.N.; Johnston, K.L.; Knight, S.; Kuhnert, N. Hierarchical scheme for LC-MSn identification of chlorogenic acids. J. Agric. Food Chem. 2003, 51, 2900–2911. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Yu, S.; Prior, R.L. LC/MS/MS characterization of phenolic constituents in dried plums. J. Agric. Food Chem. 2002, 50, 3579–3585. [Google Scholar] [CrossRef] [PubMed]
- Cong, H.-J.; Zhang, S.-W.; Shen, Y.; Zheng, Y.; Huang, Y.-J.; Wang, W.-Q.; Leng, Y.; Xuan, L.-J. Guanidine alkaloids from Plumbago zeylanica. J. Nat. Prod. 2013, 76, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Cuyckens, F.; Claeys, M. Mass spectrometry in the structural analysis of flavonoids. J. Mass Spectrom. 2004, 39, 1–15. [Google Scholar] [CrossRef]
- Kolodziejczyk-Czepas, J.; Krzyżanowska-Kowalczyk, J.; Sieradzka, M.; Nowak, P.; Stochmal, A. Clovamide and clovamide-rich extracts of three Trifolium species as antioxidants and moderate antiplatelet agents in vitro. Phytochemistry 2017, 143, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Masike, K.; Khoza, B.; Steenkamp, P.; Smit, E.; Dubery, I.; Madala, N. A metabolomics-guided exploration of the phytochemical constituents of Vernonia fastigiata with the aid of pressurized hot water extraction and liquid chromatography-mass spectrometry. Molecules 2017, 22, 1200. [Google Scholar] [CrossRef] [PubMed]
- Tava, A.; Pecio, Ł.; Lo Scalzo, R.; Stochmal, A.; Pecetti, L. Phenolic content and antioxidant activity in Trifolium germplasm from different environments. Molecules 2019, 24, 298. [Google Scholar] [CrossRef]
- Hvattum, E.; Ekeberg, D. Study of the collision-induced radical cleavage of flavonoid glycosides using negative electrospray ionization tandem quadrupole mass spectrometry. J. Mass Spectrom. 2003, 38, 43–49. [Google Scholar] [CrossRef]
- Winterhalter, P.; Schreier, P. Free and bound C13 norisoprenoids in quince (Cydonia oblonga, Mill.) fruit. J. Agric. Food Chem. 1988, 36, 1251–1256. [Google Scholar] [CrossRef]
- Fabre, N.; Rustan, I.; De Hoffmann, E.; Quetin-Leclercq, J. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef]
- Mabry, T.J.; Markham, K.R.; Thomas, M.B. The Systematic Identification of Flavonoids; Springer: Berlin/Heidelberg, Germany, 1970. [Google Scholar]
