A DNA-Based Biosensor Assay for the Kinetic Characterization of Ion-Dependent Aptamer Folding and Protein Binding
Abstract
1. Introduction
2. Results
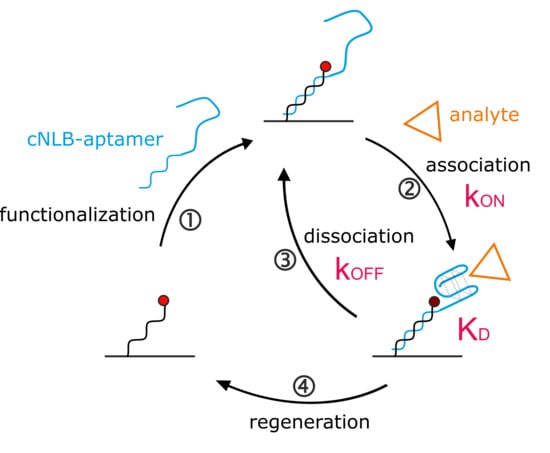
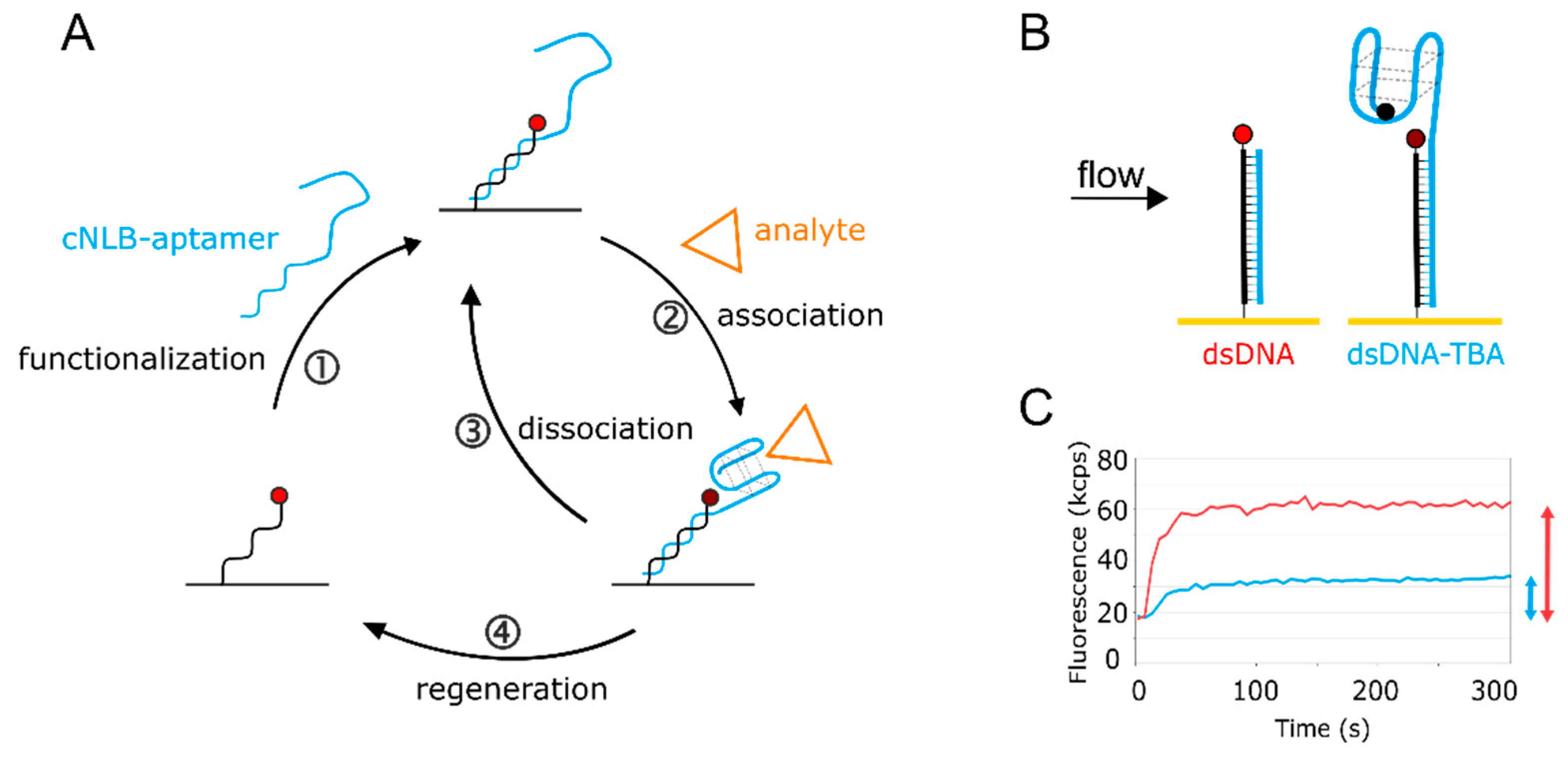
2.1. Development of the Aptasensor
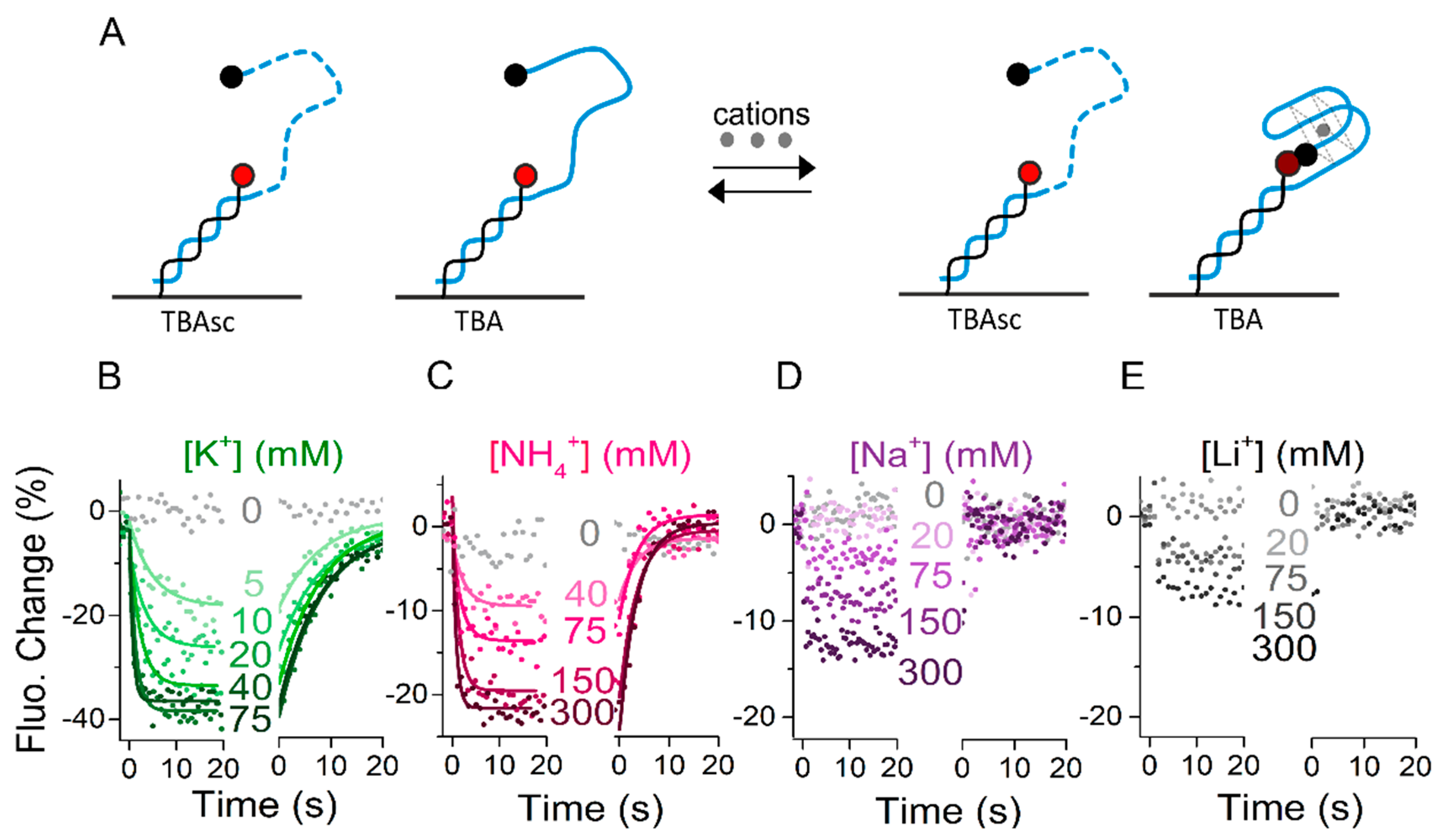
2.2. Real-Time Kinetics of Aptamer Folding
2.3. Influence of Salt Species on Thrombin-Binding Kinetics
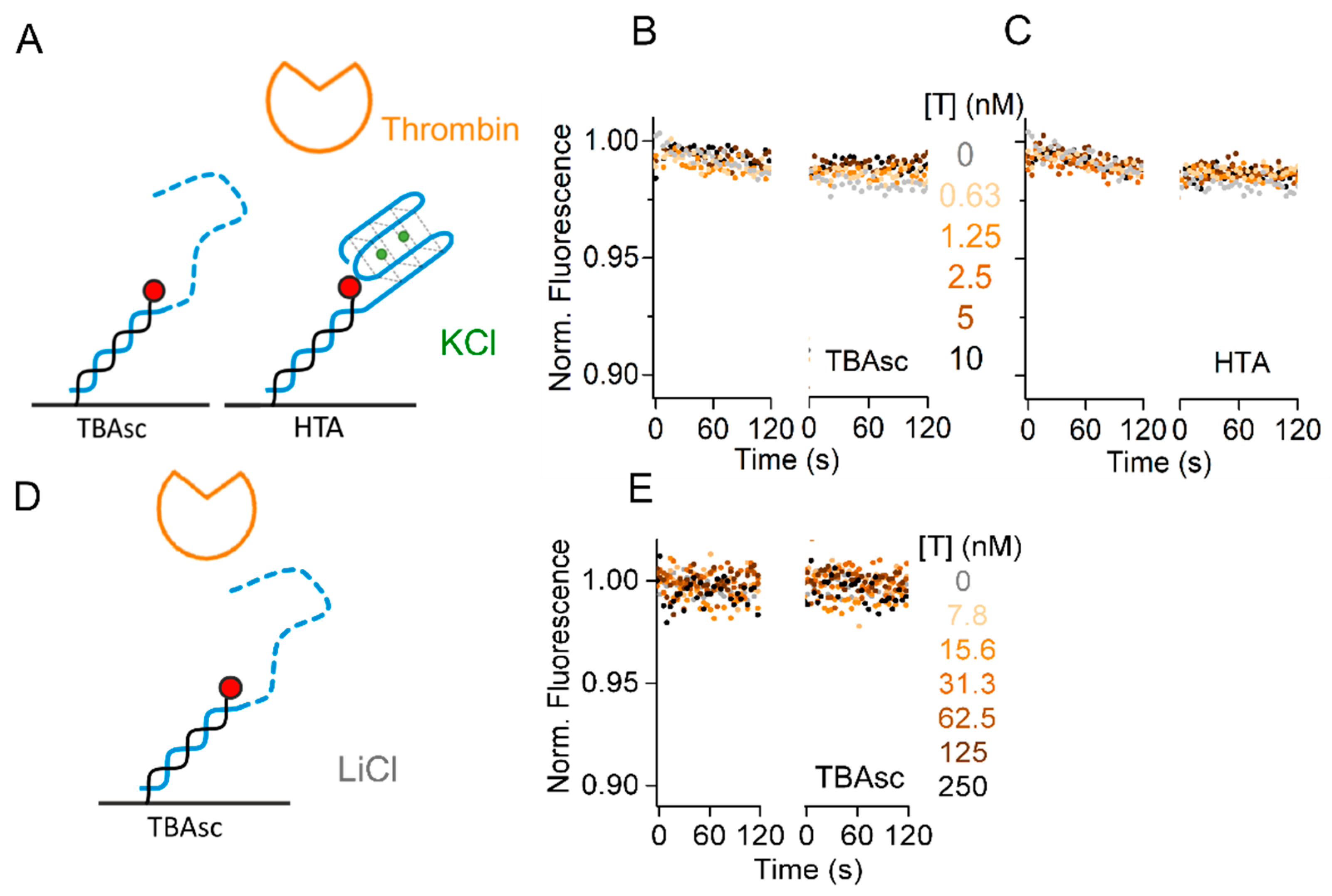
2.4. Specificity of TBA for Thrombin Interaction Allows Broad Dynamic Range
2.5. Immobilized Target for Aptamer Screening
3. Discussion
3.1. Real-Time Folding and Unfolding Rates Coupled to Ion Binding
3.2. Resolution of High-Affinity TBA–Protein Interaction
3.3. Differentiation of Prefolded Aptamer from Induced Fit
3.4. Contribution of Results to Therapeutic and Diagnostic Applications
3.5. Effect of Reversing the Assay Orientation
4. Materials and Methods
4.1. Materials
- TBA: 5′(BBQ)-GGT TGG TGT GGT TGG TTT ATC AGC GTT CGA TGC TTC CGA CTA ATC AGC CAT ATC AGC TTA CGA CTA-3′;
- TBAsc: 5′(BBQ)-GTG TGG TGT GTG TGG TTT ATC AGC GTT CGA TGC TTC CGA CTA ATC AGC CAT ATC AGC TTA CGA CTA-3′;
- HTA: 5′-TTT GGG TTA GGG TTA GGG TTA GGG TTT ATC AGC GTT CGA TGC TTC CGA CTA ATC AGC CAT ATC AGC TTA CGA CTA-3′.
4.2. Covalent Conjugation of Thrombin to cNL-B
4.3. SwitchSENSE Experiments
