Discovery of a Nitric Oxide-Responsive Protein in Arabidopsis thaliana
Abstract
1. Introduction
2. Results and Discussion
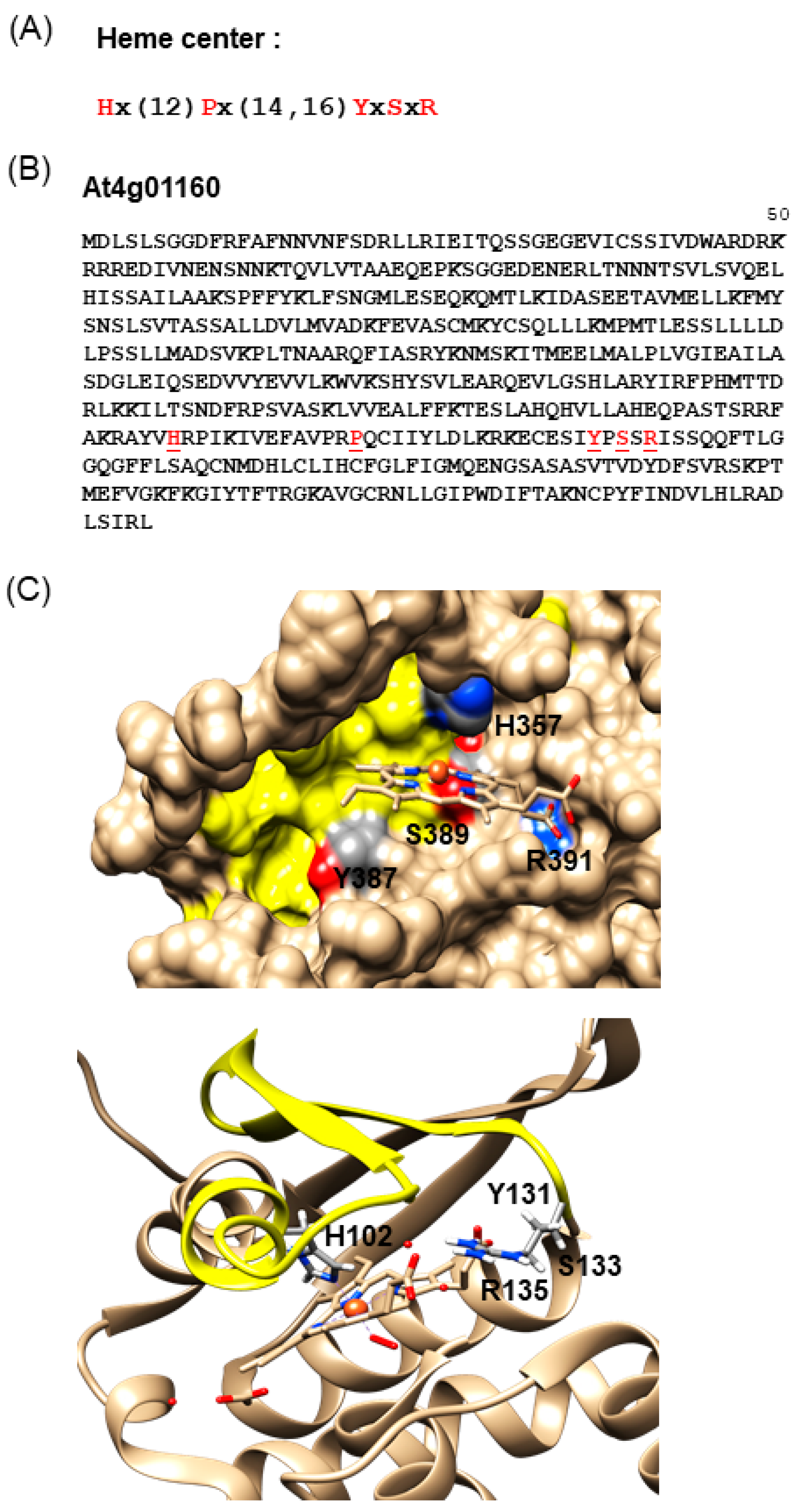
2.1. Identification of Candidate NO-Binding Proteins in Arabidopsis
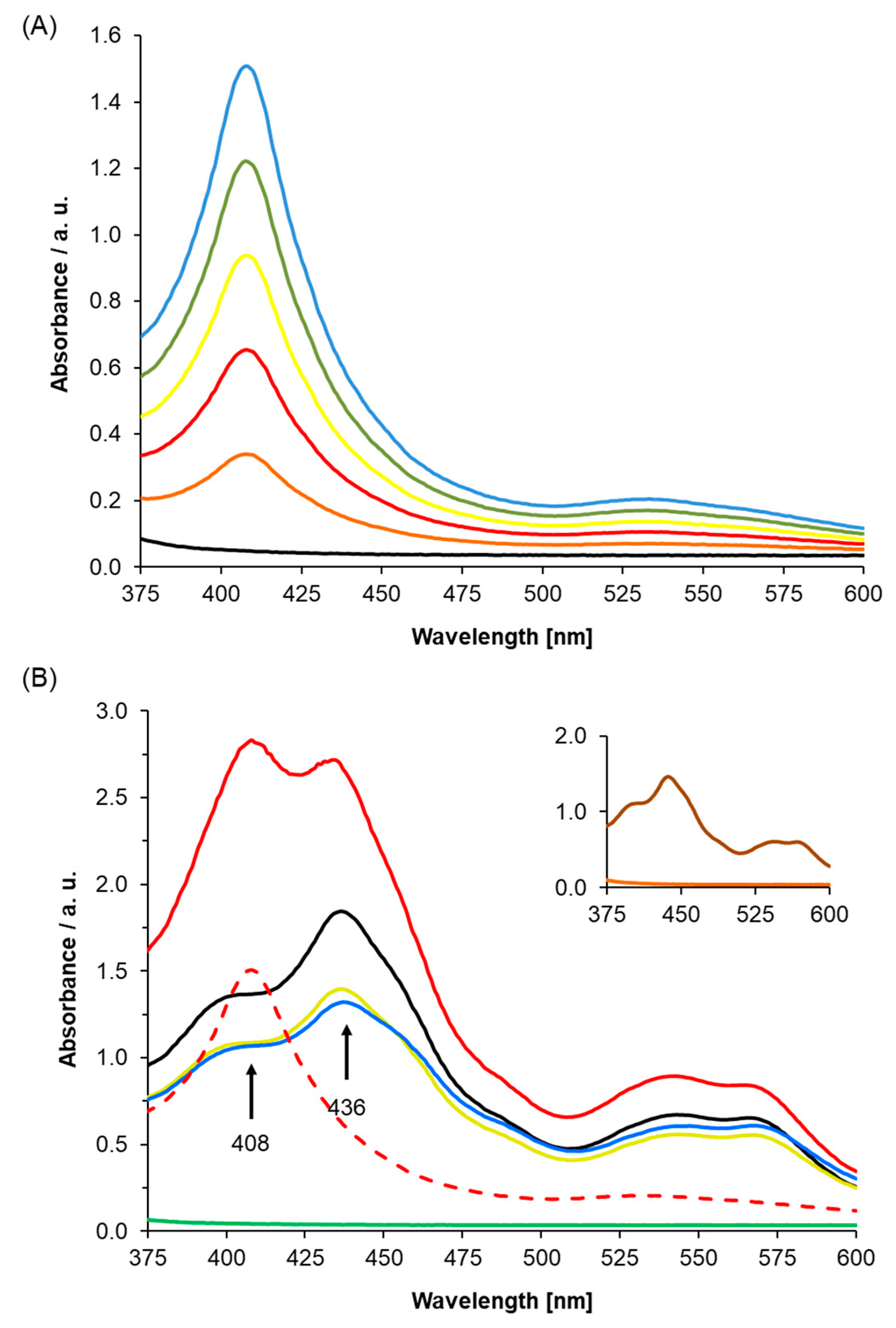
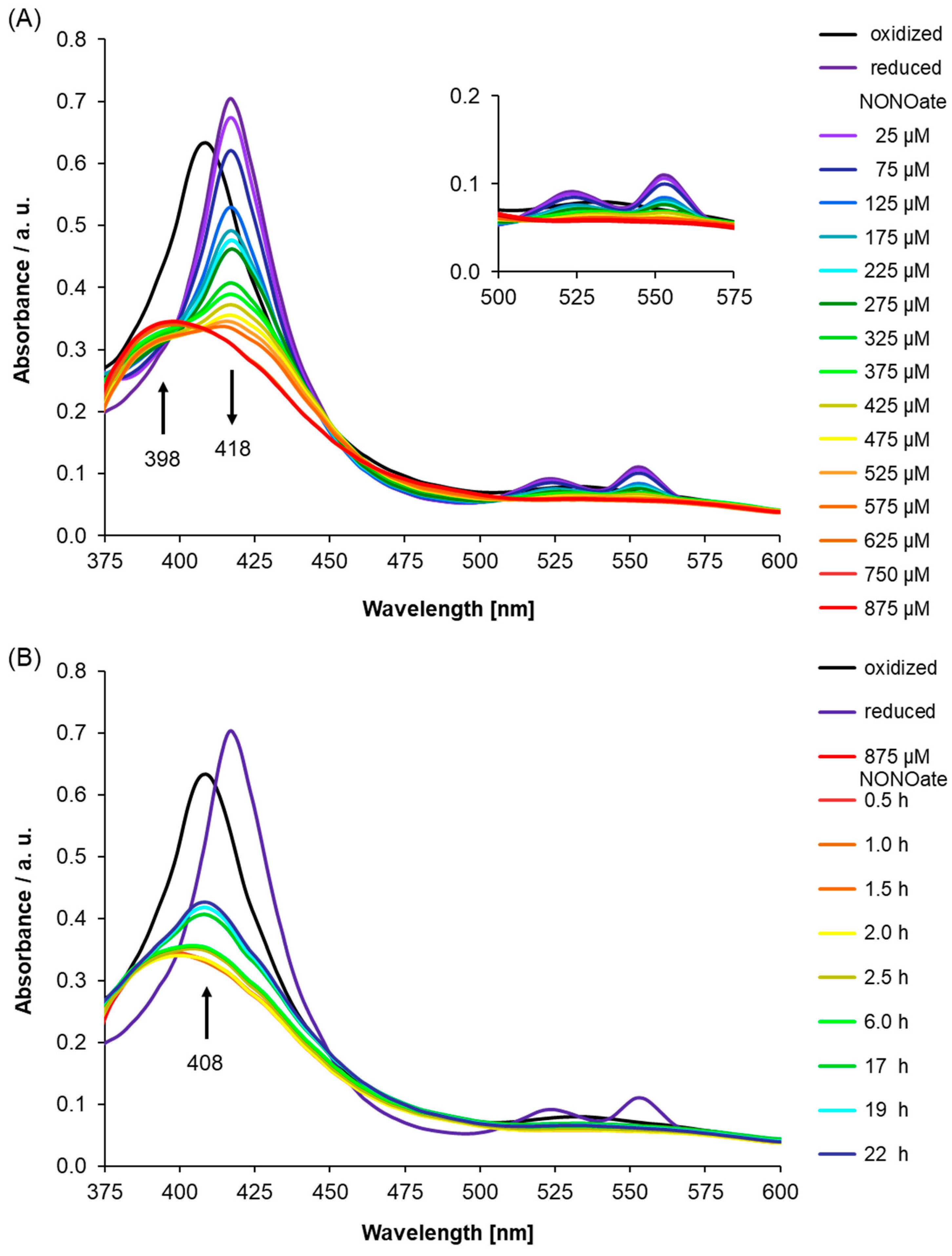
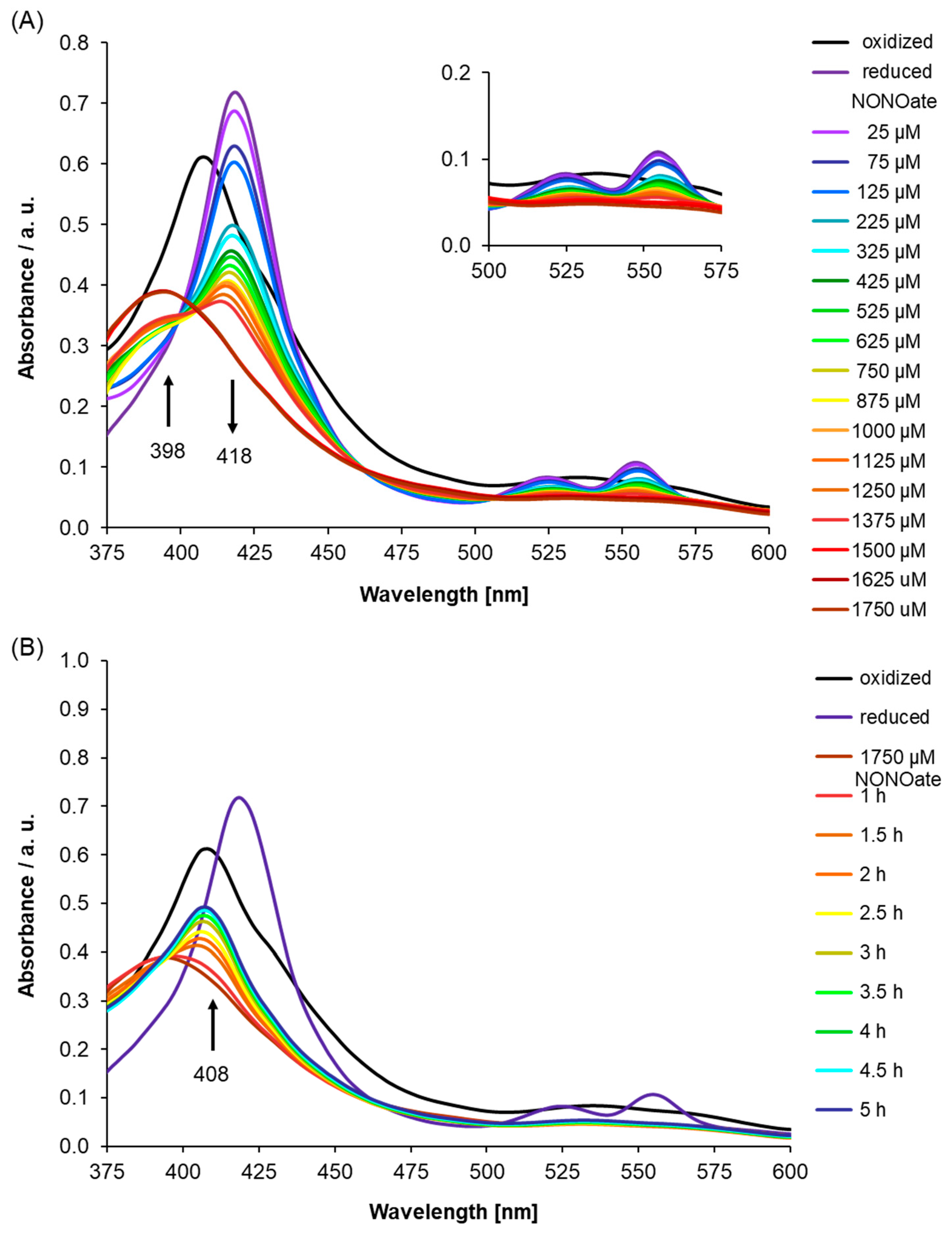
2.2. UV-Visible Spectra of Recombinant AtLRB3 in Response to NO
2.3. The Role of Light Response BTB Proteins
3. Materials and Methods
3.1. Preparation of the Recombinant AtLRB3 (At4g01160)
3.2. Computational Assessment of the AtLRB3 Heme-Binding Pocket
3.3. Determining NO Binding In Vitro
4. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Astuti, R.I.; Nasuno, R.; Takagi, H. Nitric oxide signalling in yeast. Appl. Microbiol. Biotechnol. 2016, 100, 9483–9497. [Google Scholar]
- Uehara, E.U.; Shida, B.S.; de Brito, C.A. Role of nitric oxide in immune responses against viruses: Beyond microbicidal activity. Inflamm. Res. 2015, 64, 845–852. [Google Scholar] [PubMed]
- Bogdan, C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015, 36, 161–178. [Google Scholar]
- Paskas, S.; Mazzon, E.; Basile, M.S.; Cavalli, E.; Al-Abed, Y.; He, M.; Rakocevic, S.; Nicoletti, F.; Mijatovic, S.; Maksimovic-Ivanic, D. Lopinavir-NO, a nitric oxide-releasing HIV protease inhibitor, suppresses the growth of melanoma cells in vitro and in vivo. Invest. New Drugs 2019. [Google Scholar] [CrossRef]
- Basile, M.S.; Mazzon, E.; Krajnovic, T.; Draca, D.; Cavalli, E.; Al-Abed, Y.; Bramanti, P.; Nicoletti, F.; Mijatovic, S.; Maksimovic-Ivanic, D. Anticancer and differentiation properties of the Nitric Oxide derivative of Lopinavir in human Glioblastoma cells. Molecules 2018, 23, 2463. [Google Scholar] [CrossRef]
- Tousoulis, D.; Kampoli, A.M.; Tentolouris, C.; Papageorgiou, N.; Stefanadis, C. The role of nitric oxide on endothelial function. Curr. Vasc. Pharmacol. 2012, 10, 4–18. [Google Scholar] [PubMed]
- Wendehenne, D.; Pugin, A.; Klessig, D.F.; Durner, J. Nitric oxide: Comparative synthesis and signalling in animal and plant cells. Trends Plant Sci. 2001, 6, 177–183. [Google Scholar] [PubMed]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijo, J.A. Nitric oxide: A multitasked signalling gas in plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [PubMed]
