Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters
Abstract
1. Introduction
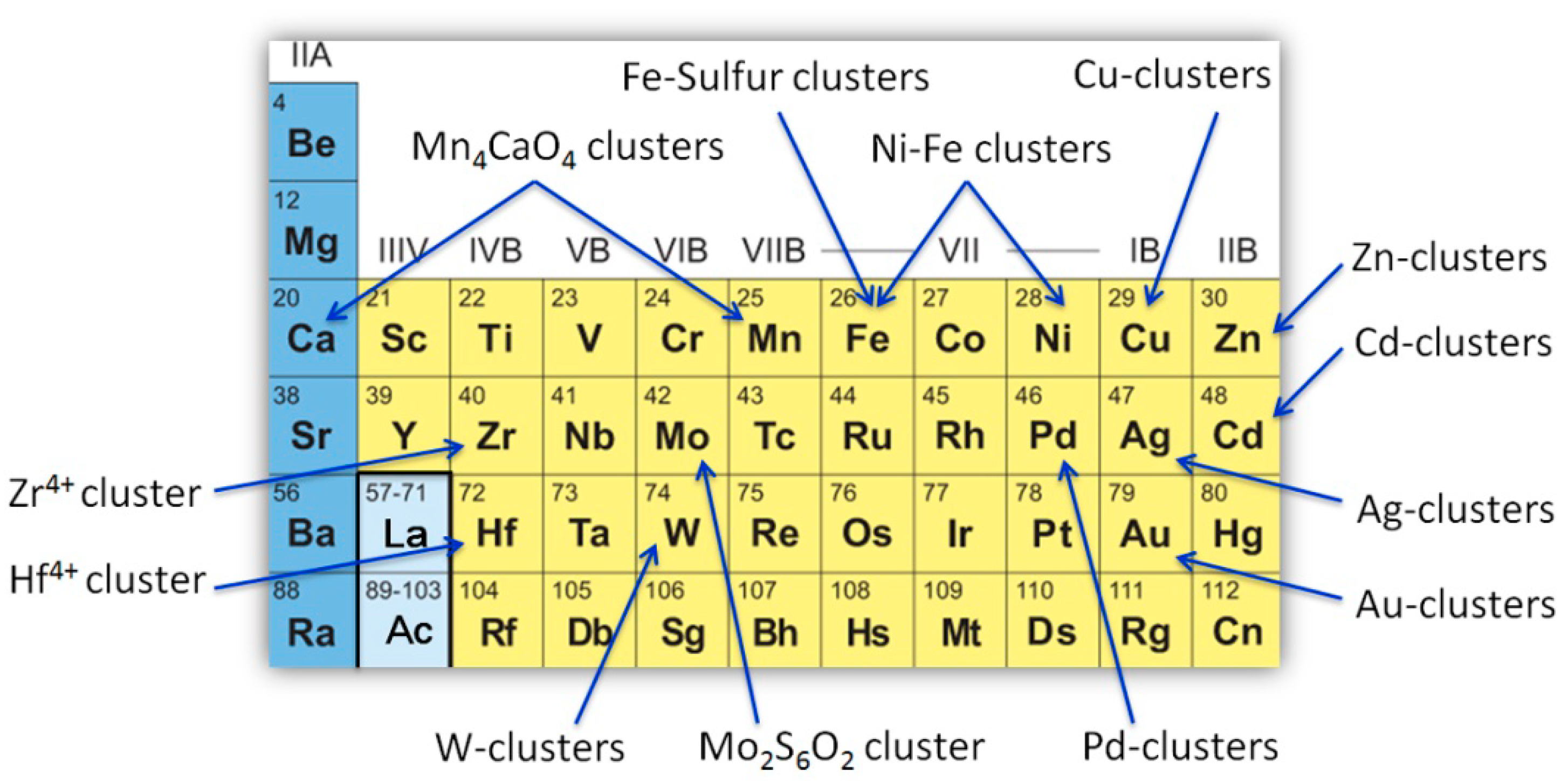
2. Artificial Metalloproteins with Metal Clusters for Structural Roles
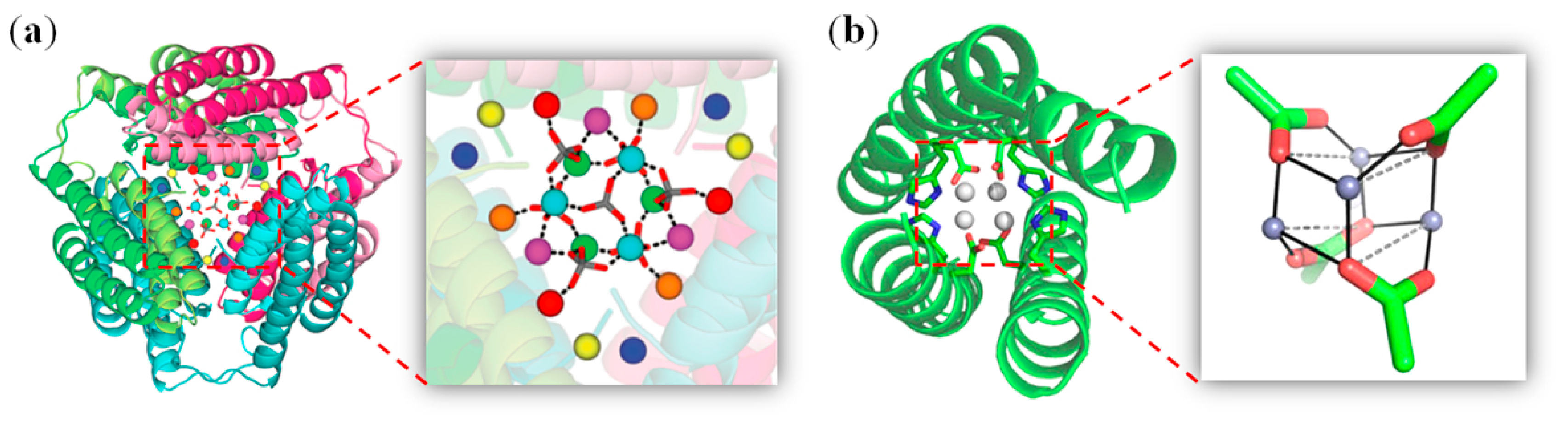
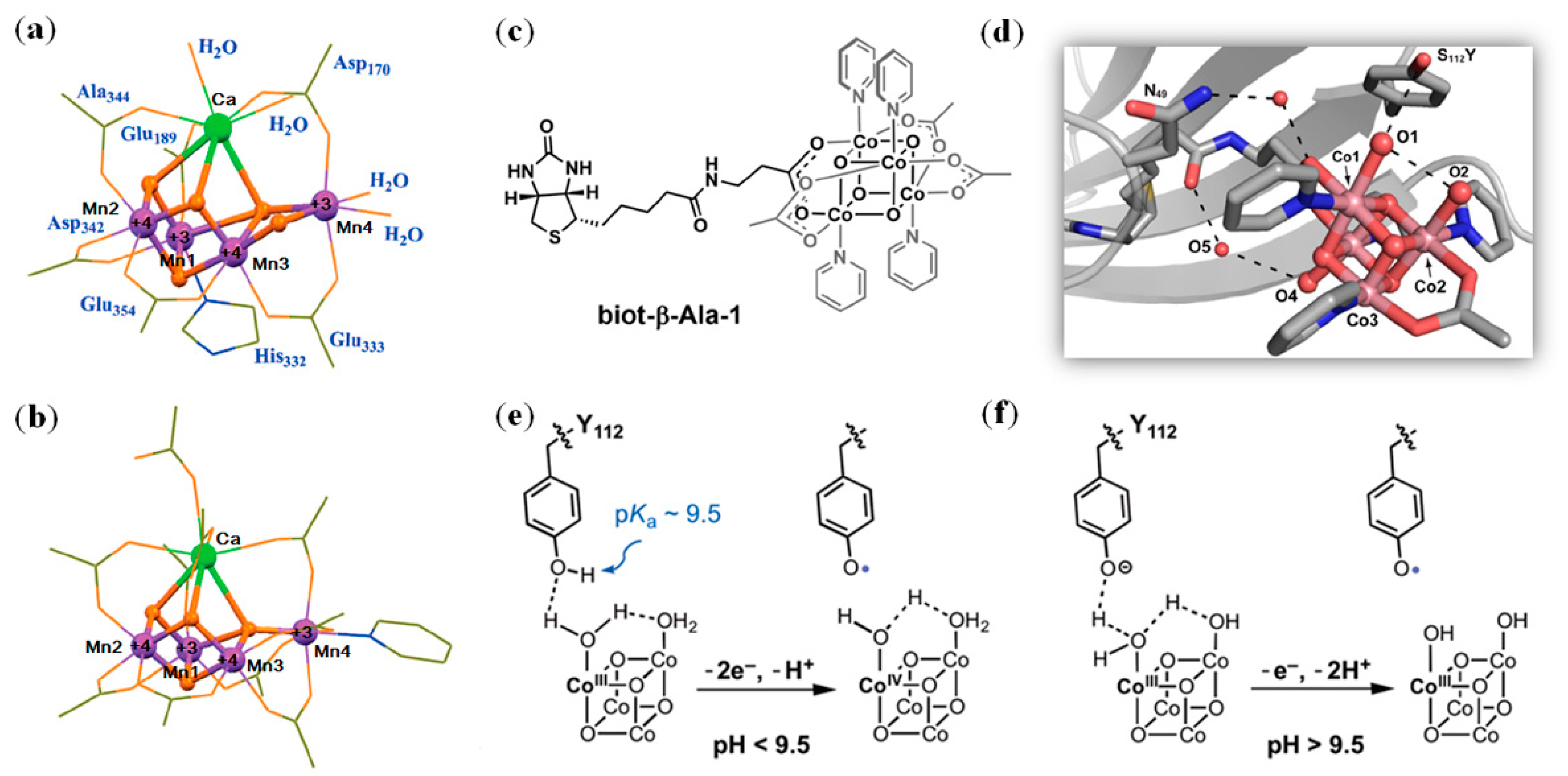
2.1. Zinc Clusters
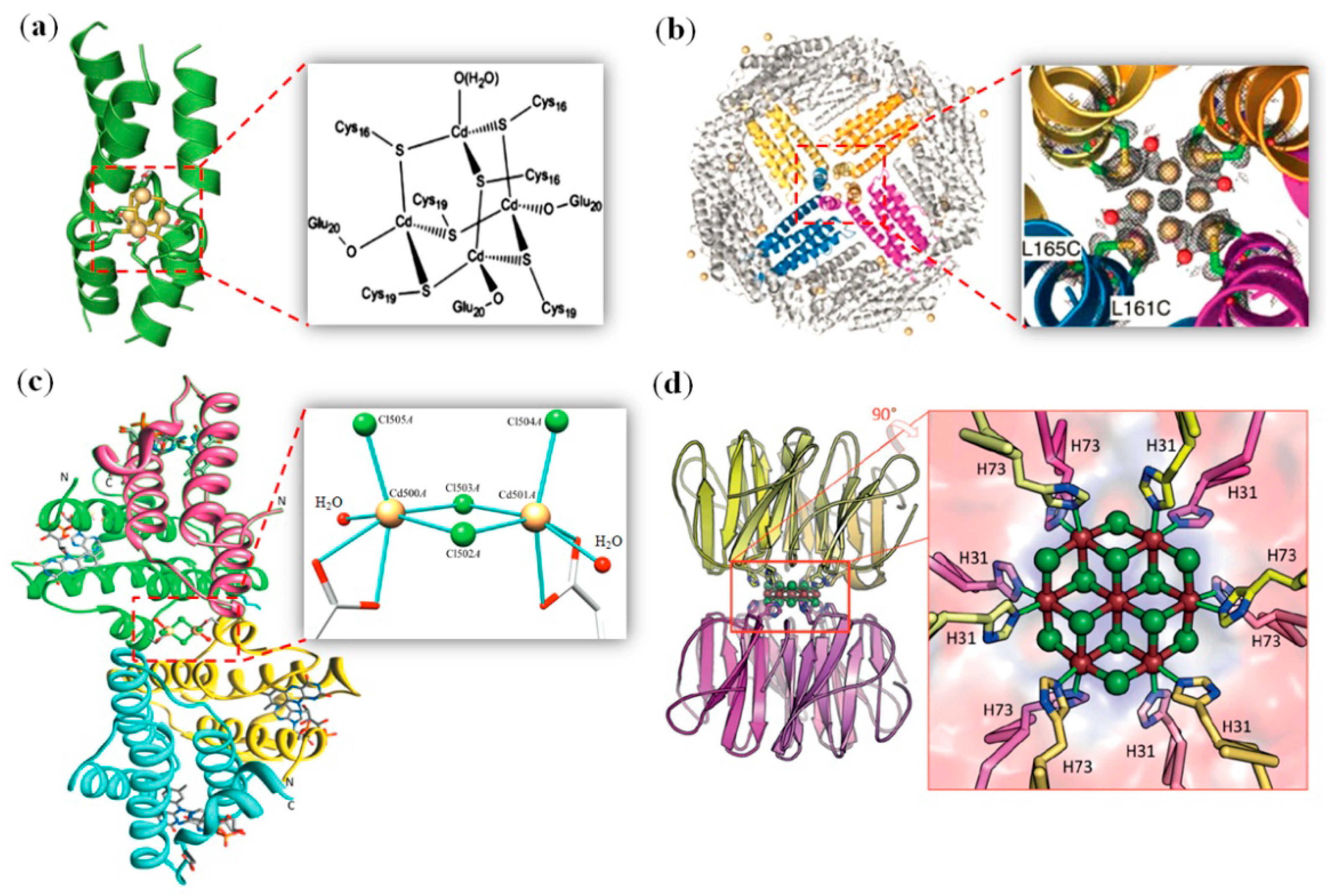
2.2. Cadmium Clusters
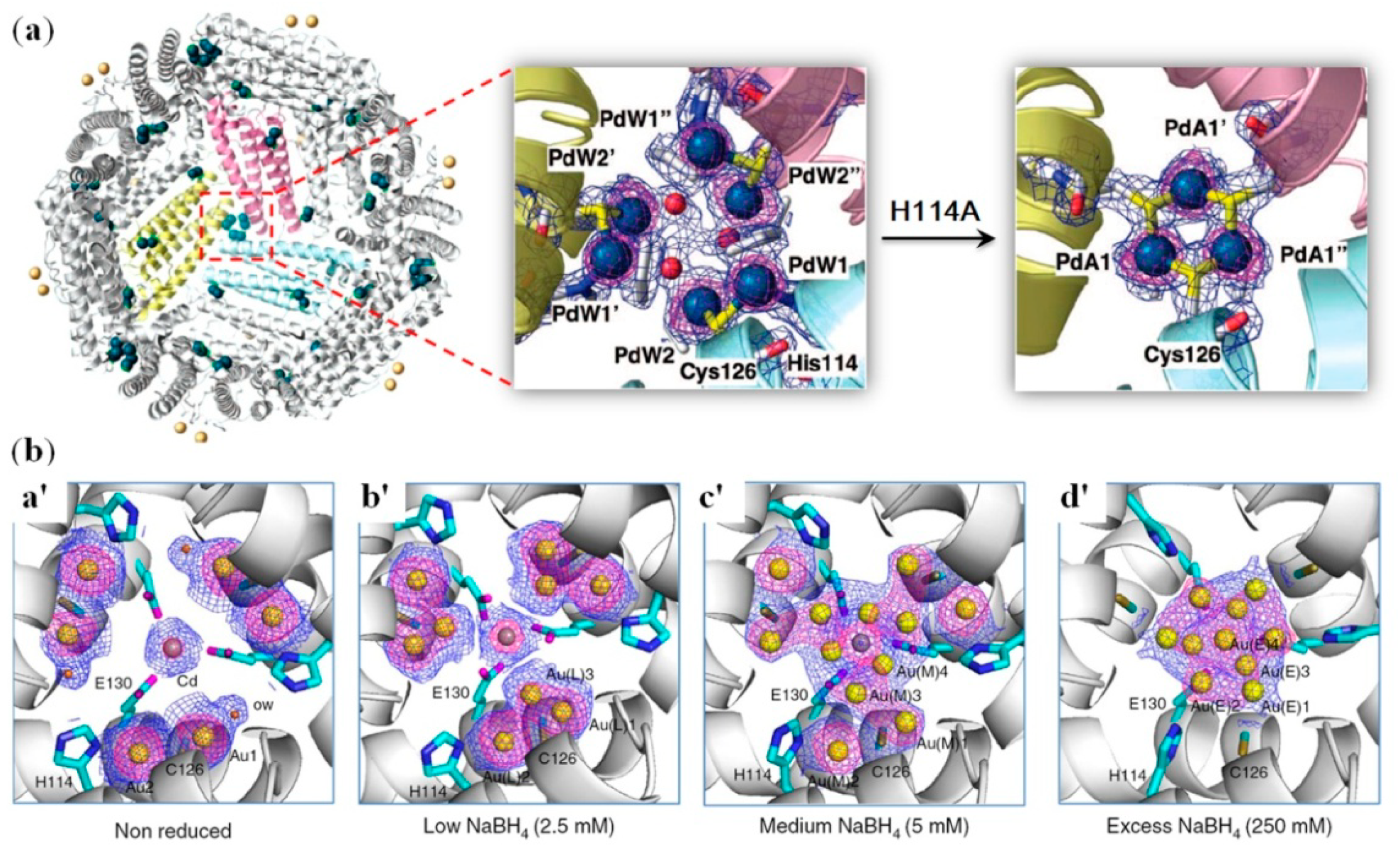
2.3. Noble Metal Clusters
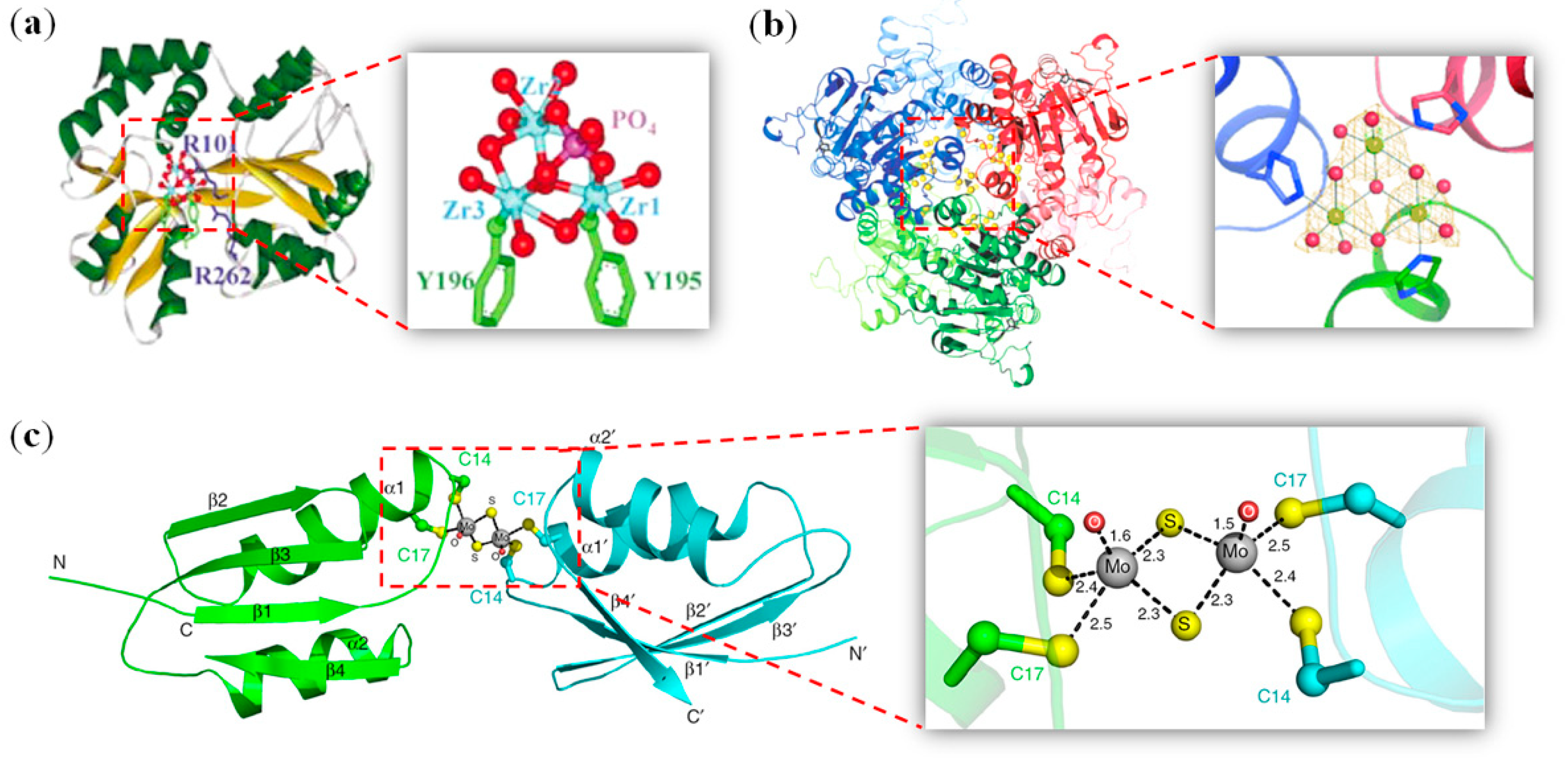
2.4. Other Metal Clusters
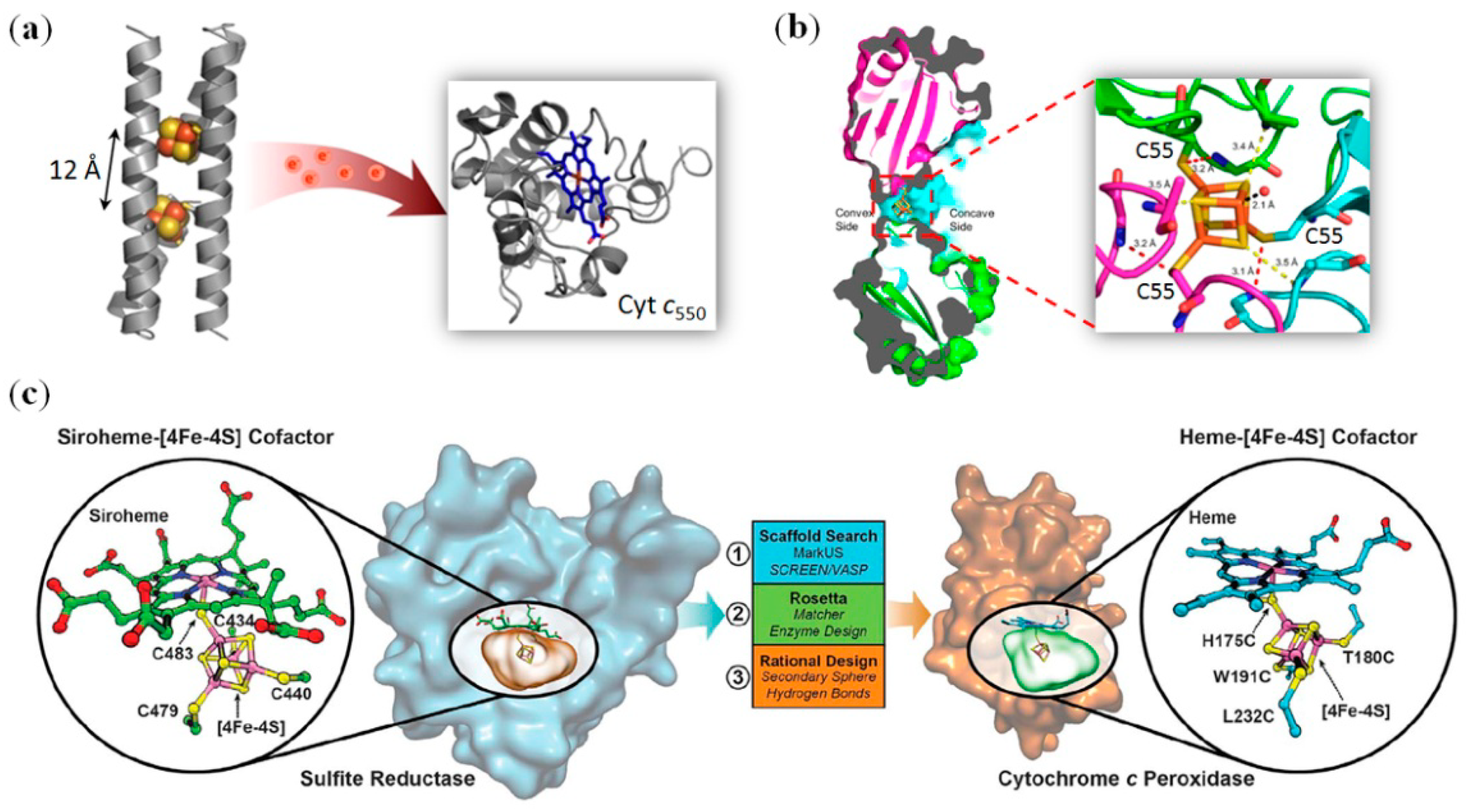
3. Artificial Metalloproteins/Metalloenzymes with Metal Clusters for ElectronTransfer
4. Artificial Metalloenzymes with Metal Clusters for Catalysis
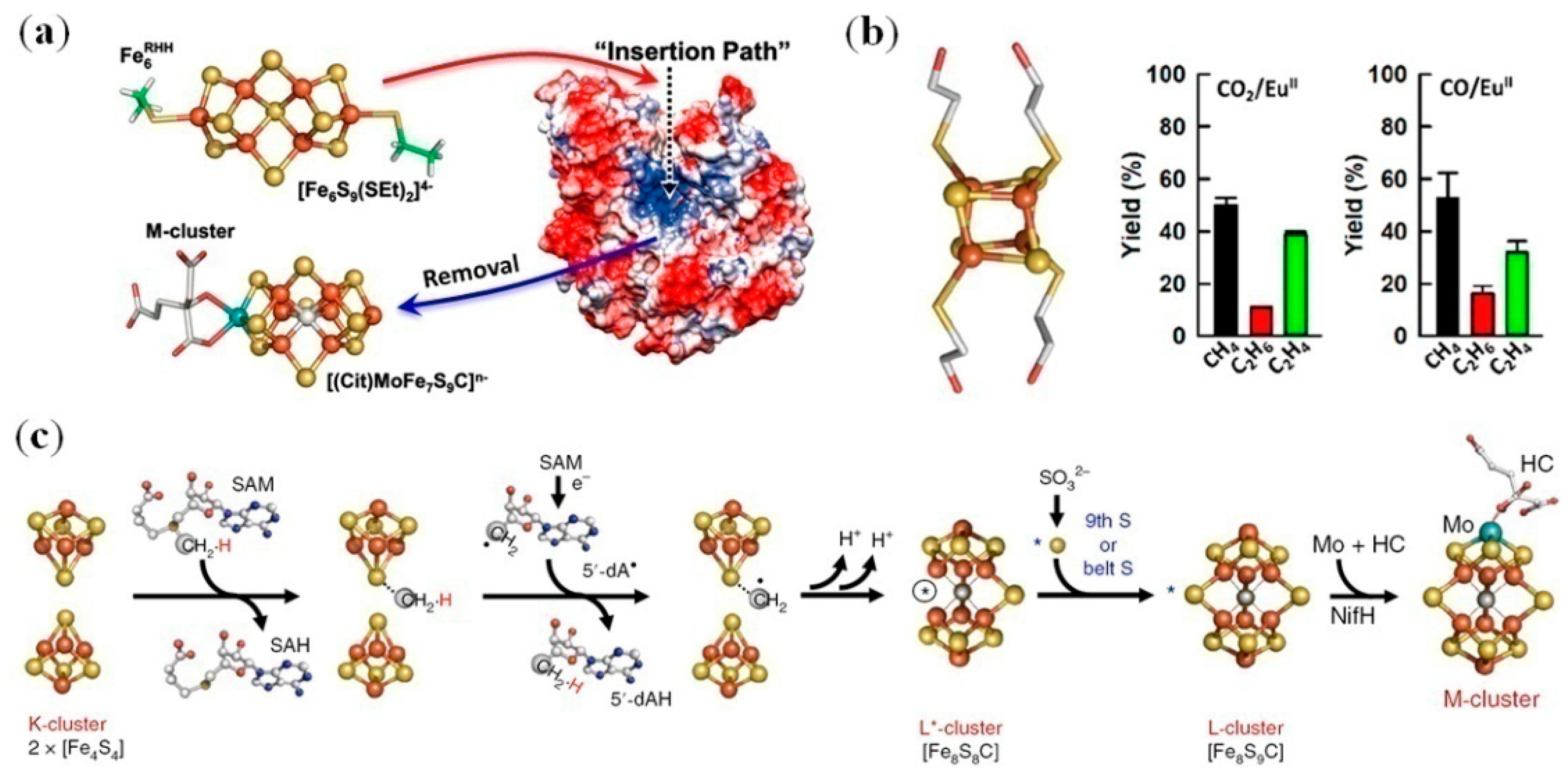
4.1. Iron–Sulfur Clusters
4.2. Copper–Sulfur Clusters
4.3. Other Metal Clusters
5. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Riordan, J.F.; Vallee, B.L. The functional roles of metals in metalloenzymes. Adv. Exp. Med. Biol. 1974, 48, 33–57. [Google Scholar] [PubMed]
- Karlin, K.D. Metalloenzymes, structural motifs, and inorganic models. Science 1993, 261, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Holm, R.H.; Kennepohl, P.; Solomon, E.I. Structural and Functional Aspects of Metal Sites in Biology. Chem. Rev. 1996, 96, 2239–2314. [Google Scholar] [CrossRef] [PubMed]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Valdez, C.E.; Smith, Q.A.; Nechay, M.R.; Alexandrova, A.N. Mysteries of metals in metalloenzymes. Acc. Chem. Res. 2014, 47, 3110–3117. [Google Scholar] [CrossRef] [PubMed]
- Poulos, T.L. Heme enzyme structure and function. Chem. Rev. 2014, 114, 3919–3962. [Google Scholar] [CrossRef]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W. Understanding the choice of copper by heme-copper oxidase using biosynthetic models in myoglobin. Inorg. Chem. Front. 2017, 4, 918–920. [Google Scholar] [CrossRef]
- Huang, X.; Groves, J.T. Oxygen Activation and Radical Transformations in Heme Proteins and Metalloporphyrins. Chem. Rev. 2018, 118, 2491–2553. [Google Scholar] [CrossRef]
- Eom, H.; Song, W.J. Emergence of metal selectivity and promiscuity in metalloenzymes. J. Biol. Inorg. Chem. 2019, 24, 517–531. [Google Scholar] [CrossRef]
- Pernil, R.; Schleiff, E. Metalloproteins in the Biology of Heterocysts. Life 2019, 9, E32. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yeung, N.; Sieracki, N.; Marshall, N.M. Design of functional metalloproteins. Nature 2009, 460, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W.; Sawyer, E.B.; Wang, J. Rational heme protein design: All roads lead to Rome. Chem. Asian. J. 2013, 8, 2534–2544. [Google Scholar] [CrossRef] [PubMed]
- Nastri, F.; Chino, M.; Maglio, O.; Bhagi-Damodaran, A.; Lu, Y.; Lombardi, A. Design and engineering of artificial oxygen-activating metalloenzymes. Chem. Soc. Rev. 2016, 45, 5020–5054. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W. Rational design of metalloenzymes: From single to multiple active sites. Coord. Chem. Rev. 2017, 336, 1–27. [Google Scholar] [CrossRef]
- Schwizer, F.; Okamoto, Y.; Heinisch, T.; Gu, Y.; Pellizzoni, M.M.; Lebrun, V.; Reuter, R.; Kohler, V.; Lewis, J.C.; Ward, T.R. Artificial Metalloenzymes: Reaction Scope and Optimization Strategies. Chem. Rev. 2018, 118, 142–231. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Griffith, W.P.; Davis, I.; Shin, I.; Wang, J.; Li, F.; Wang, Y.; Wherritt, D.J.; Liu, A. Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase. Nat. Chem. Biol. 2018, 14, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Hirota, S.; Lin, Y.-W. Design of artificial metalloproteins/metalloenzymes by tuning noncovalent interactions. J. Biol. Inorg. Chem. 2018, 23, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Chan, S.I.; Sawyer, E.B.; Yu, Y.; Wang, J. Metalloprotein design using genetic code expansion. Chem. Soc. Rev. 2014, 43, 6498–6510. [Google Scholar] [CrossRef]