- Cuyckens, F.; Rozenberg, R.; De Hoffmann, E.; Claeys, M. Structure characterization of flavonoid O-diglycosides by positive and negative nano-electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 2001, 36, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Castañar, L.; Sistaré, E.; Virgili, A.; Williamson, R.T.; Parella, T. Suppression of phase and amplitude J(HH) modulations in HSQC experiments. Magn. Reson. Chem. 2015, 53, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; van Ingen, H.; Vivekanandan, S.; Rademacher, C.; Norris, S.E.; Freedberg, D.I. More accurate 1JCH coupling measurement in the presence of 3JHH strong coupling in natural abundance. J. Magn. Reson. 2012, 215, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Żuchowski, J.; Pecio, Ł.; Marciniak, B.; Kontek, R.; Stochmal, A. Unusual isovalerylated flavonoids from the fruit of sea buckthorn (Elaeagnus rhamnoides) grown in Sokółka, Poland. Phytochemistry 2019, 163, 178–186. [Google Scholar] [CrossRef]
- Glasby, J.S. Dictionary of plants containing secondary metabolites; Taylor and Francis: London, UK, 1991; p. 1280. [Google Scholar]
- Yamaguchi, M.-A.; Terahara, N.; Shizukuishi, K.-I. Acetylated anthocyanins in Zinnia elegans flowers. Phytochemistry 1990, 29, 1269–1270. [Google Scholar] [CrossRef]
- Bohlmann, F.; Ziesche, J.; King, R.M.; Robinson, H. Eudesmanolides, guaianolides, germacranolides and elemanolides from Zinnia species. Phytochemistry 1981, 20, 1623–1630. [Google Scholar] [CrossRef]
- Zoghbi, M.d.G.B.; Andrade, E.H.A.; Carreira, L.M.M.; Mala, J.G.S. Volatile constituents of the flowers of Wulffia baccata (L. f.) Kuntze. and Zinnia elegans Jacq. (Asteraceae). J. Essent. Oil Res. 2000, 12, 415–417. [Google Scholar] [CrossRef]
- Earle, F.R.; Wolff, I.A.; Glass, C.A.; Jones, Q. Search for new industrial oils. VII. J. Am. Oil Chem. Soc. 1962, 39, 381–383. [Google Scholar] [CrossRef]
- Azimova, S.S.; Glushenkova, A.I.; Vinogradova, V.I. Lipids, Lipophilic Components and Essential Oils from Plant Sources; Springer: London, UK, 2012; p. 144. [Google Scholar]
- Christensen, L.P.; Lam, J. Acetylenes and related compounds in Heliantheae. Phytochemistry 1991, 30, 11–49. [Google Scholar] [CrossRef]
- Schroter, H. Detection of nicotine in Zinnia elegans and significance of the alkaloids for interfamilial grafting of Zinnia into Nicotiana. Arch. Pharm. Ber. Dtsch. Pharm. Ges. 1955, 288, 141–145. [Google Scholar] [PubMed]