- Klessig, D.F.; Durner, J.; Noad, R.; Navarre, D.A.; Wendehenne, D.; Kumar, D.; Zhou, J.M.; Shah, J.; Zhang, S.; Kachroo, P.; et al. Nitric oxide and salicylic acid signalling in plant defence. Proc. Natl. Acad. Sci. USA 2000, 97, 8849–8855. [Google Scholar]
- Moreau, M.; Lindermayr, C.; Durner, J.; Klessig, D.F. NO synthesis and signalling in plants--where do we stand? Physiol. Plant 2010, 138, 372–383. [Google Scholar]
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When Bad Guys Become Good Ones: The Key Role of Reactive Oxygen Species and Nitric Oxide in the Plant Responses to Abiotic Stress. Front. Plant Sci. 2016, 7, 471. [Google Scholar] [CrossRef] [PubMed]
- Boon, E.M.; Huang, S.H.; Marletta, M.A. A molecular basis for NO selectivity in soluble guanylate cyclase. Nat. Chem. Biol. 2005, 1, 53–59. [Google Scholar] [PubMed]
- Mulaudzi, T.; Ludidi, N.; Ruzvidzo, O.; Morse, M.; Hendricks, N.; Iwuoha, E.; Gehring, C. Identification of a novel Arabidopsis thaliana nitric oxide-binding molecule with guanylate cyclase activity in vitro. FEBS Lett. 2011, 585, 2693–2697. [Google Scholar] [PubMed]
- Joudoi, T.; Shichiri, Y.; Kamizono, N.; Akaike, T.; Sawa, T.; Yoshitake, J.; Yamada, N.; Iwai, S. Nitrated cyclic GMP modulates guard cell signalling in Arabidopsis. Plant Cell 2013, 25, 558–571. [Google Scholar] [PubMed]
- Su, B.; Qian, Z.; Li, T.; Zhou, Y.; Wong, A. PlantMP: A database for moonlighting plant proteins. Database 2019, 2019, baz050. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Gehring, C. The Arabidopsis thaliana proteome harbours undiscovered multi-domain molecules with functional guanylyl cyclase catalytic centres. Cell Commun. Signal. 2013, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Gehring, C.; Irving, H.R. Conserved Functional Motifs and Homology Modeling to Predict Hidden Moonlighting Functional Sites. Front. Bioeng. Biotechnol. 2015, 3, 82. [Google Scholar] [CrossRef]
- Wong, A.; Tian, X.; Gehring, C.; Marondedze, C. Discovery of Novel Functional Centres With Rationally Designed Amino Acid. Comput. Struct. Biotechnol. J. 2018, 16, 70–76. [Google Scholar]
- Ludidi, N.; Gehring, C. Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 6490–6494. [Google Scholar]
