A Novel 3-Phytosterone-9α-Hydroxylase Oxygenation Component and Its Application in Bioconversion of 4-Androstene-3,17-Dione to 9α-Hydroxy-4-Androstene-3,17-Dione Coupling with A NADH Regeneration Formate Dehydrogenase
Abstract
1. Introduction
2. Results and Discussion
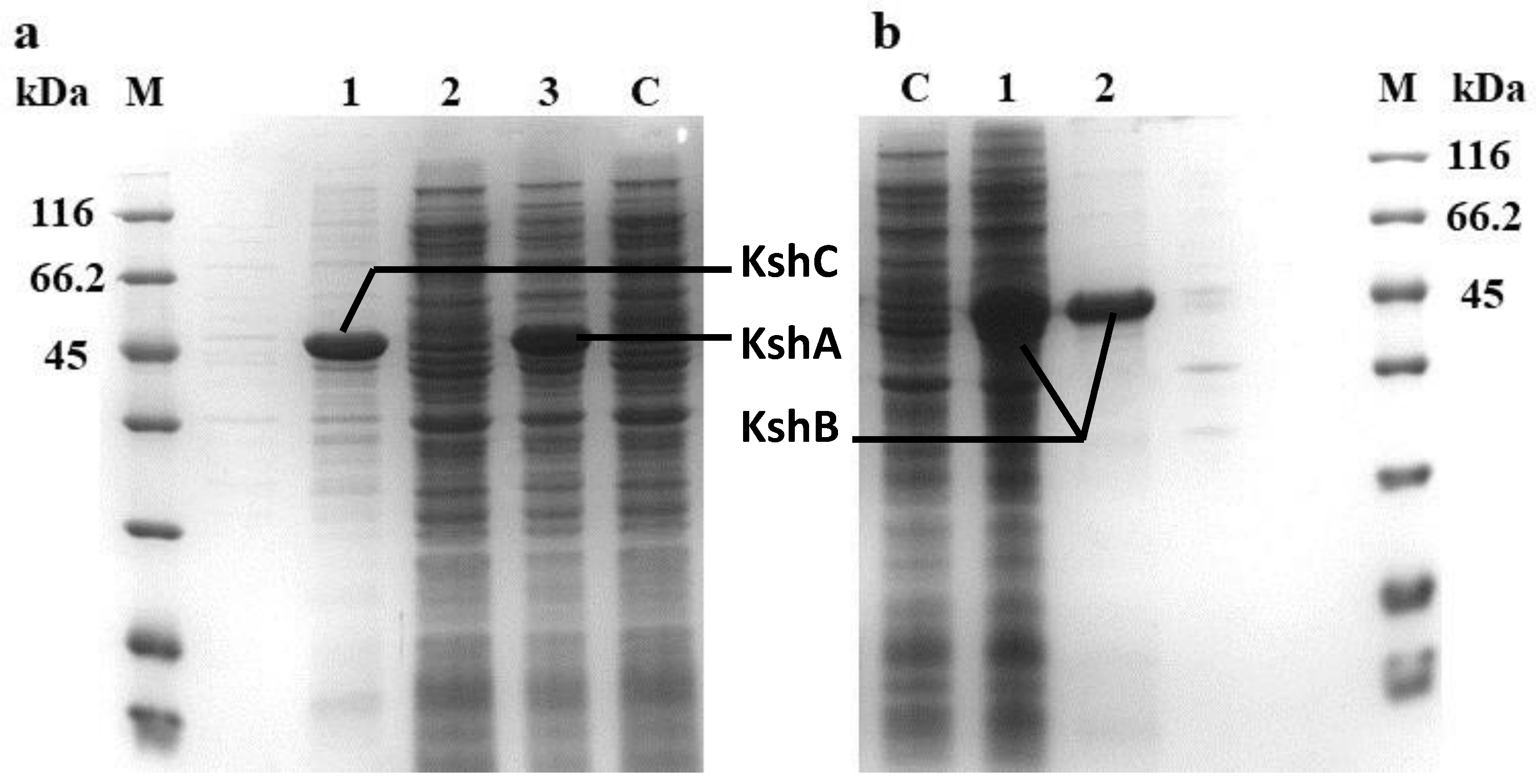
2.1. Heterologous Expression and Identification of the KshA, KshB, and KshC Proteins in E. coli
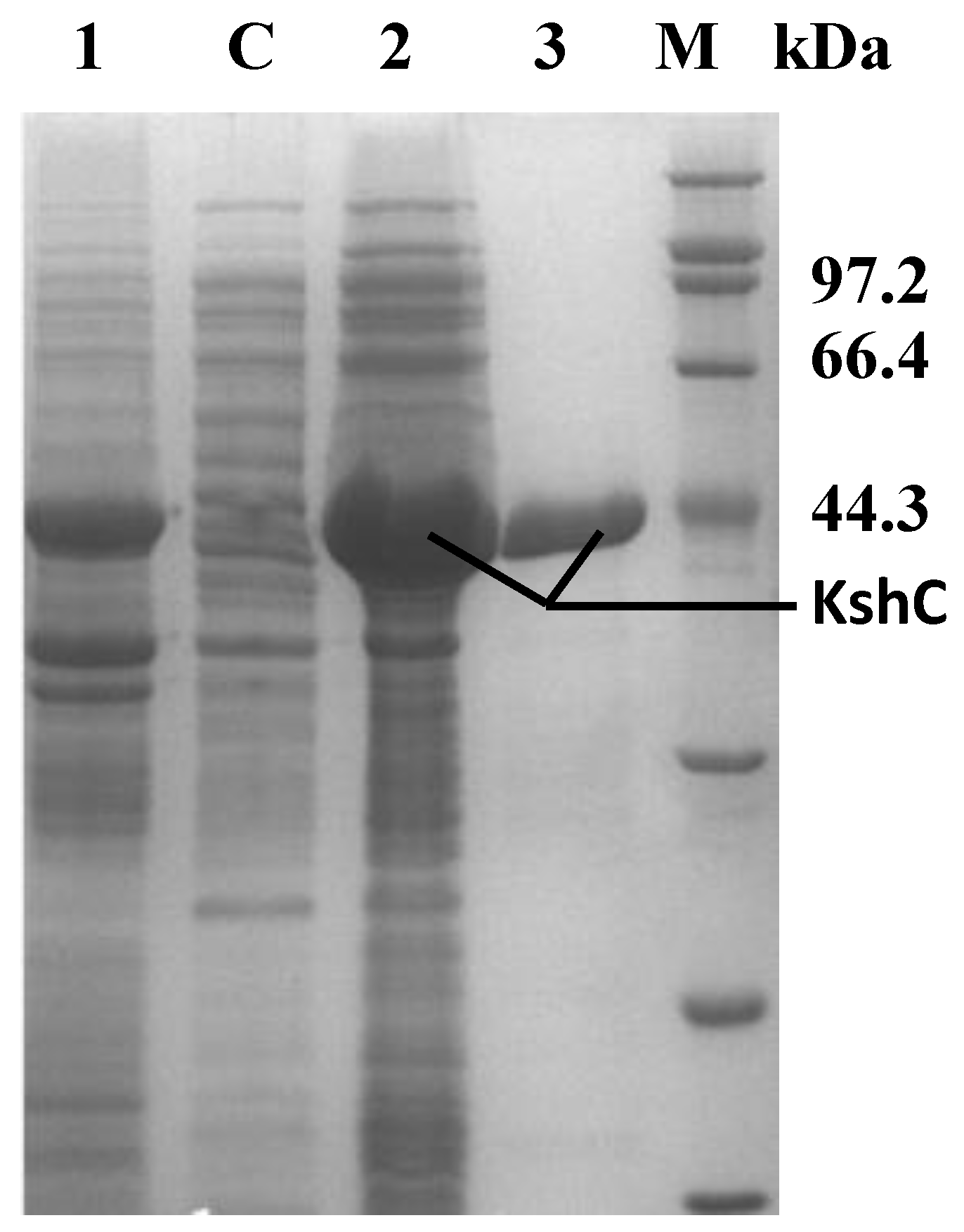
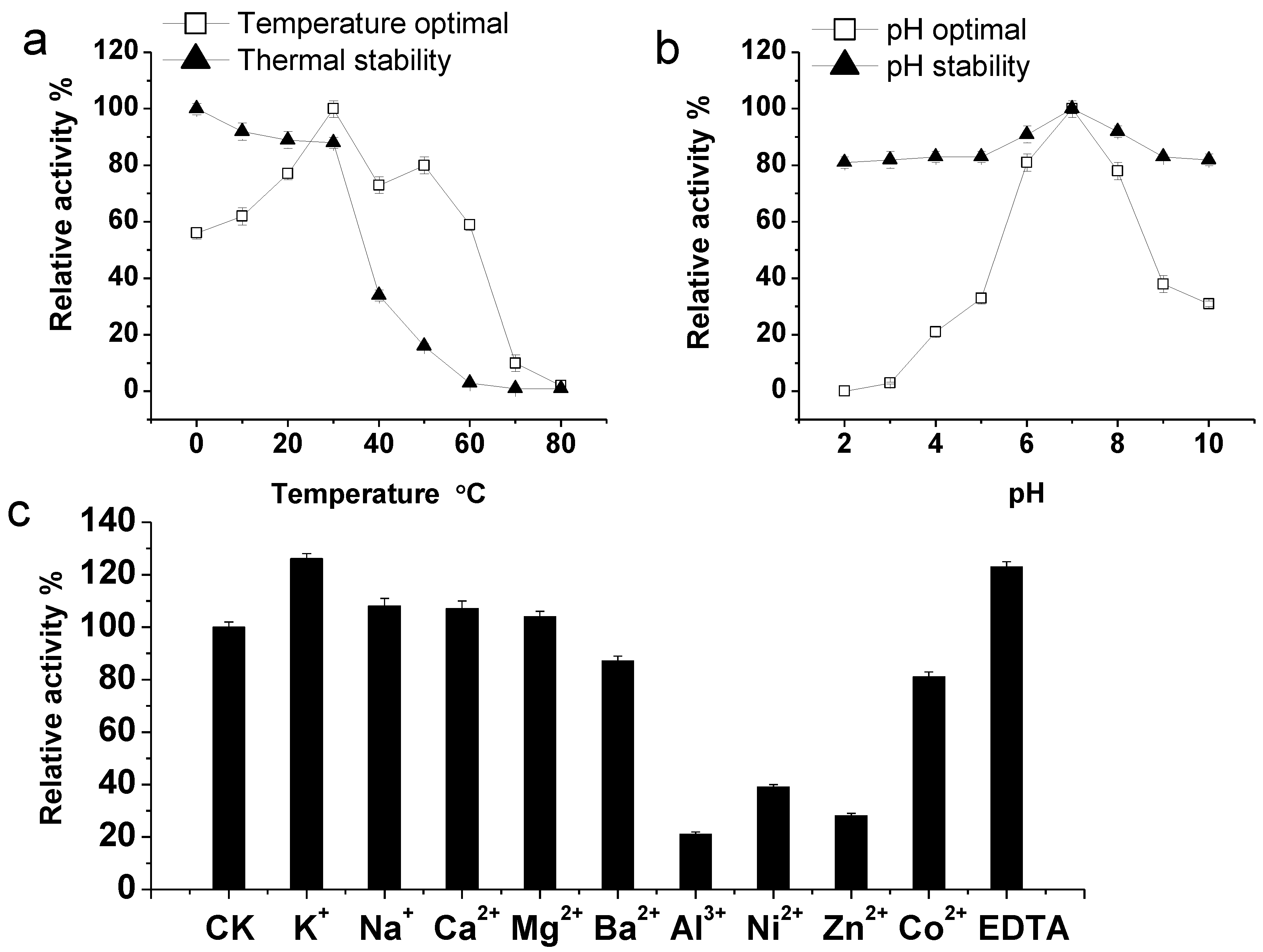
2.2. Purification and Characterization of the KshB Protein
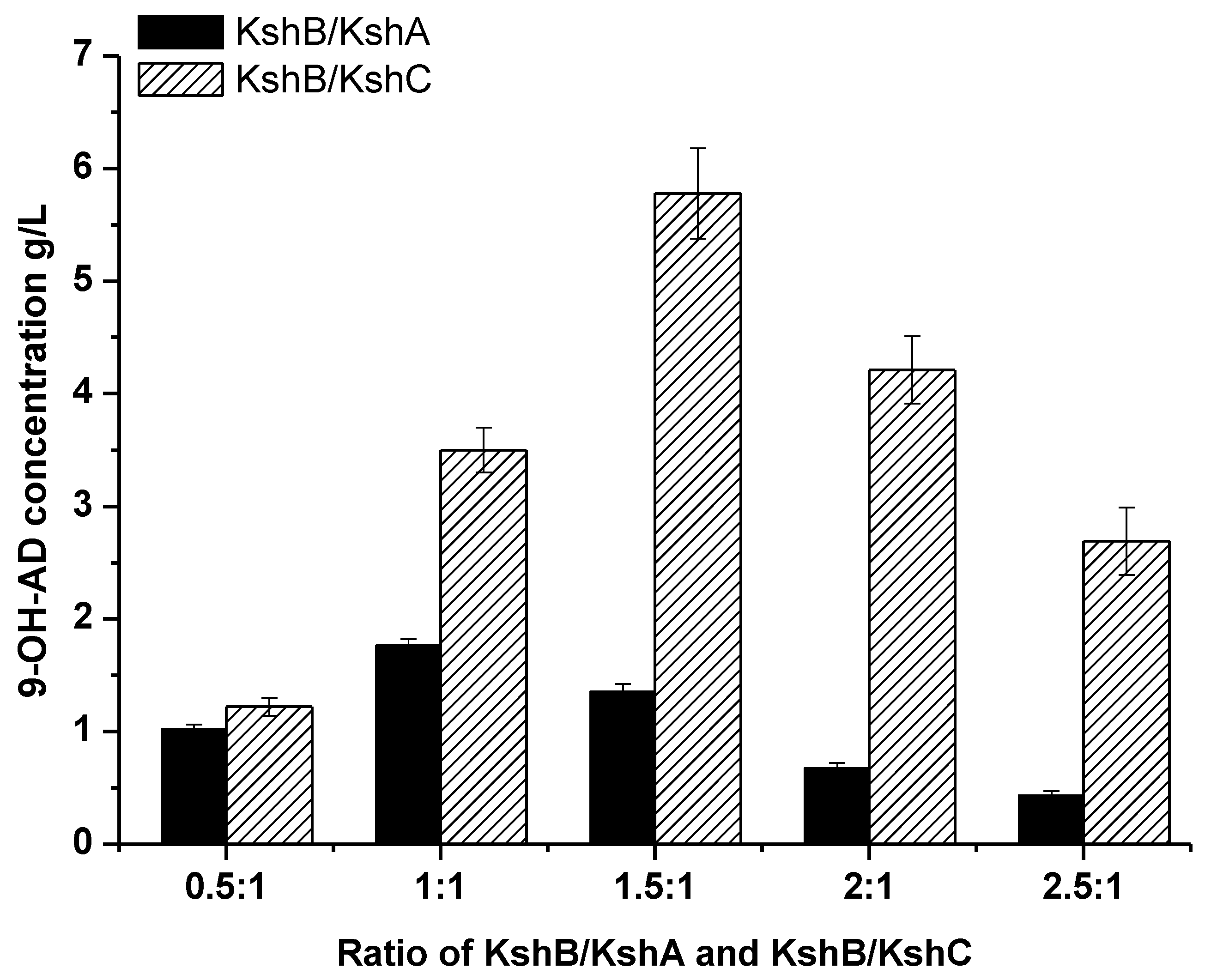
2.3. Optimization of the Terminal Oxygenase and Ferredoxin Reductase Ratio in an Enzyme-Liquid Transformation System
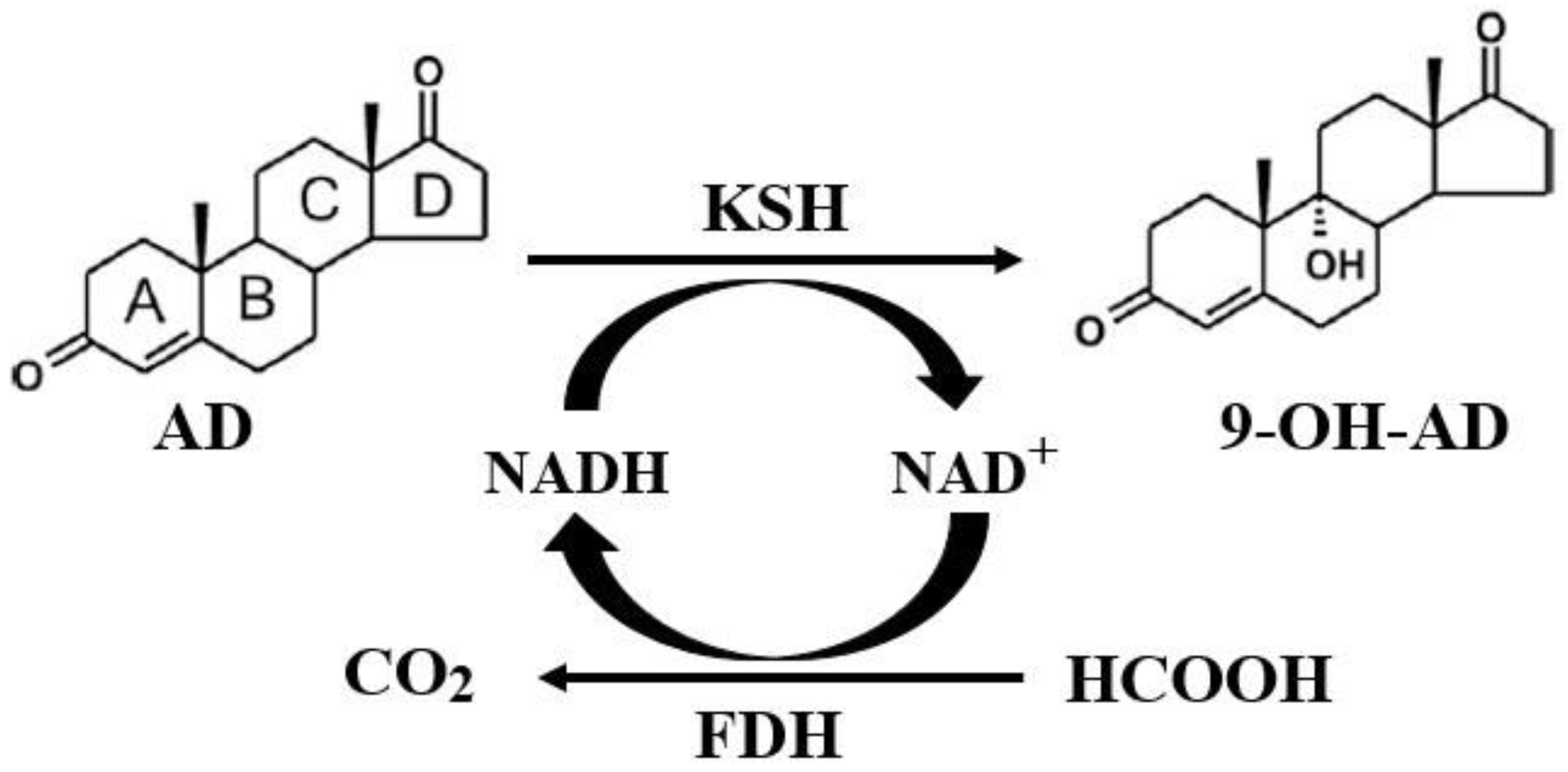
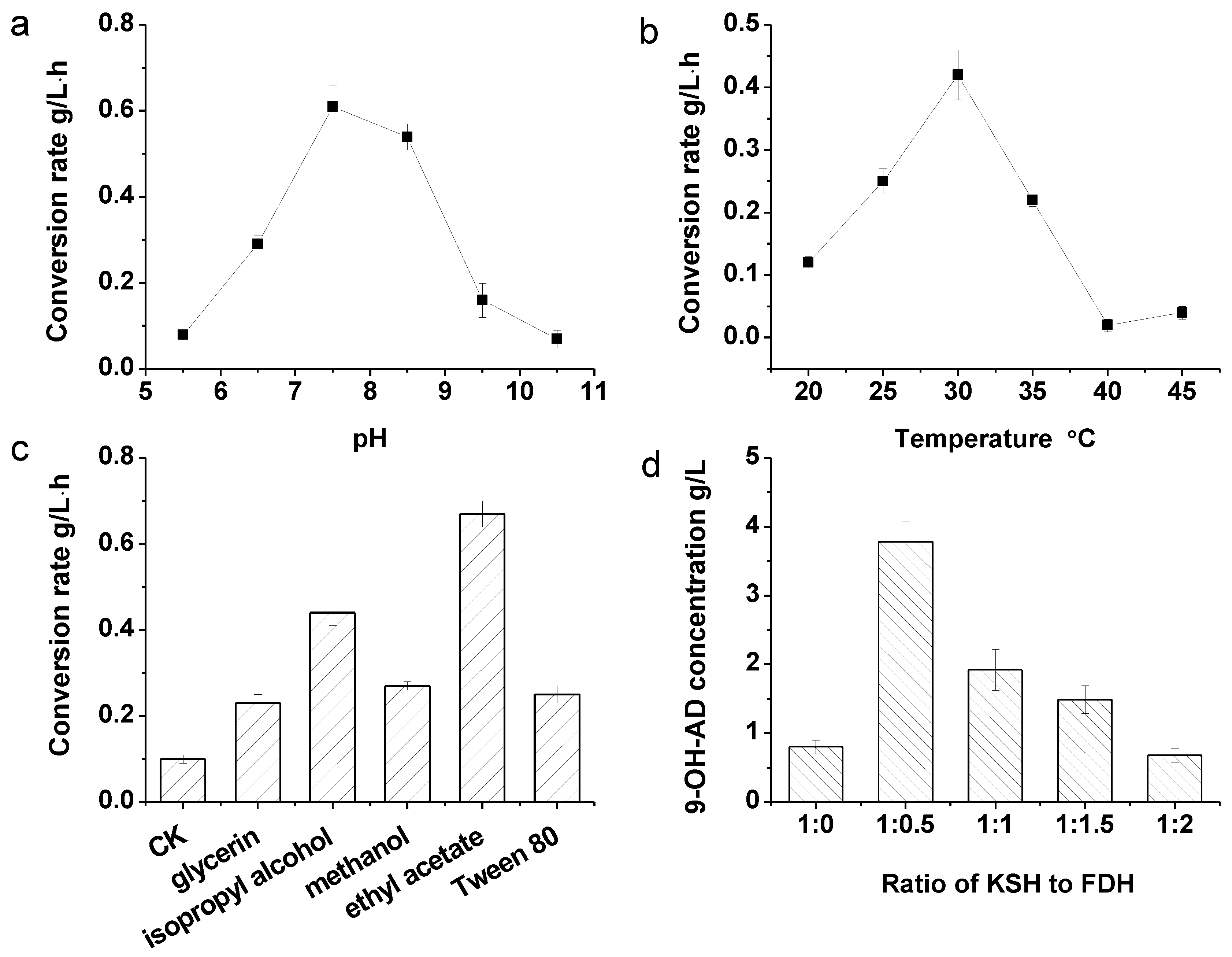
2.4. Coupled Reaction of Hydroxylation and NADH Regeneration and the Optimization of the Transformation System