- Oyenihi, A.B.; Smith, C. Are polyphenol antioxidants at the root of medicinal plant anti-cancer success? J. Ethnopharmacol. 2019, 229, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Neha, K.; Haider, M.R.; Pathak, A.; Yar, M.S. Medicinal prospects of antioxidants: A review. Eur. J. Med. Chem. 2019, 178, 687–704. [Google Scholar] [CrossRef] [PubMed]
- Belščak-Cvitanović, A.; Durgo, K.; Huđek, A.; Bačun-Družina, V.; Komes, D. Overview of polyphenols and their properties. In Polyphenols: Properties, Recovery, and Applications; Galanakis, C.M., Ed.; Elsevier: Duxford, UK, 2018; pp. 3–44. [Google Scholar]
- do Carmo, M.A.V.; Pressete, C.G.; Marques, M.J.; Granato, D.; Azevedo, L. Polyphenols as potential antiproliferative agents: Scientific trends. Curr. Opin. Food Sci. 2018, 24, 26–35. [Google Scholar] [CrossRef]
- Li, Q.-Q.; Li, Q.; Jia, J.-N.; Liu, Z.-Q.; Zhou, H.-H.; Mao, X.-Y. 12/15 lipoxygenase: A crucial enzyme in diverse types of cell death. Neurochem. Int. 2018, 118, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Van der Vlag, R.; Guo, H.; Hapko, U.; Eleftheriadis, N.; Monjas, L.; Dekker, F.J.; Hirsch, A.K.H. A combinatorial approach for the discovery of drug-like inhibitors of 15-lipoxygenase-1. Eur. J. Med. Chem. 2019, 174, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Gawlik-Dziki, U.; Świeca, M.; Dziki, D.; Kowalska, I.; Pecio, Ł.; Durak, A.; Sęczyk, Ł. Lipoxygenase inhibitors and antioxidants from green coffee—mechanism of action in the light of potential bioaccessibility. Food Res. Int. 2014, 61, 48–55. [Google Scholar] [CrossRef]
- Cretu, E.; Karonen, M.; Salminen, J.-P.; Mircea, C.; Trifan, A.; Charalambous, C.; Constantinou, A.I.; Miron, A. In vitro study on the antioxidant activity of a polyphenol-rich extract from Pinus brutia bark and its fractions. J. Med. Food 2013, 16, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Pisoschi, A.M.; Pop, A.; Cimpeanu, C.; Predoi, G. Antioxidant capacity determination in plants and plant-derived products: A review. Oxid. Med. Cell. Longev. 2016, 2016, 1–36. [Google Scholar] [CrossRef]
- Flora, S.J.S. Structural, chemical and biological aspects of antioxidants for strategies against metal and metalloid exposure. Oxid. Med. Cell. Longev. 2009, 2, 191–206. [Google Scholar] [CrossRef]