- Meier, S.; Seoighe, C.; Kwezi, L.; Irving, H.; Gehring, C. Plant nucleotide cyclases: An increasingly complex and growing family. Plant Signal. Behav. 2007, 2, 536–539. [Google Scholar]
- Gehring, C. Adenyl cyclases and cAMP in plant signalling - past and present. Cell Commun. Signal. 2010, 8, 15. [Google Scholar] [CrossRef]
- Al-Younis, I.; Wong, A.; Gehring, C. The Arabidopsis thaliana K(+)-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett. 2015, 589, 3848–3852. [Google Scholar]
- Bianchet, C.; Wong, A.; Quaglia, M.; Alqurashi, M.; Gehring, C.; Ntoukakis, V.; Pasqualini, S. An Arabidopsis thaliana leucine-rich repeat protein harbours an adenylyl cyclase catalytic centre and affects responses to pathogens. J. Plant Physiol. 2018, 232, 12–22. [Google Scholar] [PubMed]
- Al-Younis, I.; Wong, A.; Lemtiri-Chlieh, F.; Schmöckel, S.; Tester, M.; Gehring, C.; Donaldson, L. The Arabidopsis thaliana K+-Uptake Permease 5 (AtKUP5) Contains a Functional Cytosolic Adenylate Cyclase Essential for K+ Transport. Front. Plant Sci. 2018, 9, 1645. [Google Scholar] [CrossRef]
- Xu, N.; Fu, D.; Li, S.; Wang, Y.; Wong, A. GCPred: A web tool for guanylyl cyclase functional centre prediction from amino acid sequence. Bioinformatics 2018, 34, 2134–2135. [Google Scholar] [PubMed]
- Xu, N.; Zhang, C.; Lim, L.L.; Wong, A. Bioinformatic Analysis of Nucleotide Cyclase Functional Centres and Development of ACPred Webserver. In Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics; ACM: Washington, DC, USA, 2018; pp. 122–129. [Google Scholar]
- Ooi, A.; Lemtiri-Chlieh, F.; Wong, A.; Gehring, C. Direct Modulation of the Guard Cell Outward-Rectifying Potassium Channel (GORK) by Abscisic Acid. Mol. Plant 2017, 10, 1469–1472. [Google Scholar] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [PubMed]
- Garbers, D.L. The guanylyl cyclase receptor family. New Biol. 1990, 2, 499–504. [Google Scholar]
- Wedel, B.J.; Garbers, D.L. Guanylyl cyclases: Approaching year thirty. Trends Endocrinol. Metab. 1998, 9, 213–219. [Google Scholar]
- Dias, F.V.; Serrazina, S.; Vitorino, M.; Marchese, D.; Heilmann, I.; Godinho, M.; Rodrigues, M.; Malho, R. A role for diacylglycerol kinase 4 in signalling crosstalk during Arabidopsis pollen tube growth. New Phytol. 2019, 222, 1434–1446. [Google Scholar]
