Variability of Bioactive Glucosinolates, Isothiocyanates and Enzyme Patterns in Horseradish Hairy Root Cultures Initiated from Different Organs
Abstract
1. Introduction
2. Results
2.1. In Vitro Horseradish Cultures
2.2. Identification of Glucosinolates in Horseradish Hairy Roots by LC-ESI-MS/MS Analysis
2.3. Detection of Nitriles and Isothiocyanates as GLS Breakdown Products from Horseradish Hairy Roots by gas Chromatography-Mass Spectrometry (GC-MS)
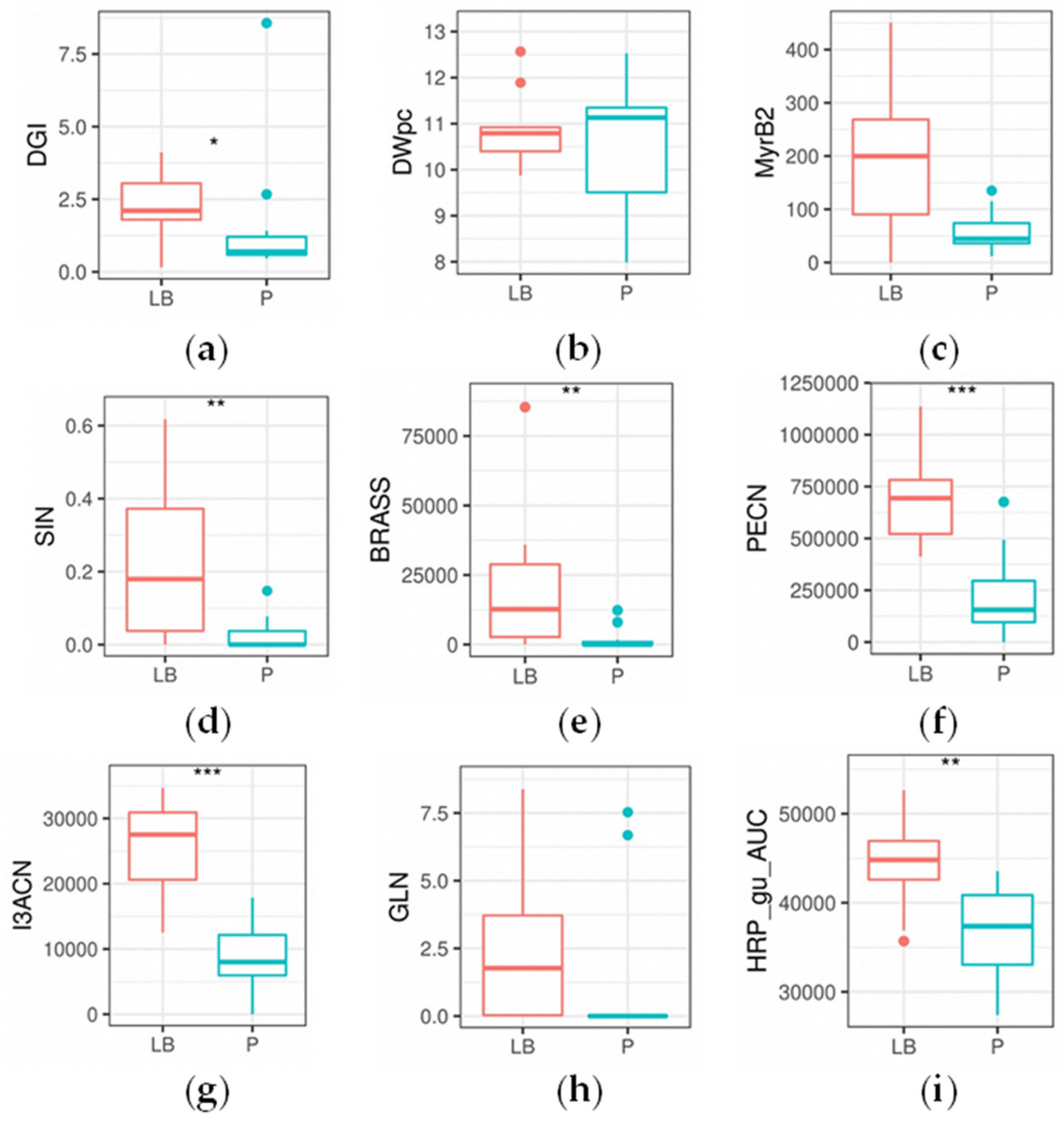
2.4. Enzyme Content of Horseradish Hairy Roots
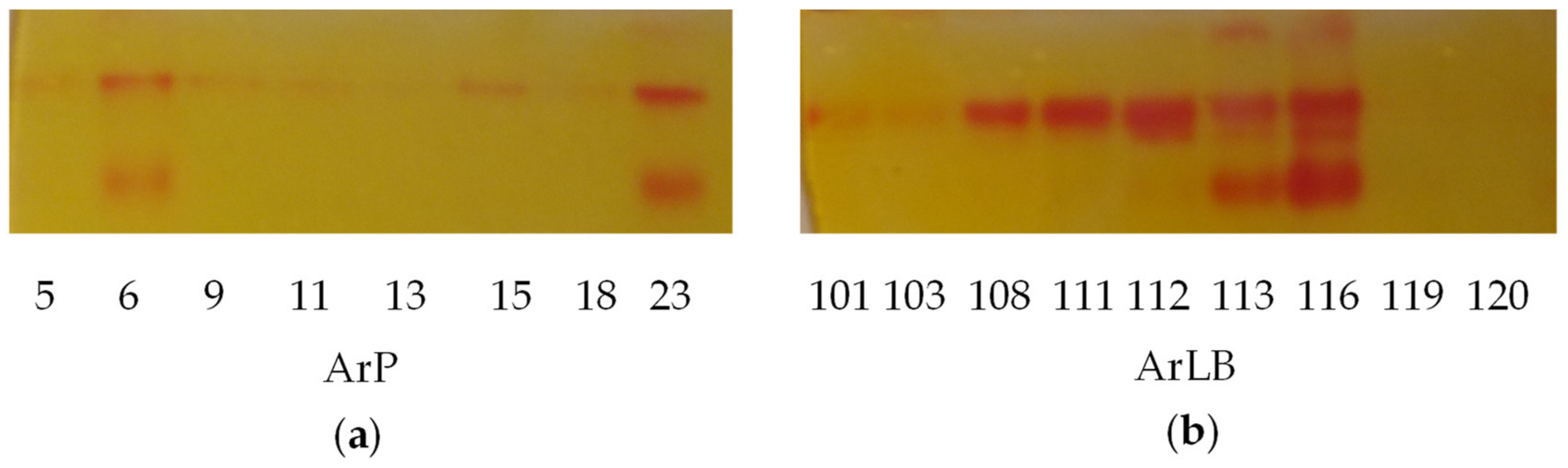
2.4.1. Myrosinase Activity
2.4.2. Peroxidase Content and Activity
2.5. Morphological Evaluations
2.6. Agrobacterial Inoculation of Different Horseradish Plant Organs Results in Different Feature Patterns in HRCs
3. Discussion
3.1. Comparison of Hairy Root and Native Root Glucosinolate Patterns
3.2. Indolic Glucosinolates and Their Breakdown Products
3.3. Agrobacterial Inoculation of Different Horseradish Plant Organs Results in Different Feature Pattern in HRCs
3.4. Likely Dependence of Biologically Active Compound Concentrations on Myrosinase Isoenzyme Pattern
4. Materials and Methods
4.1. Plant Material, In Vitro Cultures, Plant Transformation.
4.2. Confirmation of Transformation by PCR
4.3. Biomass Production
4.4. Gas Chromatography Measurements
4.5. Liquid Chromatography Measurements
4.6. Myrosinase Activity Analysis
4.7. Peroxidase Content and Activity
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shehata, A.; Mulwa, R.M.S.; Babadoost, M.; Uchanski, M.; Norton, M.A.; Skirvin, R.; Walters, S.A. Horseradish: Botany, horticulture, breeding. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 221–261. ISBN 978-0-47-059377-6. [Google Scholar]
- Agneta, R.; Möllers, C.; Rivelli, A.R. Horseradish (Armoracia rusticana), a neglected medical and condiment species with a relevant glucosinolate profile: A review. Genet. Resour. Crop Evol. 2013, 60, 1923–1943. [Google Scholar] [CrossRef]
- Nguyen, N.M.; Gonda, S.; Vasas, G. A Review on the Phytochemical Composition and Potential Medicinal Uses of Horseradish (Armoracia rusticana) Root. Food Rev. Int. 2013, 29, 261–275. [Google Scholar] [CrossRef]
- Gonda, S.; Kiss-Szikszai, A.; Szűcs, Z.; Nguyen, N.M.; Vasas, G. Myrosinase Compatible Simultaneous Determination of Glucosinolates and Allyl Isothiocyanate by Capillary Electrophoresis Micellar Electrokinetic Chromatography (CE-MEKC): Simultaneous Determination of Glucosinolates and AITC by CE-MEKC. Phytochem. Anal. 2016, 27, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Szűcs, Z.; Plaszkó, T.; Cziáky, Z.; Kiss-Szikszai, A.; Emri, T.; Bertóti, R.; Sinka, L.T.; Vasas, G.; Gonda, S. Endophytic fungi from the roots of horseradish (Armoracia rusticana) and their interactions with the defensive metabolites of the glucosinolate-myrosinase-isothiocyanate system. BMC Plant Biol. 2018, 18, 85. [Google Scholar] [CrossRef] [PubMed]