- Yu, Y.; Hu, C.; Xia, L.; Wang, J. Artificial Metalloenzyme Design with Unnatural Amino Acids and Non-Native Cofactors. ACS Catal. 2018, 8, 1851–1863. [Google Scholar] [CrossRef]
- Oohora, K.; Onoda, A.; Hayashi, T. Hemoproteins Reconstituted with Artificial Metal Complexes as Biohybrid Catalysts. Acc. Chem. Res. 2019, 52, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Shoji, O.; Aiba, Y.; Watanabe, Y. Hoodwinking Cytochrome P450BM3 into Hydroxylating Non-Native Substrates by Exploiting Its Substrate Misrecognition. Acc. Chem. Res. 2019, 52, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Directed Evolution of Artificial Metalloenzymes: A Universal Means to Tune the Selectivity of Transition Metal Catalysts? Acc. Chem. Res. 2019, 52, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Natoli, S.N.; Hartwig, J.F. Noble−Metal Substitution in Hemoproteins: An Emerging Strategy for Abiological Catalysis. Acc. Chem. Res. 2019, 52, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Mirts, E.N.; Bhagi-Damodaran, A.; Lu, Y. Understanding and Modulating Metalloenzymes with Unnatural Amino Acids, Non-Native Metal Ions, and Non-Native Metallocofactors. Acc. Chem. Res. 2019, 52, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, A.; Pirro, F.; Maglio, O.; Chino, M.; DeGrado, W.F. De Novo Design of Four-Helix Bundle Metalloproteins: One Scaffold, Diverse Reactivities. Acc. Chem. Res. 2019, 52, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, C.; Cong, Z. Strategies for Substrate-Regulated P450 Catalysis: From Substrate Engineering to Co-catalysis. Chemistry 2019, 25, 6853–6863. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W. Rational design of heme enzymes for biodegradation of pollutants toward a green future. Biotechnol. Appl. Biochem. 2019. [Google Scholar] [CrossRef]
- Nastri, F.; D’Alonzo, D.; Leone, L.; Zambrano, G.; Pavone, V.; Lombardi, A. Engineering Metalloprotein Functions in Designed and Native Scaffolds. Trends Biochem. Sci. 2019. [Google Scholar] [CrossRef]
- Sligar, S.G.; Egeberg, K.D.; Sage, J.T.; Morikis, D.; Champion, P.M. Alteration of heme axial ligands by site-directed mutagenesis: A cytochrome becomes a catalytic demethylase. J. Am. Chem. Soc. 1987, 109, 7896–7897. [Google Scholar] [CrossRef]
- Hay, M.; Richards, J.H.; Lu, Y. Construction and characterization of an azurin analog for the purple copper site in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 1996, 93, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Ying, T.; Zhong, F.; Wang, Z.H.; Li, W.; Tan, X.; Huang, Z.X. A route to novel functional metalloproteins via hybrids of cytochrome P450 and cytochrome c. Chem. Bio. Chem. 2011, 12, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Berry, S.M.; Pfister, T.D. Engineering novel metalloproteins: Design of metal-binding sites into native protein scaffolds. Chem. Rev. 2001, 101, 3047–3080. [Google Scholar] [CrossRef] [PubMed]