- Olea, C.; Boon, E.M.; Pellicena, P.; Kuriyan, J.; Marletta, M.A. Probing the function of heme distortion in the H-NOX family. ACS Chem. Biol. 2008, 3, 703–710. [Google Scholar] [PubMed]
- Hong, J.K.; Yun, B.W.; Kang, J.G.; Raja, M.U.; Kwon, E.; Sorhagen, K.; Chu, C.; Wang, Y.; Loake, G.J. Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 2008, 59, 147–154. [Google Scholar] [PubMed]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marin-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [PubMed]
- Imran, Q.M.; Hussain, A.; Lee, S.U.; Mun, B.G.; Falak, N.; Loake, G.J.; Yun, B.W. Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci. Rep. 2018, 8, 771. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.C.; Coego, A.; Costa-Broseta, Á.; León, J. Nitric oxide responses in Arabidopsis hypocotyls are mediated by diverse phytohormone pathways. J. Exp. Bot. 2018, 69, 5265–5278. [Google Scholar] [PubMed]
- Marondedze, C.; Thomas, L.; Serrano, N.L.; Lilley, K.S.; Gehring, C. The RNA-binding protein repertoire of Arabidopsis thaliana. Sci. Rep. 2016, 6, 29766. [Google Scholar] [CrossRef]
- Genschik, P.; Sumara, I.; Lechner, E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): Cellular functions and disease implications. EMBO J. 2013, 32, 2307–2320. [Google Scholar]
- Wedel, B.; Humbert, P.; Harteneck, C.; Foerster, J.; Malkewitz, J.; Bohme, E.; Schultz, G.; Koesling, D. Mutation of His-105 in the beta 1 subunit yields a nitric oxide-insensitive form of soluble guanylyl cyclase. Proc. Natl. Acad. Sci. USA 1994, 91, 2592–2596. [Google Scholar]
- Zhao, Y.; Schelvis, J.P.; Babcock, G.T.; Marletta, M.A. Identification of histidine 105 in the beta1 subunit of soluble guanylate cyclase as the heme proximal ligand. Biochemistry 1998, 37, 4502–4509. [Google Scholar]
- Stone, J.R.; Sands, R.H.; Dunham, W.R.; Marletta, M.A. Electron paramagnetic resonance spectral evidence for the formation of a pentacoordinate nitrosyl-heme complex on soluble guanylate cyclase. Biochem. Biophys. Res. Commun. 1995, 207, 572–577. [Google Scholar] [PubMed]
- Dai, Z.; Farquhar, E.R.; Arora, D.P.; Boon, E.M. Is histidine dissociation a critical component of the NO/H-NOX signalling mechanism? Insights from X-ray absorption spectroscopy. Dalton Trans. 2012, 41, 7984–7993. [Google Scholar] [PubMed]