- Amarowicz, R.; Pegg, R.B. Natural antioxidants of plant origin. In Advances in Food and Nutrition Research; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Calderon-Montano, M.J.; Burgos-Moron, E.; Perez-Guerrero, C.; Lopez-Lazaro, M. A review on the dietary flavonoid kaempferol. Mini-Reviews Med. Chem. 2011, 11, 298–344. [Google Scholar] [CrossRef]
- Neganova, M.E.; Afanas’eva, S.V.; Klochkov, S.G.; Shevtsova, E.F. Mechanisms of antioxidant effect of natural sesquiterpene lactone and alkaloid derivatives. Bull. Exp. Biol. Med. 2012, 152, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chon, J.K.; Kim, S.; Shin, W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins Struct. Funct. Bioinforma. 2008, 70, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Malterud, K.E.; Rydland, K.M. Inhibitors of 15-lipoxygenase from orange peel. J. Agric. Food Chem. 2000, 48, 5576–5580. [Google Scholar] [CrossRef] [PubMed]
- Venditti, E.; Bacchetti, T.; Tiano, L.; Carloni, P.; Greci, L.; Damiani, E. Hot vs. cold water steeping of different teas: Do they affect antioxidant activity? Food Chem. 2010, 119, 1597–1604. [Google Scholar] [CrossRef]
- Burlec, A.F.; Cioancă, O.; Mircea, C.; Arsene, C.; Tuchiluş, C.; Corciovă, A.; Hăncianu, M. Antioxidant and antimicrobial properties of Chrysanthemum and Tagetes selective extracts. Farmacia 2019, 67, 405–410. [Google Scholar] [CrossRef]
- Assaad, H.I.; Zhou, L.; Carroll, R.J.; Wu, G. Rapid publication-ready MS-Word tables for one-way ANOVA. Springerplus 2014, 3, 474. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
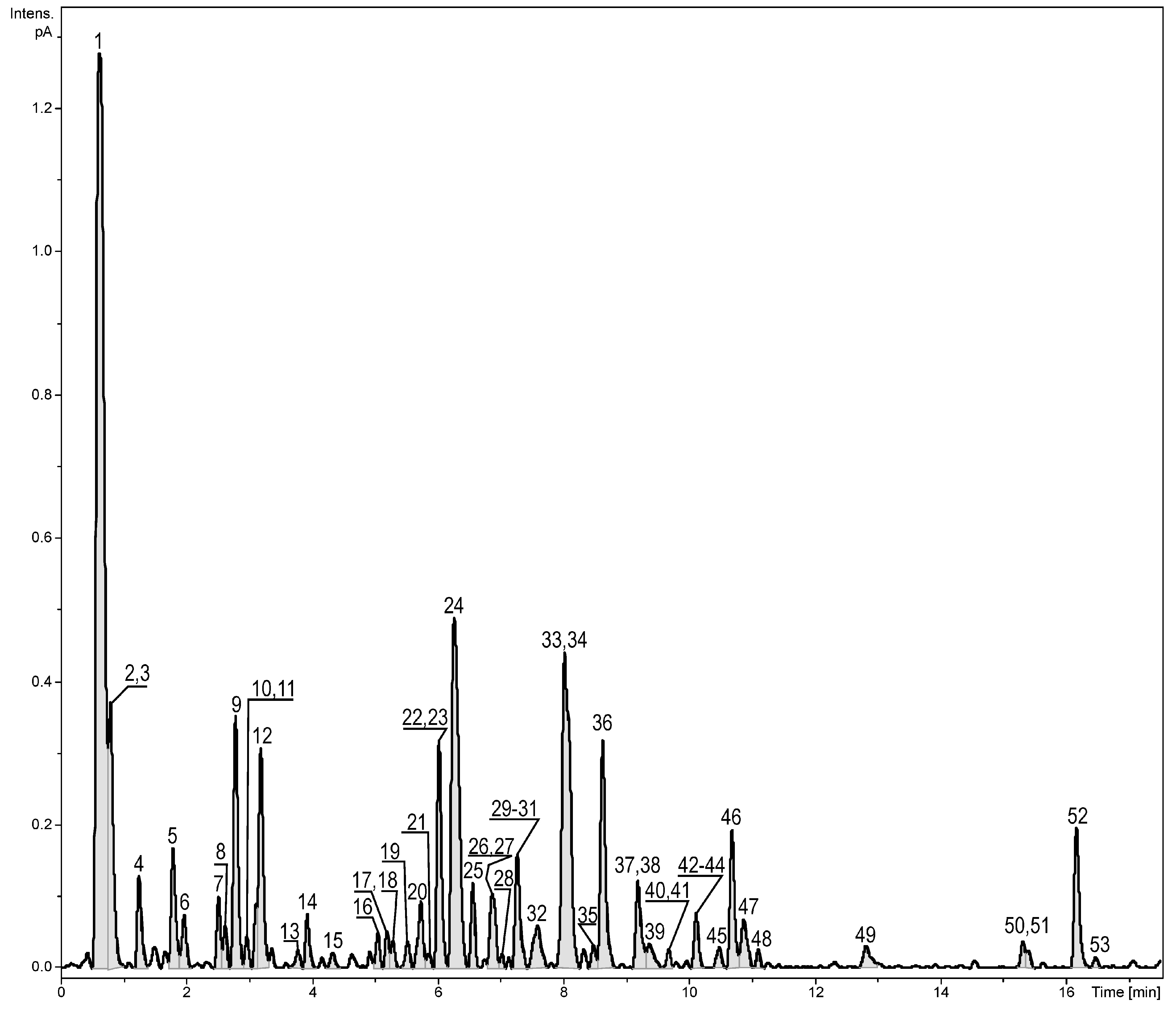
| No. | Compound Name | RT (min) | λmax (nm) | Formula | Error (ppm) ** | mσ *** | Observed [M − H]− | Major Fragments (%) | Fraction | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Quinic acid | 0.62 | - | C7H12O6 | 0.5 | 1.5 | 191.0560 | 173.0450 (2) | 1,2 | [14] |
| 2. | Hexoso(iso)leucine | 0.79 | - | C12H23NO7 | −0.7 | 1.4 | 294.1549 * | 276.1443 (100), 258.1338 (19), 230.1388 (17), 294.1548 (12), 132.1021 (5) | 1 | [15] |
| 3. | (Iso)leucine | 0.79 | - | C6H13NO2 | −0.6 | 2.8 | 132.1020 * | 132.1019 (100), 86.0967 (1) | 1 | [15] |
| 4. | Phenylalanine | 1.24 | - | C9H11NO2 | 5.3 | 15.7 | 164.0708 | - | 1 | [15] |
| 5. | 3-CQA | 1.79 | 250, 325 | C16H18O9 | −0.1 | 3.6 | 353.0878 | 191.0565 (100), 179.0354 (37), 135.0436 (16) | 1,2 | [16] |
| 6. | Tryptophan | 1.96 | 280 | C11H12N2O2 | −0.9 | 9.1 | 205.0972 * | 188.0708 (100), 146.0603 (26), 144.0811 (6), 205.0974 (6) | 1,2 | [15] |