- Yeung, N.; Lin, Y.-W.; Gao, Y.G.; Zhao, X.; Russell, B.S.; Lei, L.; Miner, K.D.; Robinson, H.; Lu, Y. Rational design of a structural and functional nitric oxide reductase. Nature 2009, 462, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Pordea, A. Metal-binding promiscuity in artificial metalloenzyme design. Curr. Opin. Chem. Biol. 2015, 25, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Peacock, A.F. Recent advances in designed coiled coils and helical bundles with inorganic prosthetic groups-from structural to functional applications. Curr. Opin. Chem. Biol. 2016, 31, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.-G.; Su, J.-H.; Du, K.-J.; You, Y.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. Rational Design of Dual Active Sites in a Single Protein Scaffold: A Case Study of Heme Protein in Myoglobin. Chem. Open 2016, 5, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Rittle, J.; Field, M.J.; Green, M.T.; Tezcan, F.A. An efficient, step-economical strategy for the design of functional metalloproteins. Nat. Chem. 2019, 11, 434–441. [Google Scholar] [CrossRef]
- Lu, Y. Design and engineering of metalloproteins containing unnatural amino acids or non-native metal-containing cofactors. Curr. Opin. Chem. Biol. 2005, 9, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.C. Metallopeptide catalysts and artificial metalloenzymes containing unnatural amino acids. Curr. Opin. Chem. Biol. 2015, 25, 27–35. [Google Scholar] [CrossRef]
- Sreenilayam, G.; Moore, E.J.; Steck, V.; Fasan, R. Stereoselective Olefin Cyclopropanation under Aerobic Conditions with an Artificial Enzyme Incorporating an Iron-Chlorin e6 Cofactor. ACS Catal. 2017, 7, 7629–7633. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed. Engl. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W. The broad diversity of heme-protein cross-links: An overview. Biochim. Biophys. Acta 2015, 1854, 844–859. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.-J.; Li, W.; Xiang, Y.; Wen, G.-B.; Lin, Y.-W.; Tan, X. A novel tyrosine-heme C-O covalent linkage in F43Y myoglobin: A new post-translational modification of heme proteins. Chem. Bio. Chem. 2015, 16, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.-J.; Yuan, H.; Li, W.; Xiang, Y.; He, B.; Nie, C.-M.; Wen, G.B.; Lin, Y.-W.; Tan, X. How a novel tyrosine-heme cross-link fine-tunes the structure and functions of heme proteins: A direct comparative study of L29H/F43Y myoglobin. Dalton Trans. 2015, 44, 18815–18822. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-L.; Yuan, H.; Liao, F.; He, B.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. Rational Design of Artificial Dye-decolorizing Peroxidases using Myoglobin by Engineering Tyr/Trp in the Heme Center. Dalton Trans. 2017, 46, 11230–11238. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-M.; Yuan, H.; Wang, X.-J.; Xu, J.-K.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. Formation of Cys-heme cross-link in K42C myoglobin under reductive conditions with molecular oxygen. J. Inorg. Biochem. 2018, 182, 141–149. [Google Scholar] [CrossRef]
- Lin, Y.-W. Structure and function of heme proteins regulated by diverse post-translational modifications. Arch. Biochem. Biophys. 2018, 641, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.-L.; Yuan, H.; Du, K.-J.; He, B.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. Regulation of both the structure and function by a de novo designed disulfide bond: A case study of heme proteins in myoglobin. Chem. Commun. 2018, 54, 4356–4359. [Google Scholar] [CrossRef]
- Liu, C.; Yuan, H.; Liao, F.; Wei, C.-W.; Du, K.-J.; Gao, S.-Q.; Tan, X.; Lin, Y.-W. Unique Tyr-heme double cross-links in F43Y/T67R myoglobin: An artificial enzyme with a peroxidase activity comparable to that of native peroxidases. Chem. Commun. 2019, 55, 6610–6613. [Google Scholar] [CrossRef]