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Nilsen-Hamilton, M. Aptamers: Multifunctional molecules for biomedical research. J. Mol. Med. 2013, 91, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar]
- Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules 2016, 21, 1613. [Google Scholar] [CrossRef]
- Drolet, D.W.; Green, L.S.; Gold, L.; Janjic, N. Fit for the Eye: Aptamers in Ocular Disorders. Nucleic Acid Ther. 2016, 26, 127–146. [Google Scholar] [CrossRef]
- Bock, C.L.; Griffin, L.C.; Latham, J.A.; Veermas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Shuman, M.A.; Majerus, P.W. The measurement of thrombin in clotting blood by radioimmunoassay. J. Clin. Investig. 1976, 58, 1249–1258. [Google Scholar] [CrossRef]
- Brummel-Ziedins, K.E.; Vossen, C.Y.; Butenas, S.; Mann, K.G.; Rosendaal, F.R. Thrombin generation profiles in deep venous thrombosis. J. Thromb. Haemost. 2005, 3, 2497–2505. [Google Scholar] [CrossRef]
- Deng, B.; Lin, Y.; Wang, C.; Li, F.; Wang, Z.; Zhang, H.; Li, X.-F.; Le, X.C. Aptamer binding assays for proteins: The thrombin example—A review. Anal. Chim. Acta 2014, 837, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, Y.; Shimizu, N.; Ogino, C.; Kondo, A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010, 38, e21. [Google Scholar] [CrossRef]
- Lin, P.-H.; Chen, R.-H.; Lee, C.-H.; Chang, Y.; Chen, C.-S.; Chen, W.-Y. Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry. Coll. Surf. B Biointerfaces 2011, 88, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.W.; Tan, Q.; Gu, L.-Q. Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res. 2009, 37, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef]