- Tomsone, L.; Kruma, Z.; Galoburda, R.; Talou, T. Composition of Volatile Compounds of Horseradish Roots (Armoracia rusticana L.) Depending on the Genotype. Proc. Latv. Univ. Agric. 2013, 29, 1–10. [Google Scholar] [CrossRef]
- Tisserand, R.; Young, R. Essential Oil Safety: A Guide for Health Care Professionals, 2nd ed.; Elsevier: Edinburgh, UK, 2013; ISBN 978-0-44-306241-4. [Google Scholar]
- Bones, A.M.; Rossiter, J.T. The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol. Plant. 1996, 97, 194–208. [Google Scholar] [CrossRef]
- Mcdanell, R.; Mclean, A.E.M.; Hanley, A.B.; Heane, R.K.; Fenwick, G.R. Chemical and biological properties of indole glucosinolates (glucobrassicins): A review. Food Chem. Toxicol. 1988, 26, 59–70. [Google Scholar] [CrossRef]
- Vig, A.P.; Rampal, G.; Thind, T.S.; Arora, S. Bio-protective effects of glucosinolates—A review. LWT Food Sci. Technol. 2009, 42, 1561–1572. [Google Scholar] [CrossRef]
- Li, X.; Kushad, M.M. Purification and characterization of myrosinase from horseradish (Armoracia rusticana) roots. Plant Physiol. Biochem. 2005, 43, 503–511. [Google Scholar] [CrossRef]
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. In Plant Molecular Evolution; Doyle, J.J., Gaut, B.S., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 93–113. ISBN 978-9-40-105833-9. [Google Scholar]
- Márton, M.-R.; Krumbein, A.; Platz, S.; Schreiner, M.; Rohn, S.; Rehmers, A.; Lavric, V.; Mersch-Sundermann, V.; Lamy, E. Determination of bioactive, free isothiocyanates from a glucosinolate-containing phytotherapeutic agent: A pilot study with in vitro models and human intervention. Fitoterapia 2013, 85, 25–34. [Google Scholar] [CrossRef]
- Angelino, D.; Dosz, E.B.; Sun, J.; Hoeflinger, J.L.; Van Tassell, M.L.; Chen, P.; Harnly, J.M.; Miller, M.J.; Jeffery, E.H. Myrosinase-dependent and -independent formation and control of isothiocyanate products of glucosinolate hydrolysis. Front. Plant Sci. 2015, 6, 831. [Google Scholar] [CrossRef] [PubMed]
- Gonda, S.; Szűcs, Z.; Plaszkó, T.; Cziáky, Z.; Kiss-Szikszai, A.; Vasas, G.; M-Hamvas, M. A Simple Method for On-Gel Detection of Myrosinase Activity. Molecules 2018, 23, 2204. [Google Scholar] [CrossRef] [PubMed]
- Akbar, H.; Sedzro, D.M.; Khan, M.; Bellah, S.F.; Billah, S.M.S. Structure, Function and Applications of a Classic Enzyme: Horseradish Peroxidase. J. Chem. Environ. Biol. Eng. 2018, 2, 52–59. [Google Scholar]