- Chen, M.; Wang, Z.; Shu, J.; Jiang, X.; Wang, W.; Shi, Z.-H.; Lin, Y.-W. Mimicking a Natural Enzyme System: Cytochrome c Oxidase-Like Activity of Cu2O Nanoparticles by Receiving Electrons from Cytochrome c. Inorg. Chem. 2017, 56, 9400–9403. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wei, C.; Su, J.-H.; He, B.; Wen, G.-B.; Lin, Y.-W.; Zhang, Y. A Chiral Ligand Assembly That Confers One-Electron O2 Reduction Activity for a Cu2+-Selective Metallohydrogel. Angew. Chem. Int. Ed. 2018, 57, 3504–3508. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes(II). Chem. Soc. Rev. 2019, 18, 1004–1076. [Google Scholar] [CrossRef]
- Fehl, C.; Davis, B.G. Proteins as templates for complex synthetic metalloclusters: Towards biologically programmed heterogeneous catalysis. Proc. Math. Phys. Eng. Sci. 2016, 472, 20160078. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Maity, B.; Ueno, T. Functionalization of protein crystals with metal ions, complexes and nanoparticles. Curr. Opin. Chem. Biol. 2018, 43, 68–76. [Google Scholar] [CrossRef]
- Song, W.J.; Sontz, P.A.; Ambroggio, X.I.; Tezcan, F.A. Metals in protein-protein interfaces. Annu. Rev. Biophys. 2014, 43, 409–431. [Google Scholar] [CrossRef] [PubMed]
- Radford, R.J.; Brodin, J.D.; Salgado, E.N.; Tezcan, F.A. Expanding the utility of proteins as platforms for coordination chemistry. Coord. Chem. Rev. 2011, 255, 790–803. [Google Scholar] [CrossRef]
- Song, W.J.; Tezcan, F.A. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 2014, 346, 1525–1528. [Google Scholar] [CrossRef]
- Miyamoto, T.; Kuribayashi, M.; Nagao, S.; Shomura, Y.; Higuchi, Y.; Hirota, S. Domain-swapped cytochrome cb562 dimer and its nanocage encapsulating a Zn-SO4 cluster in the internal cavity. Chem. Sci. 2015, 6, 7336–7342. [Google Scholar] [CrossRef]
- Tebo, A.G.; Pecoraro, V.L. Artificial metalloenzymes derived from three-helix bundles. Curr. Opin. Chem. Biol. 2015, 25, 65–70. [Google Scholar] [CrossRef]
- Cangelosi, V.M.; Deb, A.; Penner-Hahn, J.E.; Pecoraro, V.L. A de novo designed metalloenzyme for the hydration of CO2. Angew. Chem. Int. Ed. Engl. 2014, 53, 7900–7903. [Google Scholar] [CrossRef]
- Ulas, G.; Lemmin, T.; Wu, Y.; Gassner, G.T.; DeGrado, W.F. Designed metalloprotein stabilizes a semiquinone radical. Nat. Chem. 2016, 8, 354–359. [Google Scholar] [CrossRef]
- Zhang, S.-Q.; Chino, M.; Liu, L.; Tang, Y.; Hu, X.; DeGrado, W.F.; Lombardi, A. De Novo Design of Tetranuclear Transition Metal Clusters Stabilized by Hydrogen-Bonded Networks in Helical Bundles. J. Am. Chem. Soc. 2018, 140, 1294–1304. [Google Scholar] [CrossRef]
- Chino, M.; Zhang, S.Q.; Pirro, F.; Leone, L.; Maglio, O.; Lombardi, A.; DeGrado, W.F. Spectroscopic and metal binding properties of a de novo metalloprotein binding a tetrazinc cluster. Biopolymers 2018, 109, e23339. [Google Scholar] [CrossRef]
- Kharenko, O.A.; Kennedy, D.C.; Demeler, B.; Maroney, M.J.; Ogawa, M.Y. Cu(I) Luminescence from the Tetranuclear Cu4S4 Cofactor of a Synthetic 4-Helix Bundle. J. Am. Chem. Soc. 2005, 127, 7678–7679. [Google Scholar] [CrossRef]
- Xie, F.; Sutherland, D.E.K.; Stillman, M.J.; Ogawa, M.Y. Cu(I) binding properties of a designed metalloprotein. J. Inorg. Biochem. 2010, 104, 261–267. [Google Scholar] [CrossRef]
- Zaytsev, D.V.; Morozov, V.A.; Fan, J.; Zhu, X.; Mukherjee, M.; Ni, S.; Kennedy, M.A.; Ogawa, M.Y. Metal-binding properties and structural characterization of a self-assembled coiled coil: Formation of a polynuclear Cd-thiolate cluster. J. Inorg. Biochem. 2013, 119, 1–9. [Google Scholar] [CrossRef]
- Theil, E.C. Ferritin protein nanocages use ion channels, catalytic sites, and nucleation channels to manage iron/oxygen chemistry. Curr. Opin. Chem. Biol. 2011, 15, 304–311. [Google Scholar] [CrossRef]