- Henderson, E.; Walk, S.K.; Blackburn’, H. Telomeric DNA Oligonucleotides Form Novel Intramolecular StrucWes Containing Guanine=Guanine Base F%iirs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Trajkovski, M.; Šket, P.; Plavec, J. Cation localization and movement within DNA thrombin binding aptamer in solution. Org. Biomol. Chem. 2009, 7, 4677. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin Binding Aptamer, More than a Simple Aptamer: Chemically Modified Derivatives and Biomedical Applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, E.; O’Sullivan, C.K. Ability of thrombin to act as molecular chaperone, inducing formation of quadruplex structure of thrombin-binding aptamer. Anal. Biochem. 2005, 341, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.L.; McKeague, M.; Liang, J.C.; Smolke, C.D. Kinetic and Equilibrium Binding Characterization of Aptamers to Small Molecules using a Label-Free, Sensitive, and Scalable Platform. Anal. Chem. 2014, 86, 3273–3278. [Google Scholar] [CrossRef] [PubMed]
- Rant, U.; Arinaga, K.; Fujita, S.; Yokoyama, N.; Abstreiter, G.; Tornow, M. Dynamic Electrical Switching of DNA Layers on a Metal Surface. Nano Lett. 2004, 4, 2441–2445. [Google Scholar] [CrossRef]