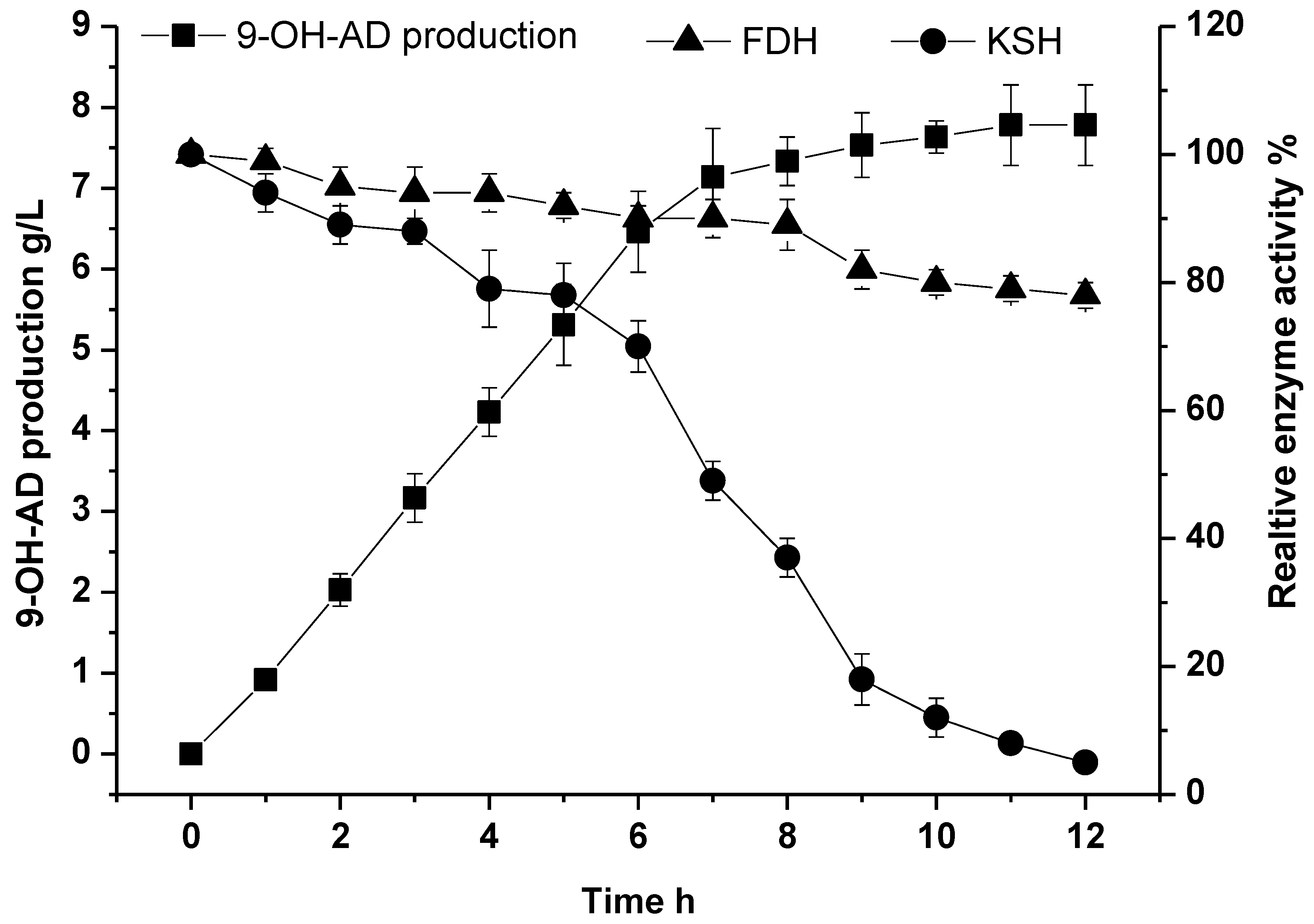
2.5. Fed-Batch Conversion under Optimal Conditions for the Biocatalysis of 9-OH-AD
3. Materials and Methods
3.1. Chemicals, Primers, Plasmids, and Bacterial Strains
3.2. Construction of Recombinant Plasmids and Strains
3.3. Expression and Purification of Enzymes
3.4. Assay of KshB, KSH, and FDH Activities
3.5. Characterization of KshB
3.6. Optimization of Conversion Conditions
3.7. Production of 9-OH-AD by Fed-Batch Conversion
3.8. Analysis of Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Donova, M.V.; Egorova, O.V. Microbial steroid transformations: Current state and prospects. Appl. Microbiol. Biotechnol. 2012, 94, 1423–1447. [Google Scholar] [CrossRef] [PubMed]
- Wovcha, M.G.; Antosz, F.J.; Knight, J.C.; Kominek, L.A.; Pyke, T.R. Bioconversion of sitosterol to useful steroidal intermediates by mutants of Mycobacterium fortuitum. Biochim. Biophys. Acta 1978, 531, 308–321. [Google Scholar] [CrossRef]