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y. Transgenic Horseradish (Armoracia rusticana). In Transgenic Crops II; Bajaj, Y.P.S., Ed.; Springer: Berlin, Germany, 2001; pp. 26–38. ISBN 978-3-64-256901-2. [Google Scholar]
- Gelvin, S.B. Agrobacterium-Mediated Plant Transformation: The Biology behind the “Gene-Jockeying” Tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Habibi, P.; Soccol, C.R.; Grossi-de-Sa, M.F. Hairy Root-Mediated Biotransformation: Recent Advances and Exciting Prospects. In Hairy Roots; Srivastava, V., Mehrotra, S., Mishra, S., Eds.; Springer: Singapore, 2018; pp. 185–211. ISBN 978-9-81-132561-8. [Google Scholar]
- Häkkinen, S.T.; Moyano, E.; Cusidó, R.M.; Oksman-Caldentey, K.-M. Exploring the Metabolic Stability of Engineered Hairy Roots after 16 Years Maintenance. Front. Plant Sci. 2016, 7, 1486. [Google Scholar] [CrossRef]
- Bourgaud, F.; Gravot, A.; Milesi, S.; Gontier, E. Production of plant secondary metabolites: A historical perspective. Plant Sci. 2001, 161, 839–851. [Google Scholar] [CrossRef]
- Srivastava, S.; Srivastava, A.K. Hairy Root Culture for Mass-Production of High-Value Secondary Metabolites. Crit. Rev. Biotechnol. 2007, 27, 29–43. [Google Scholar] [CrossRef]
- Tian, L. Using Hairy Roots for Production of Valuable Plant Secondary Metabolites. In Filaments in Bioprocesses; Krull, R., Bley, T., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 149, pp. 275–324. ISBN 978-3-31-920510-6. [Google Scholar]
- Kastell, A.; Smetanska, I.; Ulrichs, C.; Cai, Z.; Mewis, I. Effects of Phytohormones and Jasmonic Acid on Glucosinolate Content in Hairy Root Cultures of Sinapis alba and Brassica rapa. Appl. Biochem. Biotechnol. 2013, 169, 624–635. [Google Scholar] [CrossRef]
- Kastell, A.; Schreiner, M.; Knorr, D.; Ulrichs, C.; Mewis, I. Influence of nutrient supply and elicitors on glucosinolate production in E. sativa hairy root cultures. Plant Cell Tissue Organ Cult. PCTOC 2018, 132, 561–572. [Google Scholar] [CrossRef]
- Zhong, C.; Nambiar-Veetil, M.; Bogusz, D.; Franche, C. Hairy Roots as a Tool for the Functional Analysis of Plant Genes. In Hairy Roots; Srivastava, V., Mehrotra, S., Mishra, S., Eds.; Springer: Singapore, 2018; pp. 275–292. ISBN 978-9-81-132561-8. [Google Scholar]
- Nakashimada, Y.; Uozumi, N.; Kobayashi, T. Production of plantlets for use as artificial seeds from horseradish hairy roots fragmented in a blender. J. Ferment. Bioeng. 1995, 79, 458–464. [Google Scholar] [CrossRef]
- Repunte, V.P.; Kino-Oka, M.; Taya, M.; Tone, S. Reversible morphology change of horseradish hairy roots cultivated in phytohormone-containing media. J. Ferment. Bioeng. 1993, 75, 271–275. [Google Scholar] [CrossRef]