- Pellicena, P.; Karow, D.S.; Boon, E.M.; Marletta, M.A.; Kuriyan, J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc. Natl. Acad. Sci. USA 2004, 101, 12854–12859. [Google Scholar] [PubMed]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centres. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [PubMed]
- Beltran, J.; Kloss, B.; Hosler, J.P.; Geng, J.; Liu, A.; Modi, A.; Dawson, J.H.; Sono, M.; Shumskaya, M.; Ampomah-Dwamena, C. Control of carotenoid biosynthesis through a heme-based cis-trans isomerase. Nat. Chem. Biol. 2015, 11, 598–605. [Google Scholar] [PubMed]
- Zhong, F.; Wang, H.; Ying, T.; Huang, Z.X.; Tan, X. Efficient expression of human soluble guanylate cyclase in Escherichia coli and its signalling-related interaction with nitric oxide. Amino Acids 2010, 39, 399–408. [Google Scholar] [PubMed]
- Aller, I.; Rouhier, N.; Meyer, A.J. Development of roGFP2-derived redox probes for measurement of the glutathione redox potential in the cytosol of severely glutathione-deficient rml1 seedlings. Front. Plant Sci. 2013, 4, 506. [Google Scholar] [CrossRef] [PubMed]
- Hunt, A.P.; Lehnert, N. Heme-nitrosyls: Electronic structure implications for function in biology. Acc. Chem. Res. 2015, 48, 2117–2125. [Google Scholar] [PubMed]
- Brandish, P.E.; Buechler, W.; Marletta, M.A. Regeneration of the ferrous heme of soluble guanylate cyclase from the nitric oxide complex: Acceleration by thiols and oxyhemoglobin. Biochemistry 1998, 37, 16898–16907. [Google Scholar] [PubMed]
- Tsai, A.L.; Berka, V.; Martin, F.; Ma, X.; van den Akker, F.; Fabian, M.; Olson, J.S. Is Nostoc H-NOX a NO sensor or redox switch? Biochemistry 2010, 49, 6587–6599. [Google Scholar] [PubMed]
- Rao, M.; Herzik MA, Jr.; Iavarone, A.T.; Marletta, M.A. Nitric Oxide-Induced Conformational Changes Govern H-NOX and Histidine Kinase Interaction and Regulation in Shewanella oneidensis. Biochemistry 2017, 56, 1274–1284. [Google Scholar] [PubMed]
- Olea, C.Jr.; Herzik, M.A.Jr.; Kuriyan, J.; Marletta, M.A. Structural insights into the molecular mechanism of H-NOX activation. Protein Sci. 2010, 19, 881–887. [Google Scholar] [PubMed]
- Herzik, M.A.Jr.; Jonnalagadda, R.; Kuriyan, J.; Marletta, M.A. Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E4156–E4164. [Google Scholar] [PubMed]