- Joh, N.H.; Wang, T.; Bhate, M.P.; Acharya, R.; Wu, Y.; Grabe, M.; Hong, M.; Grigoryan, G.; DeGrado, W.F. De novo design of a transmembrane Zn(2)(+)-transporting four-helix bundle. Science 2014, 346, 1520–1524. [Google Scholar] [CrossRef]
- Abe, S.; Ito, N.; Maity, B.; Lu, C.; Lu, D.; Ueno, T. Coordination design of cadmium ions at the 4-fold axis channel of the apo-ferritin cage. Dalton Trans. 2019, 48, 9759–9764. [Google Scholar] [CrossRef]
- Florence, Q.; Wu, C.K.; Habel, J.; Swindell, J.T., 2nd; Wang, B.C.; Rose, J.P. The structure of augmenter of liver regeneration crystallized in the presence of 50 mM CdCl2 reveals a novel Cd2Cl4O6 cluster that aids in crystal packing. Acta Crystallogr. D Biol. Crystallogr. 2012, 68 Pt 9, 1128–1133. [Google Scholar] [CrossRef]
- Voet, A.R.; Noguchi, H.; Addy, C.; Zhang, K.Y.; Tame, J.R. Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein. Angew. Chem. Int. Ed. Engl. 2015, 54, 9857–9860. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.; Pradeep, T. Noble Metal Clusters: Applications in Energy, Environment, and Biology. Part. Part. Syst. Charact. 2014, 31, 1017–1053. [Google Scholar] [CrossRef]
- Morozov, V.A.; Ogawa, M.Y. Controlled formation of emissive silver nanoclusters using rationally designed metal-binding proteins. Inorg. Chem. 2013, 52, 9166–9168. [Google Scholar] [CrossRef] [PubMed]
- Aires, A.; Llarena, I.; Moller, M.; Castro-Smirnov, J.; Cabanillas-Gonzalez, J.; Cortajarena, A.L. A Simple Approach to Design Proteins for the Sustainable Synthesis of Metal Nanoclusters. Angew. Chem. Int. Ed. 2019, 58, 6214–6219. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Niemeyer, J.; Abe, M.; Takezawa, Y.; Ueno, T.; Hikage, T.; Erker, G.; Watanabe, Y. Control of the coordination structure of organometallic palladium complexes in an apo-ferritin cage. J. Am. Chem. Soc. 2008, 130, 10512–10514. [Google Scholar] [CrossRef] [PubMed]
- Maity, B.; Abe, S.; Ueno, T. Observation of gold sub-nanocluster nucleation within a crystalline protein cage. Nat.Commun. 2017, 8, 14820. [Google Scholar] [CrossRef]
- Alexeev, D.; Zhu, H.; Guo, M.; Zhong, W.; Hunter, D.J.B.; Yang, W.; Campopiano, D.J.; Sadler, P.J. A novel protein–mineral interface. Nat. Struct. Mol. Biol. 2003, 10, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Alexeev, D.; Harvey, I.; Guo, M.; Hunter, D.J.; Zhu, H.; Campopiano, D.J.; Sadler, P.J. Assembly of an oxo-zirconium(IV) cluster in a protein cleft. Angew. Chem. Int. Ed. Engl. 2004, 43, 5914–5918. [Google Scholar] [CrossRef]
- Schemberg, J.; Schneider, K.; Demmer, U.; Warkentin, E.; Muller, A.; Ermler, U. Towards biological supramolecular chemistry: A variety of pocket-templated, individual metal oxide cluster nucleations in the cavity of a mo/w-storage protein. Angew. Chem. Int. Ed. Engl. 2007, 46, 2408–2413. [Google Scholar] [CrossRef]
- Fang, T.; Chen, W.; Sheng, Y.; Yuan, S.; Tang, Q.; Li, G.; Huang, G.; Su, J.; Zhang, X.; Zang, J.; et al. Tetrathiomolybdate induces dimerization of the metal-binding domain of ATPase and inhibits platination of the protein. Nat.Commun. 2019, 10, 186. [Google Scholar] [CrossRef]
- Fontecave, M. Iron-sulfur clusters: Ever-expanding roles. Nat. Chem. Biol. 2006, 2, 171–174. [Google Scholar] [CrossRef]
- Nanda, V.; Senn, S.; Pike, D.H.; Rodriguez-Granillo, A.; Hansen, W.A.; Khare, S.D.; Noy, D. Structural principles for computational and de novo design of 4Fe-4S metalloproteins. Biochim. Biophys. Acta 2016, 1857, 531–538. [Google Scholar] [CrossRef]
- Coldren, C.D.; Hellinga, H.W.; Caradonna, J.P. The rational design and construction of a cuboidal iron-sulfur protein. Proc. Natl. Acad. Sci. USA 1997, 94, 6635–6640. [Google Scholar] [CrossRef]