- Fernandes, P.; Cruz, A.; Angelova, B.; Pinheiro, H.M.; Cabral, J.M.S. Microbial conversion of steroid compounds: Recent developments. Enzym. Microb. Technol. 2003, 32, 688–705. [Google Scholar] [CrossRef]
- Sedlaczek, L. Biotransformations of steroids. Crit. Rev. Biotechnol. 1988, 7, 187–236. [Google Scholar] [CrossRef] [PubMed]
- Mutafov, S.; Angelova, B.; Avramova, T.; Boyadjieva, L.; Dimova, I. The inducibility of 9 alpha-steroid hydroxylating activity in resting Rhodococcus sp. cells. Process Biochem. 1997, 32, 585–589. [Google Scholar] [CrossRef]
- Petrusma, M.; Dijkhuizen, L.; van der Geize, R. Rhodococcus rhodochrous DSM 43269 3-Ketosteroid 9 alpha-Hydroxylase, a Two-Component Iron-Sulfur-Containing Monooxygenase with Subtle Steroid Substrate Specificity. Appl. Environ. Microbiol. 2009, 75, 5300–5307. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brzostek, A.; Sliwinski, T.; Rumijowska-Galewicz, A.; Korycka-Machala, M.; Dziadek, J. Identification and targeted disruption of the gene encoding the main 3-ketosteroid dehydrogenase in Mycobacterium smegmatis. Microbiology 2005, 151, 2393–2402. [Google Scholar] [CrossRef]
- Strijewski, A. The steroid-9 alpha-hydroxylation system from Nocardia species. Eur. J. Biochem. 1982, 128, 125–135. [Google Scholar] [CrossRef]
- Dutta, R.K.; Roy, M.K.; Singh, H.D. Role of Plasmid Pjl1 Of Arthrobacter-Oxydans 317 In the Degradation of Beta-Sitosterol. J. Basic Microbiol. 1992, 32, 317–324. [Google Scholar] [CrossRef]
- van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, R.; Dijkhuizen, L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9 alpha-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 2002, 45, 1007–1018. [Google Scholar] [CrossRef]