- Karow, D.S.; Pan, D.; Tran, R.; Pellicena, P.; Presley, A.; Mathies, R.A.; Marletta, M.A. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry 2004, 43, 10203–10211. [Google Scholar] [PubMed]
- Praneeth, V.K. Spectroscopic properties and electronic structure of five- and six-coordinate iron(II) porphyrin NO complexes Effect of the axial N-donor. Inorg. Chem. 2006, 45. [Google Scholar]
- Russwurm, M.; Koesling, D. NO activation of guanylyl cyclase. EMBO J. 2004, 23, 4443–4450. [Google Scholar] [PubMed]
- Soldatova, A.V.; Ibrahim, M.; Olson, J.S.; Czernuszewicz, R.S.; Spiro, T.G. New light on NO bonding in Fe(III) heme proteins from resonance Raman spectroscopy and DFT modeling. J. Am. Chem. Soc. 2010, 132, 4614–4625. [Google Scholar]
- Goodrich, L.E.; Paulat, F.; Praneeth, V.K.; Lehnert, N. Electronic structure of heme-nitrosyls and its significance for nitric oxide reactivity, sensing, transport, and toxicity in biological systems. Inorg. Chem. 2010, 49, 6293–6316. [Google Scholar]
- Canning, P.; Bullock, A.N. New strategies to inhibit KEAP1 and the Cul3-based E3 ubiquitin ligases. Biochem. Soc. Trans. 2014, 42, 103–107. [Google Scholar]
- Bartberger, M.D.; Liu, W.; Ford, E.; Miranda, K.M.; Switzer, C.; Fukuto, J.M.; Farmer, P.J.; Wink, D.A.; Houk, K.N. The reduction potential of nitric oxide (NO) and its importance to NO biochemistry. Proc. Natl. Acad. Sci. USA 2002, 99, 10958–10963. [Google Scholar]
- Zhao, X.J.; Sampath, V.; Caughey, W.S. Infrared characterization of nitric oxide bonding to bovine heart cytochrome c oxidase and myoglobin. Biochem. Biophys. Res. Commun. 1994, 204, 537–543. [Google Scholar] [PubMed]
- Shiro, Y.; Fujii, M.; Iizuka, T.; Adachi, S.; Tsukamoto, K.; Nakahara, K.; Shoun, H. Spectroscopic and kinetic studies on reaction of cytochrome P450nor with nitric oxide. Implication for its nitric oxide reduction mechanism. J. Biol. Chem. 1995, 270, 1617–1623. [Google Scholar] [PubMed]
- Yi, J.; Campbell, A.L.; Richter-Addo, G.B. Nitric oxide coupling to generate N2O promoted by a single-heme system as examined by density functional theory. Nitric Oxide 2016, 60, 69–75. [Google Scholar] [PubMed]