- Knezevic, J.; Langer, A.; Hampel, P.A.; Kaiser, W.; Strasser, R.; Rant, U. Quantitation of Affinity Avidity and Binding Kinetics of Protein Analytes with a Dynamically Switchable Biosurface. J. Am. Chem. Soc. 2012, 134, 15225–15228. [Google Scholar] [CrossRef] [PubMed]
- Cléry, A.; Sohier, T.J.M.; Welte, T.; Langer, A.; Allain, F.H.T. switchSENSE: A new technology to study protein-RNA interactions. Methods 2017, 118–119, 137–145. [Google Scholar]
- Kankia, B.I.; Marky, L.A. Folding of the Thrombin Aptamer into a G-Quadruplex with Sr 2+: Stability, Heat, and Hydration. J. Am. Chem. Soc. 2001, 123, 10799–10804. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Maiti, S. Quadruplex to Watson–Crick duplex transition of the thrombin binding aptamer: A fluorescence resonance energy transfer study. Biochem. Biophys. Res. Commun. 2004, 319, 759–767. [Google Scholar] [CrossRef]
- Joachimi, A.; Benz, A.; Hartig, J.S. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009, 17, 6811–6815. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Hardin, C.C.; Watson, T.; Corregan, M.; Bailey, C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry 1992, 31, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Phan, A.-T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef]