| 7. | 3-pCoQA | 2.51 | 310 | C16H18O8 | −0.4 | 9.0 | 337.0930 | 163.0399 (100), 119.0495 (37), 191.0556 (19) | 1,2 | [16] |
| 8. | 1-pCoQA | 2.62 | 305 | C16H18O8 | −1.0 | 17.1 | 337.0932 | 191.0567 (100), 163.0400 (34), 119.0492 (9) | 1,2 | [16] |
| 9. | 5-CQA | 2.78 | 245, 325 | C16H18O9 | −0.4 | 3.4 | 353.0880 | 191.0563 (100), 179.0265 (1), 135.0433 (1), 173.0438 (1) | 1,2,3 | [16] |
| 10. | Caffeic acid | 2.96 | 320 | C9H8O4 | −0.3 | 1.8 | 179.0353 | 135.0441 (100), 179.0359 (42) | 2,3,4 | [17] |
| 11. | 4-CQA | 3.11 | 325 | C16H18O9 | −0.7 | 17.0 | 353.0880 | 191.0567 (100), 179.0351 (48), 173.0454 (40), 135.0439 (31) | 2,3 | [16] |
| 12. | Plumbagine B | 3.18 | - | C11H17N3O2 | −0.3 | 1.1 | 222.1251 | 222.1251 (100), 178.1351 (12) | 1 | [18] |
| 13. | Plumbagine B - isomer | 3.77 | - | C11H17N3O2 | −0.4 | 6.0 | 224.1395 * | - | 1 | [18] |
| 14. | 5-pCoQA epimer | 3.92 | 310 | C16H18O8 | −2.5 | 2.0 | 337.0937 | 191.0567 (100), 163.0402 (8) | 1,2 | [16] |
| 15. | p-Coumaric acid | 4.32 | 310 | C9H8O3 | 0.0 | 3.6 | 165.0546 * | 147.0441 (100), 119.0491 (11), 165.0543 (9) | 2 | [17] |
| 16. | Quercetin-3-O-(hexosyl-hexuronide) | 5.05 | 255, 345 | C27H28O18 | 0.8 | 5.6 | 641.1343 * | 303.0499 (100), 479.0821 (40), 301.0344 (12) | 2,3,4 | [19] |
| 17. | Clovamide | 5.19 | 290, 320 | C18H17NO7 | −1.9 | 40.4 | 358.0939 | 178.0498 (100), 179.0346 (61), 161.0247 (30) | 2,3,4 | [20,21] |
| 18. | 5-pCoQA epimer | 5.29 | 300 | C16H18O8 | −2.6 | 26.4 | 337.0938 | 191.0566 (100) | 1,2 | [16] |
| 19. | Methyl-Plumbagine B | 5.52 | - | C12H19N3O2 | 1.9 | 10.1 | 238.1550 * | 238.1546 (100), 178.1333 (1), 196.1326 (0.3), 136.1117 (0.2), 110.0959 (0.1) | 1,2 | [18] |
| 20. | Quercetin-3-O-(pentosyl-hexoside) | 5.73 | 265, 350 | C26H28O16 | −2.0 | 5.6 | 595.1316 | 300.0288 (100), 271.0248 (30) | 2,3,4 | [21] |
| 21. | Plantagoguanidinic acid isomer | 5.87 | - | C11H19N3O2 | 2.2 | 9.6 | 226.1550 * | 226.1544 (100), 208.1437 (5), 180.1488 (1), 149.0955 (1) | 1 | [18] |
| 22. | Kaempferol 3-O-[β-glucopyranosyl-(1→2)-β-glucuronopyranoside] | 6.02 | 265, 345 | C27H28O17 | −1.9 | 3.9 | 623.1266 | 285.0409 (100), 229.0510 (21), 257.0461 (15), 241.0508 (3) | 2,3 | - |
| 23. | Kaempferol 3-O-(hexosyl-hexoside) | 6.06 | 260, 345 | C27H30O16 | 3.4 | 24.7 | 611.1586 * | 287.0545 (100), 449.1073 (92), 611.1597 (38), 226.1533 (4) | 3 | [22] |