- Lee, T.Y.; Chen, Y.J.; Lu, T.C.; Huang, H.D.; Chen, Y.J. SNOSite: Exploiting maximal dependence decomposition to identify cysteine S-nitrosylation with substrate site specificity. PLoS ONE 2011, 6, e21849. [Google Scholar] [CrossRef]
- Chen, H.Y.; Chen, R.H. Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications. Front. Oncol. 2016, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [PubMed]
- Furukawa, M.; He, Y.J.; Borchers, C.; Xiong, Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 2003, 5, 1001–1007. [Google Scholar] [PubMed]
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.H.; Vidal, M.; Elledge, S.J.; Harper, J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003, 425, 316–321. [Google Scholar] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.; Nandra, S.K.; Prive, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef]
- Christians, M.J.; Gingerich, D.J.; Hua, Z.; Lauer, T.D.; Vierstra, R.D. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signalling in Arabidopsis. Plant Physiol. 2012, 160, 118–134. [Google Scholar]
- McMahon, M.; Lamont, D.J.; Beattie, K.A.; Hayes, J.D. Keap1 perceives stress via three sensors for the endogenous signalling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA 2010, 107, 18838–18843. [Google Scholar] [PubMed]
- Hayes, J.D.; McMahon, M.; Chowdhry, S.; Dinkova-Kostova, A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010, 13, 1713–1748. [Google Scholar] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [PubMed]
- Wheeler, J.I.; Wong, A.; Marondedze, C.; Groen, A.J.; Kwezi, L.; Freihat, L.; Vyas, J.; Raji, M.A.; Irving, H.R.; Gehring, C. The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signalling. Plant. J. 2017, 91, 590–600. [Google Scholar] [PubMed]
- Corwin, A.H.; Chivvis, A.B.; Poor, R.W.; Whitten, D.G.; Baker, E.W. Porphyrin studies. XXXVII. The interpretation of porphyrin and metalloporphyrin spectra. J. Am. Chem. Soc. 1968, 90, 6577–6583. [Google Scholar]
- Zhang, A.; Kwan, L.; Stillman, M.J. The spectroscopic impact of interactions with the four Gouterman orbitals from peripheral decoration of porphyrins with simple electron withdrawing and donating groups. Org. Biomol. Chem. 2017, 15, 9081–9094. [Google Scholar] [PubMed]
Sample Availability: Not available. |

| ATG number | Annotation | NO |
|---|---|---|