- Roy, A.; Sarrou, I.; Vaughn, M.D.; Astashkin, A.V.; Ghirlanda, G. De novo design of an artificial bis [4Fe-4S] binding protein. Biochemistry 2013, 52, 7586–7594. [Google Scholar] [CrossRef]
- Roy, A.; Sommer, D.J.; Schmitz, R.A.; Brown, C.L.; Gust, D.; Astashkin, A.; Ghirlanda, G. A de novo designed 2[4Fe-4S] ferredoxin mimic mediates electron transfer. J. Am. Chem. Soc. 2014, 136, 17343–17349. [Google Scholar] [CrossRef]
- Sommer, D.J.; Roy, A.; Astashkin, A.; Ghirlanda, G. Modulation of cluster incorporation specificity in a de novo iron-sulfur cluster binding peptide. Biopolymers 2015, 104, 412–418. [Google Scholar] [CrossRef]
- Mutter, A.C.; Tyryshkin, A.M.; Campbell, I.J.; Poudel, S.; Bennett, G.N.; Silberg, J.J.; Nanda, V.; Falkowski, P.G. De novo design of symmetric ferredoxins that shuttle electrons in vivo. Proc. Natl. Acad. Sci. USA 2019, 116, 14557–14562. [Google Scholar] [CrossRef]
- Aussignargues, C.; Pandelia, M.E.; Sutter, M.; Plegaria, J.S.; Zarzycki, J.; Turmo, A.; Huang, J.; Ducat, D.C.; Hegg, E.L.; Gibney, B.R.; et al. Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster. J. Am. Chem. Soc. 2016, 138, 5262–5270. [Google Scholar] [CrossRef]
- Gibney, B.R.; Mulholland, S.E.; Rabanal, F.; Dutton, P.L. Ferredoxin and ferredoxin-heme maquettes. Proc. Natl. Acad. Sci. USA 1996, 93, 15041–15046. [Google Scholar] [CrossRef]
- Ferguson-Miller, S.; Babcock, G.T. Heme/Copper Terminal Oxidases. Chem. Rev. 1996, 96, 2889–2908. [Google Scholar] [CrossRef]
- Sigman, J.A.; Kwok, B.C.; Lu, Y. From Myoglobin to Heme-Copper Oxidase: Design and Engineering of a CuB Center into Sperm Whale Myoglobin. J. Am. Chem. Soc. 2000, 122, 8192–8196. [Google Scholar] [CrossRef]
- Bhagi-Damodaran, A.; Michael, M.A.; Zhu, Q.; Reed, J.; Sandoval, B.A.; Mirts, E.N.; Chakraborty, S.; Moënne-Loccoz, P.; Zhang, Y.; Lu, Y. Why copper is preferred over iron for oxygen activation and reduction in haem-copper oxidases. Nat. Chem. 2017, 9, 257–263. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Yeung, N.; Gao, Y.-G.; Miner, K.D.; Lei, L.; Robinson, H.; Lu, Y. Introducing a 2-His-1-Glu nonheme iron center into myoglobin confers nitric oxide reductase activity. J. Am. Chem. Soc. 2010, 132, 9970–9972. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Yeung, N.; Gao, Y.-G.; Miner, K.D.; Tian, S.; Robinson, H.; Lu, Y. Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin. Proc. Natl. Acad. Sci. USA 2010, 107, 8581–8586. [Google Scholar] [CrossRef]
- Hino, T.; Matsumoto, Y.; Nagano, S.; Sugimoto, H.; Fukumori, Y.; Murata, T.; Iwata, S.; Shiro, Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 2010, 330, 1666–1670. [Google Scholar] [CrossRef]
- Crane, B.R.; Siegel, L.M.; Getzoff, E.D. Sulfite reductase structure at 1.6 A: Evolution and catalysis for reduction of inorganic anions. Science 1995, 270, 59–67. [Google Scholar] [CrossRef]
- Mirts, E.N.; Petrik, I.D.; Hosseinzadeh, P.; Nilges, M.J.; Lu, Y. A designed heme-[4Fe-4S] metalloenzyme catalyzes sulfite reduction like the native enzyme. Science 2018, 361, 1098–1101. [Google Scholar] [CrossRef]
- Sickerman, N.S.; Hu, Y.; Ribbe, M.W. Nitrogenases. Methods Mol. Biol. 2019, 1876, 3–24. [Google Scholar]
- Lee, S.C.; Lo, W.; Holm, R.H. Developments in the biomimetic chemistry of cubane-type and higher nuclearity iron-sulfur clusters. Chem. Rev. 2014, 114, 3579–3600. [Google Scholar] [CrossRef]
- Paech, C.; Reynolds, J.G.; Singer, T.P.; Holm, R.H. Structural identification of iron-sulfur clusters of the respiratory chain-linked NADH dehydrogenase. J. Biol. Chem. 1981, 256, 3167–3170. [Google Scholar]