- Huber, C.; Bartha, B.; Harpaintner, R.; Schröder, P. Metabolism of acetaminophen (paracetamol) in plants—Two independent pathways result in the formation of a glutathione and a glucose conjugate. Environ. Sci. Pollut. Res. 2009, 16, 206. [Google Scholar] [CrossRef]
- Huber, C.; Bartha, B.; Schröder, P. Metabolism of diclofenac in plants—Hydroxylation is followed by glucose conjugation. J. Hazard. Mater. 2012, 243, 250–256. [Google Scholar] [CrossRef]
- Parkinson, M.; Cotter, T.; Dix, P.J. Peroxidase production by cell suspension and hairy root cultures of horseradish (Armoracia rusticana). Plant Sci. 1990, 66, 271–277. [Google Scholar] [CrossRef]
- Krsnik-Rasol, M. Peroxidase as a developmental marker in plant tissue culture. Int. J. Dev. Biol. 2002, 35, 259–263. [Google Scholar]
- Saitou, T.; Kamada, H.; Harada, H. Isoperoxidase in hairy roots and regenerated plants of horseradish (Armoracia lapathifolia). Plant Sci. 1991, 75, 195–201. [Google Scholar] [CrossRef]
- Soudek, P.; Podlipna, R.; Marsik, P.; Vanek, T. Optimalization of the peroxidase production by tissue cultures of horseradish in vitro. Biol. Plant. 2005, 49, 487–492. [Google Scholar] [CrossRef]
- Flocco, C.G.; Giulietti, M. Effect of Chitosan on Peroxidase Activity and Isoenzyme Profile in Hairy Root Cultures of Armoracia lapathifolia. Appl. Biochem. Biotechnol. 2003, 110, 175–183. [Google Scholar] [CrossRef]
- Flocco, C.G.; Alvarez, M.A.; Giulietti, A.M. A Peroxidase production in vitro by Armoracia lapathifolia (horseradish)-transformed root cultures: Effect of elicitation on level and profile of isoenzymes. Biotechnol. Appl. Biochem. 1998, 28, 33–38. [Google Scholar]
- Taya, M.; Yoyama, A.; Nomura, R.; Kondo, O.; Matsui, C.; Kobayashi, T. Production of peroxidase with horseradish hairy root cells in a two step culture system. J. Ferment. Bioeng. 1989, 67, 31–34. [Google Scholar] [CrossRef]
- Alnsour, M.; Kleinwächter, M.; Böhme, J.; Selmar, D. Sulfate determines the glucosinolate concentration of horseradish in vitro plants (Armoracia rusticana Gaertn., Mey. & Scherb.): Sulfate increases the glucosinolate concentration of horseradish in vitro plants. J. Sci. Food Agric. 2013, 93, 918–923. [Google Scholar]
- Wielanek, M.; Królicka, A.; Bergier, K.; Gajewska, E.; Skłodowska, M. Transformation of Nasturtium officinale, Barbarea verna and Arabis caucasica for hairy roots and glucosinolate-myrosinase system production. Biotechnol. Lett. 2009, 31, 917–921. [Google Scholar] [CrossRef]
- Wielanek, M.; Urbanek, H. Enhanced glucotropaeolin production in hairy root cultures of Tropaeolum majus L. by combining elicitation and precursor feeding. Plant Cell Tissue Organ Cult. 2006, 86, 177–186. [Google Scholar] [CrossRef]