| At1G08630.7 | Threonine aldolase 1 | |
| At1G13940.1 TF | T-box transcription factor, putative (DUF863) | |
| At1G55870.1 | ABA-hypersensitive germination 2 | +1 |
| At1G74800.1 | Galactosyltransferase 5, GALT5 | +2 |
| At2G28250.1 | Protein kinase superfamily protein; | |
| At2G42280.1 TF | ABA-resp. kinase substrate 3—(bHLH) DNA-binding fam. prot. | +3 |
| At2G44710.4 RBP | RNA-binding protein (RRM/RBD/RNP motifs) | |
| At3G06260.1 | Galactinol synthase 9 | +4 |
| At3G29170.1 | Transmembrane protein (DUF872) | |
| At3G57420.1 | Regulates assembly & trafficking of cellulose and synthase complexes | |
| At4G01160.2 | Encodes a member of LRB BTB family | ++ |
| At4G17250.3 | Transmembrane protein | |
| At4G26330.2 * | Subtilisin-like serine endopeptidase family protein | +5 |
| At4G31570.3 | Nucleoporin | |
| At4G35830.1 RBP | Aconitase 1 | +6 |
| At4G36195.1 | Serine carboxypeptidase s28 family protein | |
| At4G39290.1 | Galactose oxidase/KELCH repeat superfamily protein | |
| At5G02310.2 * | Proteolysis 6, PRT6 subtilase family protein | |
| At5G11940.1 * | Subtilase family protein | |
| At5G52850.1 | Pentatricopeptide repeat (PPR) superfamily protein | |
| At5G53280.1 | PVD1, plastid division1 | |
| At5G67170.1 * | SEC-C motif-containing protein/OTU-like cysteine protease family |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarban, R.; Vogler, M.; Wong, A.; Eppinger, J.; Al-Babili, S.; Gehring, C. Discovery of a Nitric Oxide-Responsive Protein in Arabidopsis thaliana. Molecules 2019, 24, 2691. https://doi.org/10.3390/molecules24152691
Zarban R, Vogler M, Wong A, Eppinger J, Al-Babili S, Gehring C. Discovery of a Nitric Oxide-Responsive Protein in Arabidopsis thaliana. Molecules. 2019; 24(15):2691. https://doi.org/10.3390/molecules24152691
Chicago/Turabian StyleZarban, Randa, Malvina Vogler, Aloysius Wong, Joerg Eppinger, Salim Al-Babili, and Chris Gehring. 2019. "Discovery of a Nitric Oxide-Responsive Protein in Arabidopsis thaliana" Molecules 24, no. 15: 2691. https://doi.org/10.3390/molecules24152691
APA StyleZarban, R., Vogler, M., Wong, A., Eppinger, J., Al-Babili, S., & Gehring, C. (2019). Discovery of a Nitric Oxide-Responsive Protein in Arabidopsis thaliana. Molecules, 24(15), 2691. https://doi.org/10.3390/molecules24152691