- Tanifuji, K.; Lee, C.C.; Ohki, Y.; Tatsumi, K.; Hu, Y.; Ribbe, M.W. Combining a Nitrogenase Scaffold and a Synthetic Compound into an Artificial Enzyme. Angew. Chem. Int. Ed. Engl. 2015, 54, 14022–14025. [Google Scholar] [CrossRef]
- Tanifuji, K.; Lee, C.C.; Sickerman, N.S.; Tatsumi, K.; Ohki, Y.; Hu, Y.; Ribbe, M.W. Tracing the ‘ninth sulfur’ of the nitrogenase cofactor via a semi-synthetic approach. Nat. Chem. 2018, 10, 568–572. [Google Scholar] [CrossRef]
- Stiebritz, M.T.; Hiller, C.J.; Sickerman, N.S.; Lee, C.C.; Tanifuji, K.; Ohki, Y.; Hu, Y. Ambient conversion of CO2 to hydrocarbons by biogenic and synthetic [Fe4S4] clusters. Nat. Catal. 2018, 1, 444–451. [Google Scholar] [CrossRef]
- Lee, C.C.; Stiebritz, M.T.; Hu, Y. Reactivity of [Fe4S4] Clusters toward C1 Substrates: Mechanism, Implications, and Potential Applications. Acc. Chem. Res. 2019, 52, 1168–1176. [Google Scholar] [CrossRef]
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [CrossRef]
- Wilson, T.D.; Yu, Y.; Lu, Y. Understanding copper-thiolate containing electron transfer centers by incorporation of unnatural amino acids and the CuA center into the type 1 copper protein azurin. Coord. Chem. Rev. 2013, 257, 260–276. [Google Scholar] [CrossRef]
- Marshall, N.M.; Garner, D.K.; Wilson, T.D.; Gao, Y.G.; Robinson, H.; Nilges, M.J.; Lu, Y. Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature 2009, 462, 113–116. [Google Scholar] [CrossRef]
- Hosseinzadeh, P.; Marshall, N.M.; Chacon, K.N.; Yu, Y.; Nilges, M.J.; New, S.Y.; Tashkov, S.A.; Blackburn, N.J.; Lu, Y. Design of a single protein that spans the entire 2-V range of physiological redox potentials. Proc. Natl. Acad. Sci. USA 2016, 113, 262–267. [Google Scholar] [CrossRef]
- Miner, K.D.; Mukherjee, A.; Gao, Y.G.; Null, E.L.; Petrik, I.D.; Zhao, X.; Yeung, N.; Robinson, H.; Lu, Y. A designed functional metalloenzyme that reduces O2 to H2O with over one thousand turnovers. Angew. Chem. Int. Ed. Engl. 2012, 51, 5589–5592. [Google Scholar] [CrossRef]
- Liu, X.; Yu, Y.; Hu, C.; Zhang, W.; Lu, Y.; Wang, J. Significant increase of oxidase activity through the genetic incorporation of a tyrosine-histidine cross-link in a myoglobin model of heme-copper oxidase. Angew. Chem. Int. Ed. Engl. 2012, 51, 4312–4316. [Google Scholar] [CrossRef]
- Brown, K.; Tegoni, M.; Prudencio, M.; Pereira, A.S.; Besson, S.; Moura, J.J.; Moura, I.; Cambillau, C. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat. Struct. Biol. 2000, 7, 191–195. [Google Scholar] [CrossRef]
- Dell’Acqua, S.; Pauleta, S.R.; Moura, I.; Moura, J.J. The tetranuclear copper active site of nitrous oxide reductase: The CuZ center. J. Biol. Inorg. Chem. 2011, 16, 183–194. [Google Scholar] [CrossRef]
- Johnston, E.M.; Dell’Acqua, S.; Ramos, S.; Pauleta, S.R.; Moura, I.; Solomon, E.I. Determination of the active form of the tetranuclear copper sulfur cluster in nitrous oxide reductase. J. Am. Chem. Soc. 2014, 136, 614–617. [Google Scholar] [CrossRef]
- Timmons, A.J.; Symes, M.D. Converting between the oxides of nitrogen using metal-ligand coordination complexes. Chem. Soc. Rev. 2015, 44, 6708–6722. [Google Scholar] [CrossRef]
- Lehnert, N.; Dong, H.T.; Harland, J.B.; Hunt, A.P.; White, C.J. Reversing nitrogen fixation. Nat. Rev. Chem. 2018, 2, 278–289. [Google Scholar] [CrossRef]
- Bar-Nahum, I.; Gupta, A.K.; Huber, S.M.; Ertem, M.Z.; Cramer, C.J.; Tolman, W.B. Reduction of nitrous oxide to dinitrogen by a mixed valent tricopper-disulfido cluster. J. Am. Chem. Soc. 2009, 131, 2812–2814. [Google Scholar] [CrossRef]
- Esmieu, C.; Orio, M.; Torelli, S.; Le Pape, L.; Pécaut, J.; Lebrun, C.; Ménage, S. N2O reduction at a dissymmetric Cu2S-containing mixed-valent center. Chem. Sci. 2014, 5, 4774–4784. [Google Scholar] [CrossRef]