| 24. | Plantagoguanidinic acid | 6.26 | - | C11H19N3O2 | −1.4 | 5.1 | 224.1408 | 141.0913 (100), 224.1404 (44), 180.1510 (32) | 1 | [18] |
| 25. | Quercetin 3-O-hexoside | 6.56 | 255, 350 | C21H20O12 | −1.3 | 1.9 | 463.0888 | 300.0285 (100) | 3,4,5 | [23] |
| 26. | Kaempferol 3-O-(pentosyl-hexoside) | 6.87 | 265, 345 | C26H28O15 | 2.1 | 14.3 | 581.1501 * | 287.0542 (100), 449.1067 (9), 163.0601 (1), 145.0495 (1) | 2,3,4 | [19] |
| 27. | Kaempferol 3-O-(pentosyl-hexuronide) | 6.87 | 265, 345 | C26H26O16 | 2.4 | 19.4 | 595.1279 * | 287.0546 (100), 463.0866 (42), 273.0748 (12) | 2,3,4 | [19] |
| 28. | Kaempferol 3-O-(pentosyl-hexoside) | 7.02 | 265, 345 | C26H28O15 | -1.8 | 7.5 | 579.1355 | 284.0330 (100), 255.0311 (37), 227.0344 (17) | 2,3,4 | [19] |
| 29. | Kaempferol 3-O-pentoside-7-O-hexuronide | 7.27 | 265, 345 | C26H26O16 | 1.0 | 13.1 | 595.1288 * | 287.0541 (100), 463.0866 (33), 433.1130 (12), 271.0587 (10) | 2,3,4 | [19] |
| 30. | Kaempferol 3-O-hexoside | 7.27 | 265, 345 | C21H20O11 | 2.1 | 3.5 | 449.1069 * | 287.0545 (100), 449.1060 (15) | 4,5 | [19] |
| 31. | Apigenin 7-O-dihexoside | 7.27 | 265, 345 | C27H30O15 | −0.7 | 8.4 | 593.1516 | 269.0462 (100) | 2,3 | [19] |
| 32. | Kaempferol 3-O-hexoside | 7.59 | 260, 335 | C21H20O11 | −0.9 | 11.3 | 447.0937 | 284.0340 (100), 255.0312 (55) | 2,3,4,5 | [22] |
| 33. | Kaempferol-3-O-hexuronide | 8.02 | 245, 325 | C21H18O12 | −0.9 | 13.0 | 461.0725 | 285.0409 (100), 229.0502 (22), 257.0458 (10) | 1,2,3,4 | [19] |
| 34. | 1,5-diCQA/3,5-diCQA | 8.02 | 245, 325 | C25H24O12 | 0.1 | 1.7 | 515.1195 | 191.0562 (100), 179.0349 (10), 173.0450 (3)/191.0564 (100), 179.0349 (34), 135.0445 (16)353.0876 (12) | 3,4,5 | [16] |
| 35. | Kaempferol-3-O-hexoside | 8.47 | 265, 340 | C21H20O11 | 0.1 | 2.7 | 447.0932 | 285.0404 (100), 257.0456 (1), 241.0497 (1) | 2,3,4,5 | [19] |
| 36. | Resokaempferol 3-O-hexoside | 8.63 | 265, 335 | C21H20O10 | 0.6 | 5 | 431.0981 | 268.0379 (100) | 3,4,5 | [19] |
| 37. | 3,4-diCQA | 9.19 | 325 | C25H24O12 | 0.7 | 5.8 | 515.1191 | 191.0558 (100), 173.0451 (77) | 3,4,5 | [16] |
| 38. | Kaempferol-3-O-(malonyl-hexoside) | 9.19 | 325 | C24H22O14 | 1.1 | 21.7 | 535.1076 * | 287.0545 (100), 535.1076 (60), 285.0388 (12), 257.0442 (6), 449.1067 (3) | 3,4,5 | [19] |
| 39. | pCo,CQA isomer | 9.37 | 320 | C25H24O11 | 1.9 | 8.6 | 501.1382 * | 163.0383 (100), 147.0436 (99), 483.1278 (32), 337.0924 (5) | 3 | [16] |