- Cho, M.-S.; Kim, Y.-W.; Han, S.-Y.; Min, K.-I.; Rahman, M.D.A.; Shim, Y.-B.; Ban, C.-I. Detection for folding of the thrombin binding aptamer using label-free electrochemical methods. BMB Rep. 2008, 41, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.S.; Yoon, H.-J.; Kim, B.; Yim, Y.-H.; So, H.-Y.; Shin, S.K. Mass spectrometric studies of alkali metal ion binding on thrombin-binding aptamer DNA. J. Am. Soc. Mass Spectrom. 2010, 21, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Freitag, D.; Mayer, G.; Pötzsch, B. Anticoagulant characteristics of HD1-22, a bivalent aptamer that specifically inhibits thrombin and prothrombinase. J. Thromb. Haemost. 2008, 6, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.-A.; Leclerc, M. Optical Sensors Based on Hybrid Aptamer/Conjugated Polymer Complexes. J. Am. Chem. Soc. 2004, 126, 1384–1387. [Google Scholar] [CrossRef]
- Nagatoishi, S.; Tanaka, Y.; Tsumoto, K. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007, 352, 812–817. [Google Scholar] [CrossRef]
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer Beacons for the Direct Detection of Proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef]
- Torimura, M.; Kurata, S.; Yamada, K.; Yokomaku, T.; Kamagata, Y.; Kanagawa, T.; Kurane, R. Fluorescence-Quenching Phenomenon by Photoinduced Electron Transfer between a Fluorescent Dye and a Nucleotide Base. Anal. Sci. 2001, 17, 155–160. [Google Scholar] [CrossRef]
- D.B.G. Compatibility Sheet. Available online: https://www.dynamic-biosensors.com/project/switchsense-compatibility-sheet/ (accessed on 1 May 2017).