- Tong, W.Y.; Dong, X. Microbial biotransformation: Recent developments on steroid drugs. Recent Pat. Biotechnol. 2009, 3, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Peart, P.C.; McCook, K.P.; Russell, F.A.; Reynolds, W.F.; Reese, P.B. Hydroxylation of steroids by Fusarium oxysporum, Exophiala jeanselmei and Ceratocystis paradoxa. Steroids 2011, 76, 1317–1330. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.M.; Ge, W.; Huang, L.; Shan, L. Synthesis of 7alpha-hydroxy-dehydroepiandrosterone and 7beta-hydroxy-dehydroepiandrosterone. Steroids 2005, 70, 970–973. [Google Scholar] [CrossRef] [PubMed]
- Lamm, A.S.; Chen, A.R.; Reynolds, W.F.; Reese, P.B. Steroid hydroxylation by Whetzelinia sclerotiorum, Phanerochaete chrysosporium and Mucor plumbeus. Steroids 2007, 72, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, R. Neuroactive steroids: mechanisms of action and neuropsychopharmacological properties. Psychoneuroendocrinology 2003, 28, 139–168. [Google Scholar] [CrossRef]
- Baker, M.E. Origin and diversification of steroids: Co-evolution of enzymes and nuclear receptors. Mol. Cell. Endocrinol. 2011, 334, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Schule, C.; Eser, D.; Baghai, T.C.; Nothdurfter, C.; Kessler, J.S.; Rupprecht, R. Neuroactive steroids in affective disorders: Target for novel antidepressant or anxiolytic drugs. Neuroscience 2011, 191, 55–77. [Google Scholar] [CrossRef]
- Angelova, B.; Mutafov, S.; Avramova, T.; Dimova, I.; Boyadjieva, L. 9 alpha-hydroxylation of 4-androstene-3,17-dione by resting Rhodococcus sp. cells. Process. Biochem. 1996, 31, 179–184. [Google Scholar] [CrossRef]
- Donova, M.V.; Gulevskaya, S.A.; Dovbnya, D.V.; Puntus, I.F. Mycobacterium sp. mutant strain producing 9alpha-hydroxyandrostenedione from sitosterol. Appl. Microbiol. Biotechnol. 2005, 67, 671–678. [Google Scholar] [CrossRef]