- Johnson, B.J.; Antholine, W.E.; Lindeman, S.V.; Graham, M.J.; Mankad, N.P. A One-Hole Cu4S Cluster with N2O Reductase Activity: A Structural and Functional Model for CuZ. J. Am. Chem. Soc. 2016, 138, 13107–13110. [Google Scholar] [CrossRef]
- Kanady, J.S.; Tsui, E.Y.; Day, M.W.; Agapie, T. A synthetic model of the Mn(3)Ca subsite of the oxygen-evolving complex in photosystem II. Science 2011, 333, 733–736. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, C.; Dong, H.; Shen, J.R.; Dau, H.; Zhao, J. Inorganic chemistry. A synthetic Mn4Ca-cluster mimicking the oxygen-evolving center of photosynthesis. Science 2015, 348, 690–693. [Google Scholar] [CrossRef]
- Chen, C.; Chen, Y.; Yao, R.; Li, Y.; Zhang, C. Artificial Mn4Ca Clusters with Exchangeable Solvent Molecules Mimicking the Oxygen-Evolving Center in Photosynthesis. Angew. Chem. Int. Ed. Engl. 2019, 58, 3939–3942. [Google Scholar] [CrossRef]
- Olshansky, L.; Huerta-Lavorie, R.; Nguyen, A.I.; Vallapurackal, J.; Furst, A.; Tilley, T.D.; Borovik, A.S. Artificial Metalloproteins Containing Co4O4 Cubane Active Sites. J. Am. Chem. Soc. 2018, 140, 2739–2742. [Google Scholar] [CrossRef]
- Liao, F.; Yuan, H.; Du, K.J.; You, Y.; Gao, S.Q.; Wen, G.-B.; Lin, Y.-W.; Tan, X. Distinct roles of a tyrosine-associated hydrogen-bond network in fine-tuning the structure and function of heme proteins: Two cases designed for myoglobin. Mol. Biosyst. 2016, 12, 3139–3145. [Google Scholar] [CrossRef]
- Liu, C.; Xu, J.; Gao, S.-Q.; He, B.; Wei, C.-W.; Wang, X.-J.; Wang, Z.; Lin, Y.-W. Green and efficient biosynthesis of indigo from indole by engineered myoglobins. RSC Adv. 2018, 8, 33325–33330. [Google Scholar] [CrossRef]
- Yu, Y.; Cui, C.; Liu, X.; Petrik, I.D.; Wang, J.; Lu, Y. A Designed Metalloenzyme Achieving the Catalytic Rate of a Native Enzyme. J. Am. Chem. Soc. 2015, 137, 11570–11573. [Google Scholar] [CrossRef]
- Dydio, P.; Key, H.M.; Nazarenko, A.; Rha, J.Y.E.; Seyedkazemi, V.; Clark, D.S.; Hartwig, J.F. An artificial metalloenzyme with the kinetics of native enzymes. Science 2016, 354, 102–106. [Google Scholar] [CrossRef]
- Yin, L.-L.; Yuan, H.; Liu, C.; He, B.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. A Rationally Designed Myoglobin Exhibits a Catalytic Dehalogenation Efficiency More than 1000-Fold That of a Native Dehaloperoxidase. ACS Catal. 2018, 8, 9619–9624. [Google Scholar] [CrossRef]
- Kan, S.B.; Lewis, R.D.; Chen, K.; Arnold, F.H. Directed evolution of cytochrome c for carbon-silicon bond formation: Bringing silicon to life. Science 2016, 354, 1048–1051. [Google Scholar] [CrossRef]
- Martinez, A.T.; Ruiz-Duenas, F.J.; Camarero, S.; Serrano, A.; Linde, D.; Lund, H.; Vind, J.; Tovborg, M.; Herold-Majumdar, O.M.; Hofrichter, M.; et al. Oxidoreductases on their way to industrial biotransformations. Biotechnol. Adv. 2017, 35, 815–831. [Google Scholar] [CrossRef]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-W. Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters. Molecules 2019, 24, 2743. https://doi.org/10.3390/molecules24152743
Lin Y-W. Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters. Molecules. 2019; 24(15):2743. https://doi.org/10.3390/molecules24152743
Chicago/Turabian StyleLin, Ying-Wu. 2019. "Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters" Molecules 24, no. 15: 2743. https://doi.org/10.3390/molecules24152743
APA StyleLin, Y.-W. (2019). Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters. Molecules, 24(15), 2743. https://doi.org/10.3390/molecules24152743