- Lee, S.Y.; Bong, S.J.; Kim, J.K.; Park, S.U. Glucosinolate biosynthesis as influenced by growth media and auxin in hairy root cultures of kale (brassica oleracea var. acephala). Emir. J. Food Agric. 2016, 28, 277–282. [Google Scholar] [CrossRef]
- Chung, I.-M.; Rekha, K.; Rajakumar, G.; Thiruvengadam, M. Production of glucosinolates, phenolic compounds and associated gene expression profiles of hairy root cultures in turnip (Brassica rapa ssp. rapa). 3 Biotech 2016, 6, 175. [Google Scholar] [CrossRef]
- Kastell, A.; Zrenner, R.; Schreiner, M.; Kroh, L.; Ulrichs, C.; Smetanska, I.; Mewis, I. Metabolic Engineering of Aliphatic Glucosinolates in Hairy Root Cultures of Arabidopsis thaliana. Plant Mol. Biol. Rep. 2015, 33, 598–608. [Google Scholar] [CrossRef]
- Bennett, R.N.; Mellon, F.A.; Kroon, P.A. Screening Crucifer Seeds as Sources of Specific Intact Glucosinolates Using Ion-Pair High-Performance Liquid Chromatography Negative Ion Electrospray Mass Spectrometry. J. Agric. Food Chem. 2004, 52, 428–438. [Google Scholar] [CrossRef]
- Argentieri, M.; Accogli, R.; Fanizzi, F.; Avato, P. Glucosinolates Profile of “Mugnolo”, a Variety of Brassica oleracea L. Native to Southern Italy (Salento). Planta Med. 2011, 77, 287–292. [Google Scholar] [CrossRef]
- Sansom, C.E.; Jones, V.S.; Joyce, N.I.; Smallfield, B.M.; Perry, N.B.; van Klink, J.W. Flavor, Glucosinolates, and Isothiocyanates of Nau (Cook’s Scurvy Grass, Lepidium oleraceum) and Other Rare New Zealand Lepidium Species. J. Agric. Food Chem. 2015, 63, 1833–1838. [Google Scholar] [CrossRef]
- Fabre, N.; Poinsot, V.; Debrauwer, L.; Vigor, C.; Tulliez, J.; Fourasté, I.; Moulis, C. Characterisation of glucosinolates using electrospray ion trap and electrospray quadrupole time-of-flight mass spectrometry. Phyochem. Anal. 2007, 18, 306–319. [Google Scholar] [CrossRef]
- Rochfort, S.J.; Trenerry, V.C.; Imsic, M.; Panozzo, J.; Jones, R. Class targeted metabolomics: ESI ion trap screening methods for glucosinolates based on MSn fragmentation. Phytochemistry 2008, 69, 1671–1679. [Google Scholar] [CrossRef]
- Agneta, R.; Rivelli, A.R.; Ventrella, E.; Lelario, F.; Sarli, G.; Bufo, S.A. Investigation of Glucosinolate Profile and Qualitative Aspects in Sprouts and Roots of Horseradish (Armoracia rusticana) Using LC-ESI–Hybrid Linear Ion Trap with Fourier Transform Ion Cyclotron Resonance Mass Spectrometry and Infrared Multiphoton Dissociation. J. Agric. Food Chem. 2012, 60, 7474–7482. [Google Scholar]