- Su, C.T.-T.; Ling, W.-L.; Lua, W.-H.; Poh, J.-J.; Gan, S.K.-E. The role of Antibody Vκ Framework 3 region towards Antigen binding: Effects on recombinant production and Protein L binding. Sci. Rep. 2017, 7, 3766. [Google Scholar] [CrossRef]
Sample Availability: Sources for Thrombin-binding aptamer and thrombin are stated in Materials & Methods, these samples are not available from the authors. |


| Ion | kON (M−1s−1) | kOFF = kU (10−1 s−1) | KD (mM) | kF (s−1) | KF (No Unit) |
|---|---|---|---|---|---|
| NH4+ | 2.96 ± 0.8 | 3.07 ± 0.3 | 104 ± 40 | 1.18 ± 0.23 | 3.84 ± 0.8 |
| K+ | 15.2 ± 1.7 | 1.20 ± 0.01 | 7.89 ± 1.3 | 1.26 ± 0.14 | 10.4 ± 1.1 |
| Na+ | - | - | - | - | - |
| Li+ | - | - | - | - | - |
| Buffer | kON (107 M−1s−1) | kOFF (10−2 s−1) | KD (nM) |
|---|---|---|---|
| TE140-KCl | 38 ± 1.3 | 5.8 ± 0.1 | 0.15 ± 0.01 |
| TE140-NH4Cl | 17 ± 1.0 | 4.8 ± 0.1 | 0.28 ± 0.02 |
| TE140-NaCl | 3.3 ± 0.2 | 4.3 ± 0.1 | 1.31 ± 0.12 |
| TE140-LiCl | 0.03 ± 0.01 | 6.3 ± 0.4 | 245 ± 94 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponzo, I.; Möller, F.M.; Daub, H.; Matscheko, N. A DNA-Based Biosensor Assay for the Kinetic Characterization of Ion-Dependent Aptamer Folding and Protein Binding. Molecules 2019, 24, 2877. https://doi.org/10.3390/molecules24162877
Ponzo I, Möller FM, Daub H, Matscheko N. A DNA-Based Biosensor Assay for the Kinetic Characterization of Ion-Dependent Aptamer Folding and Protein Binding. Molecules. 2019; 24(16):2877. https://doi.org/10.3390/molecules24162877
Chicago/Turabian StylePonzo, Irene, Friederike M. Möller, Herwin Daub, and Nena Matscheko. 2019. "A DNA-Based Biosensor Assay for the Kinetic Characterization of Ion-Dependent Aptamer Folding and Protein Binding" Molecules 24, no. 16: 2877. https://doi.org/10.3390/molecules24162877
APA StylePonzo, I., Möller, F. M., Daub, H., & Matscheko, N. (2019). A DNA-Based Biosensor Assay for the Kinetic Characterization of Ion-Dependent Aptamer Folding and Protein Binding. Molecules, 24(16), 2877. https://doi.org/10.3390/molecules24162877