| 40. | C13-norisoprenoid hexoside | 9.68 | - | C19H32O7 | 2.6 | 14.9 | 373.2211 * | 211.1690 (100), 193.1585 (25), 135.1162 (12), 175.1473 (11) | 1 | [24] |
| 41. | Kaempferol 3-O-(caffeoyl-pentoside)-7-O-hexuronide | 9.68 | 330 | C35H32O19 | 2.4 | 12.7 | 757.1592 * | 287.0546 (100), 277.0704 (36), 463.0868 (35), 163.0388 (22), 295.0807 (11) | 5 | [19] |
| 42. | Methyl-plantagoguanidinic acid | 10.11 | - | C12H21N3O2 | 2.2 | 4.1 | 240.1701 * | 240.1704 (100), 208.1439 (4), 181.1214 (0.6) | 1 | [18] |
| 43. | Apigenin 7-O-(malonyl-hexoside) | 10.11 | 330 | C24H22O13 | 2.0 | 9.6 | 519.1123 * | 271.0596 (100), 519.1127 (27) | 3,4 | [19] |
| 44. | Kaempferol-3-O-(acetyl-hexoside) | 10.11 | 330 | C23H22O12 | 1.7 | 54.6 | 491.1176 * | 287.0544 (100) | 3 | [19] |
| 45. | Kaempferol-3-O-(malonyl-hexoside) | 10.48 | 265, 330 | C24H22O14 | 2.0 | 7.0 | 535.1072 * | 287.0550 (100), 535.1076 (91) | 3,4,5 | [19] |
| 46. | Apigenin 7-O-(malonyl-hexoside) | 10.69 | 265, 335 | C24H22O13 | 1.3 | 4.7 | 519.1126 * | 271.0601 (100), 519.1133 (54) | 3,4 | [22] |
| 47. | Apigenin 7-O-(malonyl-hexoside) | 10.87 | 270, 330 | C24H22O13 | 1.4 | 3.4 | 519.1126 * | 519.1129 (100), 271.0599 (93) | 3,4 | [19] |
| 48. | Kaempferol 3-O-(p-coumaroyl-pentoside)-7-O-hexuronide | 11.10 | 265, 320 | C35H32O18 | 1.0 | 27.3 | 741.1654 * | 287.0550 (100), 261.0761 (53), 463.0873 (35), 147.0441 (21), 279.0862 (16) | 3,4 | [19] |
| 49. | Apigenin | 12.82 | 265, 335 | C15H10O5 | 1.1 | 0.8 | 269.0452 | 225.0559 (6), 117.0326 (2) | 3,4,5 | [25] |
| 50. | Di-p-coumaroyl-caffeoyl-C18H36O6 | 15.31 | 295, 310 | C45H54O13 | 2.4 | 23.6 | 803.3618 * | 147.0441 (100), 657.3271 (44), 163.0388 (27), 641.3336 (14), 275.1751 (13), 204.1017 (11), 511.2924 (11), 495.2958 (10), 119.0490 (8), 291.1697 (6), 655.3094 (5) | 2,3 | - |
| 51. | Di-p-coumaroyl-caffeoyl-C18H36O6 | 15.40 | 295, 310 | C45H54O13 | 1.2 | 31.6 | 803.3628 * | 147.0441 (100), 657.3277 (47), 163.0388 (41), 204.1016 (22), 275.1750 (20), 641.3322 (20), 495.2967 (16), 511.2907 (9) | 2,3 | - |
| 52. | Tri-p-coumaroyl-C18H36O6 | 16.17 | 300 | C45H54O12 | 0.9 | 21.3 | 787.3681 * | 147.043 (100), 641.3324 (53), 204.1017 (20), 275.1750 (15), 495.2962 (15), 119.0487 (7), 477.2854 (4), 349.2598 (1) | 2,3 | - |
| 53. | 3-O-Methyl-kaempferol | 16.47 | - | C16H12O6 | 1.0 | 8.9 | 301.0704 * | 301.0704 (100) | 5 | [19] |
| Position. | δH (J in Hz) | δC, Type |