- Radojcic Redovnikovic, I.; Gliveti, T.; Delonga, K.; Vorkapi, J. Glucosinolates and their potential role in plant. Period. Biol. 2008, 110, 297–309. [Google Scholar]
- Kissen, R.; Bones, A.M. Nitrile-specifier Proteins Involved in Glucosinolate Hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 12057–12070. [Google Scholar] [CrossRef]
- Kim, M.; Chiu, Y.-C.; Kim, N.; Park, H.; Lee, C.; Juvik, J.; Ku, K.-M. Cultivar-Specific Changes in Primary and Secondary Metabolites in Pak Choi (Brassica Rapa, Chinensis Group) by Methyl Jasmonate. Int. J. Mol. Sci. 2017, 18, 1004. [Google Scholar] [CrossRef]
- Kastell, A.; Smetanska, I.; Schreiner, M.; Mewis, I. Hairy roots, callus, and mature plants of Arabidopsis thaliana exhibit distinct glucosinolate and gene expression profiles. Plant Cell Tissue Organ Cult. PCTOC 2013, 115, 45–54. [Google Scholar] [CrossRef]
- Cuong, D.M.; Kim, J.K.; Bong, S.J.; Baek, S.A.; Jeon, J.; Park, J.S.; Park, S.U. Comparative analysis of glucosinolates and metabolite profiling of green and red mustard (brassica juncea) hairy roots. 3 Biotech 2018, 8, 382. [Google Scholar] [CrossRef]
- Kim, Y.B.; Li, X.; Kim, S.-J.; Kim, H.H.; Lee, J.; Kim, H.; Park, S.U. MYB Transcription Factors Regulate Glucosinolate Biosynthesis in Different Organs of Chinese Cabbage (Brassica rapa ssp. pekinensis). Molecules 2013, 18, 8682–8695. [Google Scholar] [CrossRef]
- Petrović, S.; Drobac, M.; Ušjak, L.; Filipović, V.; Milenković, M.; Niketić, M. Volatiles of roots of wild-growing and cultivated Armoracia macrocarpa and their antimicrobial activity, in comparison to horseradish, A. rusticana. Ind. Crops Prod. 2017, 109, 398–403. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Nycholat, C.M.; Montaut, S.; Xu, Y.; Khan, A.Q. Chemical defenses of crucifers: Elicitation and metabolism of phytoalexins and indole-3-acetonitrile in brown mustard and turnip. Phytochemistry 2002, 59, 611–625. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996, 42, 207–220. [Google Scholar]
- Alpizar, E.; Dechamp, E.; Espeout, S.; Royer, M.; Lecouls, A.C.; Nicole, M.; Bertrand, B.; Lashermes, P.; Etienne, H. Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell Rep. 2006, 25, 959–967. [Google Scholar] [CrossRef]
- Doran, P.M. Hairy Roots; CRC Press: Boca Raton, FL, USA, 1997; ISBN 978-9-05-702117-6. [Google Scholar]
- Noda, T.; Tanaka, N.; Mano, Y.; Nabeshima, S.; Ohkawa, H.; Matsui, C. Regeneration of horseradish hairy roots incited by Agrobacterium rhizogenes infection. Plant Cell Rep. 1987, 6, 283–286. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.; Hoffmann, N.; Neidermeyer, J.; Rogers, S.G.; Fraley, R.T. Leaf disc transformation. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Verma, D.P.S., Eds.; Springer: Dordrecht, The Netherlands, 1989; pp. 63–71. ISBN 978-9-40-090951-9. [Google Scholar]
- Ha, P.C.; Su, K.N.; Ji, Y.H.; Ju, B.S.; Seok, P.J.P.N., II; Un, P.S. Effects of Culture Medium on Growth and Glucosinolate Accumulation in the Hairy Root Cultures of Watercress. Res. J. Biotechnol. 2019, 14, 61–66. [Google Scholar]
- Saitou, T.; Hashizume, A.; Kamada, H. Genes for phytochrome A in horseradish: Isolation of cDNAs and analysis of expression during light-induced formation of adventitious shoots from hairy roots. Plant Cell Rep. 2000, 19, 1212–1218. [Google Scholar]
- Wittstock, U.; Meier, K.; Dörr, F.; Ravindran, B.M. NSP-Dependent Simple Nitrile Formation Dominates upon Breakdown of Major Aliphatic Glucosinolates in Roots, Seeds, and Seedlings of Arabidopsis thaliana Columbia-0. Front. Plant Sci. 2016, 7, 1821. [Google Scholar] [CrossRef]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef]
- Kuchernig, J.C.; Burow, M.; Wittstock, U. Evolution of specifier proteins in glucosinolate-containing plants. BMC Evol. Biol. 2012, 12, 127. [Google Scholar] [CrossRef]
- Li, X.; Kushad, M.M. Correlation of Glucosinolate Content to Myrosinase Activity in Horseradish (Armoracia rusticana). J. Agric. Food Chem. 2004, 52, 6950–6955. [Google Scholar] [CrossRef]
- Eriksson, S.; Ek, B.; Xue, J.; Rask, L.; Meijer, J. Identification and characterization of soluble and insoluble myrosinase isoenzymes in different organs of Sinapis alba. Physiol. Plant. 2001, 111, 353–364. [Google Scholar] [CrossRef]
- Andersson, D.; Chakrabarty, R.; Bejai, S.; Zhang, J.; Rask, L.; Meijer, J. Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry 2009, 70, 1345–1354. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bertoli, A.; Giovannini, A.; Ruffoni, B.; Guardo, A.D.; Spinelli, G.; Mazzetti, M.; Pistelli, L. Bioactive Constituent Production in St. John’s Wort in Vitro Hairy Roots. Regenerated Plant Lines †. J. Agric. Food Chem. 2008, 56, 5078–5082. [Google Scholar] [CrossRef]
- M.-Hamvas, M.; Máthé, C.; Vasas, G.; Jámbrik, K.; Papp, M.; Beyer, D.; Mészáros, I.; Borbély, G. Cylindrospermopsin and microcystin-LR alter the growth, development and peroxidase enzyme activity of white mustard (Sinapis alba L.) seedlings, a comparative analysis. Acta Biol. Hung. 2010, 61, 35–48. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Wickham, H. ggplot2—Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- ChemAxon MarvinSketch 16.5.2.0. 2016. Available online: http://www.chemaxon.com (accessed on 1 August 2019).
Sample Availability: Samples are available from the authors upon reasonable request. |
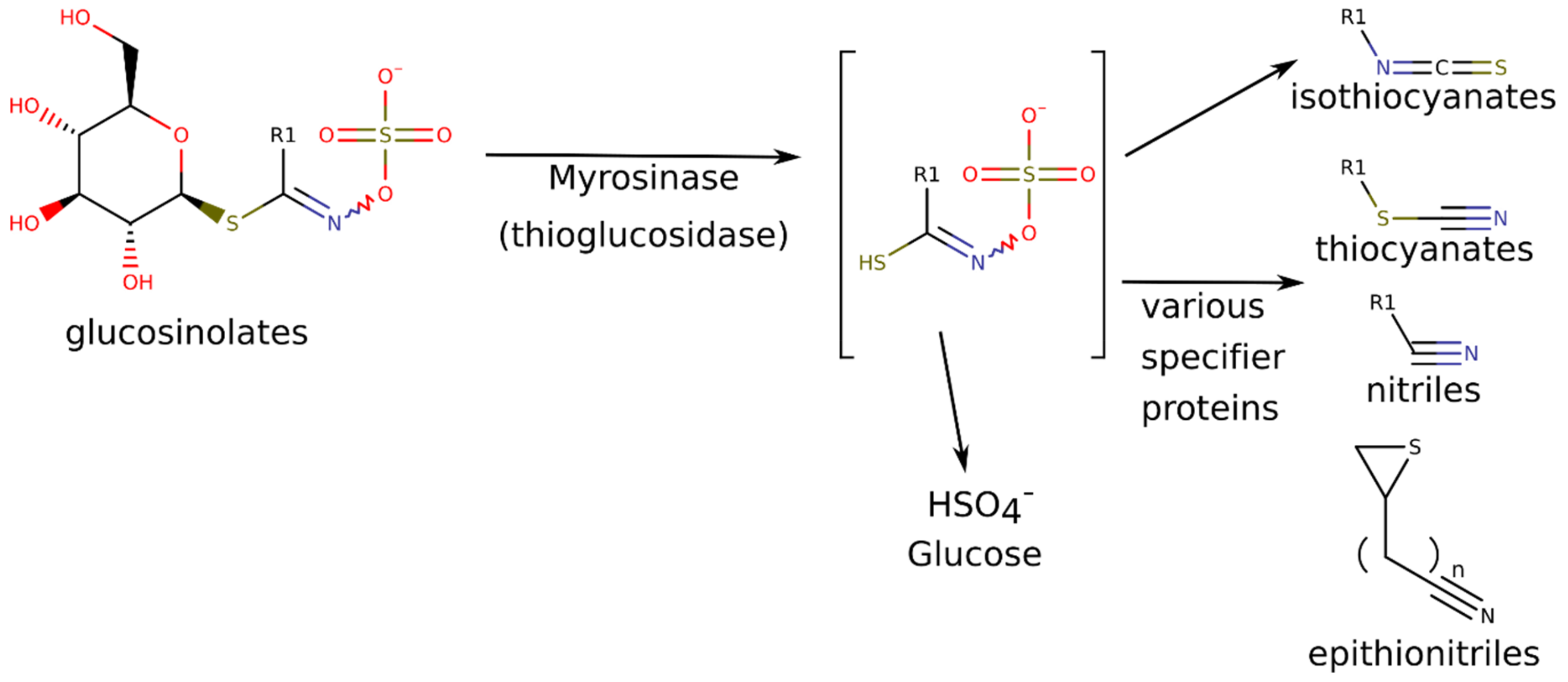