|---|---|---|
| 2 | 159.0, C | |
| 3 | 134.7, C | |
| 4 | 179.4, C | |
| 5 | 163.1, C | |
| 6 | 6.20 d (2.0) | 99.9, CH |
| 7 | 165.9, C | |
| 8 | 6.39 d (2.0) | 94.8, CH |
| 9 | 158.5, C | |
| 10 | 105.8, C | |
| 1′ | 122.7, C | |
| 2′/6′ | 8.02 d (8.5) | 132.3, CH |
| 3′/5′ | 6.90 d (8.5) | 116.3, CH |
| 4′ | 161.5, C | |
| 1′’ | 5.56 d (7.4) | 101.1, CH |
| 2′’ | 3.81 dd (9.0, 7.4) | 81.9, CH |
| 3′’ | 3.67 t (9.0) | 77.3, CH |
| 4′’ | 3.64 t (9.0) | 72.6, CH |
| 5′’ | 3.76 d (9.0) | 76.8, CH |
| 6′’ | 171.7, C | |
| 1′’’ | 4.75 d (7.3) | 104.6, CH |
| 2′’’ | 3.35 dd (9.5, 7.3) | 75.5, CH |
| 3′’’ | 3.38 overlap | 77.9, CH |
| 4′’’ | 3.37 overlap | 71.3, CH |
| 5′’’ | 3.28 ddd (8.4, 5.0, 2.2) | 78.2, CH |
| 6′’’ | 3.78 dd (12.0, 2.2)3.68 dd (12.0, 5.0) | 62.6, CH2 |
| Lipoxygenase Inhibition | Iron-Chelating Activity | |
|---|---|---|
| Sample | IC50 (μg/mL Final Solution) | EC50 (mg/mL Final Solution) |
| Fr 1 | 65.65 ± 0.50 a * | 0.714 ± 0.001 e |
| Fr 2 | 18.98 ± 0.22 d | 1.037 ± 0.003 d |
| Fr 3 | 30.25 ± 0.73 c | 1.620 ± 0.006 c |
| Fr 4 | 41.67 ± 1.46 b | 1.919 ± 0.011 a |
| Fr 5 | 69.37 ± 6.71 a | 1.664 ± 0.011 b |
| Initial extract | 69.21 ± 0.89 ** a | 0.615 ± 0.001 f |
| Positive control | 20.25 ± 0.44 d | 0.068 ± 0.003 g |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burlec, A.F.; Pecio, Ł.; Mircea, C.; Cioancă, O.; Corciovă, A.; Nicolescu, A.; Oleszek, W.; Hăncianu, M. Chemical Profile and Antioxidant Activity of Zinnia elegans Jacq. Fractions. Molecules 2019, 24, 2934. https://doi.org/10.3390/molecules24162934
Burlec AF, Pecio Ł, Mircea C, Cioancă O, Corciovă A, Nicolescu A, Oleszek W, Hăncianu M. Chemical Profile and Antioxidant Activity of Zinnia elegans Jacq. Fractions. Molecules. 2019; 24(16):2934. https://doi.org/10.3390/molecules24162934
Chicago/Turabian StyleBurlec, Ana Flavia, Łukasz Pecio, Cornelia Mircea, Oana Cioancă, Andreia Corciovă, Alina Nicolescu, Wiesław Oleszek, and Monica Hăncianu. 2019. "Chemical Profile and Antioxidant Activity of Zinnia elegans Jacq. Fractions" Molecules 24, no. 16: 2934. https://doi.org/10.3390/molecules24162934
APA StyleBurlec, A. F., Pecio, Ł., Mircea, C., Cioancă, O., Corciovă, A., Nicolescu, A., Oleszek, W., & Hăncianu, M. (2019). Chemical Profile and Antioxidant Activity of Zinnia elegans Jacq. Fractions. Molecules, 24(16), 2934. https://doi.org/10.3390/molecules24162934