| GLS | GLS Breakdown Products | |
|---|---|---|
| ITC | Nitrile [43,44] | |
| Sinigrin (SIN) a | Allyl ITC (AITC) a | allyl cyanide |
| Glucoiberverin (IBER) a | 3-(methylthio)propyl ITC (MeSPITC) a | 4-(methylthio)-butanenitrile |
| Glucoibarin (GIB) a | 7-(methylsulfinyl)heptyl ITC | NA |
| Glucobrassicin (BRASS) a | 3-indolylmethyl ITC | indol-3-acetonitrile (I3ACN) a |
| Gluconasturtiin (GLN) a | 2-phenylethyl ITC (PEITC) a | 3-phenylpropionitrile (PECN) a |
| 4-methoxy -or neoglucobrassicin (NEO) a | 4-methoxyindol-3-ylmethylor 1-methoxyindol-3-ylmethyl ITC | 1-methoxyindol-3-acetonitrile |
| Glucoarabishirsutain (ARAB) a | 7-(methylthio)-heptyl ITC | NA |
| Variable | p Value | Significance |
|---|---|---|
| Branching | 0.06823 | |
| AdShoots | 0.43899 | |
| PinkExtract | 0.05616 | |
| DWpc | 0.83269 | |
| DGI | 0.04864 | * |
| MyrB1 | 0.76511 | |
| MyrB2 | 0.08308 | |
| MyrB3 | 0.66652 | |
| SIN | 0.00730 | ** |
| IBER | 0.03864 | * |
| GIB | 0.00277 | ** |
| BRASS | 0.00291 | ** |
| GLN | 0.09426 | |
| NEO | 0.00153 | ** |
| ARAB | 0.03068 | * |
| AITC | 0.10321 | |
| PECN | 0.00072 | *** |
| MeSPITC | 0.34163 | |
| PEITC | 0.48132 | |
| I3ACN | 0.00025 | *** |
| ProtContent | 0.94386 | |
| HRP_pg | 0.62207 | |
| HRP_gu | 0.23127 | |
| HRP_pg_AUC | 0.01124 | * |
| HRP_gu_AUC | 0.00745 | ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertóti, R.; Böszörményi, A.; Alberti, Á.; Béni, S.; M-Hamvas, M.; Szőke, É.; Vasas, G.; Gonda, S. Variability of Bioactive Glucosinolates, Isothiocyanates and Enzyme Patterns in Horseradish Hairy Root Cultures Initiated from Different Organs. Molecules 2019, 24, 2828. https://doi.org/10.3390/molecules24152828
Bertóti R, Böszörményi A, Alberti Á, Béni S, M-Hamvas M, Szőke É, Vasas G, Gonda S. Variability of Bioactive Glucosinolates, Isothiocyanates and Enzyme Patterns in Horseradish Hairy Root Cultures Initiated from Different Organs. Molecules. 2019; 24(15):2828. https://doi.org/10.3390/molecules24152828
Chicago/Turabian StyleBertóti, Regina, Andrea Böszörményi, Ágnes Alberti, Szabolcs Béni, Márta M-Hamvas, Éva Szőke, Gábor Vasas, and Sándor Gonda. 2019. "Variability of Bioactive Glucosinolates, Isothiocyanates and Enzyme Patterns in Horseradish Hairy Root Cultures Initiated from Different Organs" Molecules 24, no. 15: 2828. https://doi.org/10.3390/molecules24152828
APA StyleBertóti, R., Böszörményi, A., Alberti, Á., Béni, S., M-Hamvas, M., Szőke, É., Vasas, G., & Gonda, S. (2019). Variability of Bioactive Glucosinolates, Isothiocyanates and Enzyme Patterns in Horseradish Hairy Root Cultures Initiated from Different Organs. Molecules, 24(15), 2828. https://doi.org/10.3390/molecules24152828