- Xian, Z.; Zhiming, R.; Zhang, L.; Meijuan, X.; Taowei, Y. Efficient 9α-hydroxy-4-androstene-3,17-dione production by engineered Bacillus subtilis co-expressing Mycobacterium neoaurum 3-ketosteroid 9α-hydroxylase and B. subtilis glucose 1-dehydrogenase with NADH regeneration. SpringerPlus 2016, 5, 1207. [Google Scholar]
- Yao, K.; Xu, L.Q.; Wang, F.Q.; Wei, D.Z. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9alpha-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9alpha-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab. Eng. 2014, 24, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Bragin, E.Y.; Shtratnikova, V.Y.; Dovbnya, D.V.; Schelkunov, M.I.; Pekov, Y.A.; Malakho, S.G.; Egorova, O.V.; Ivashina, T.V.; Sokolov, S.L.; Ashapkin, V.V.; et al. Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. strains. J. Steroid Biochem. Mol. Biol. 2013, 138, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Schroder, I.; Steckhan, E.; Liese, A. In situ NAD(+) regeneration using 2,2 ‘-azinobis(3-ethylbenzothiazoline-6-sulfonate) as an electron transfer mediator. J. Electroanal. Chem. 2003, 541, 109–115. [Google Scholar] [CrossRef]
- van der Donk, W.A.; Zhao, H. Recent developments in pyridine nucleotide regeneration. Curr. Opin. Biotechnol. 2003, 14, 421–426. [Google Scholar] [CrossRef]
- Mesentsev, A.V.; Lamzin, V.S.; Tishkov, V.I.; Ustinnikova, T.B.; Popov, V.O. Effect of pH on kinetic parameters of NAD+-dependent formate dehydrogenase. Biochem. J. 1997, 321, 475–480. [Google Scholar] [CrossRef]
- Matsuyama, A.; Yamamoto, H.; Kobayashi, Y. Practical application of recombinant whole-cell biocatalysts for the manufacturing of pharmaceutical intermediates such as chiral alcohols. Org. Process. Res. Dev. 2002, 6, 558–561. [Google Scholar] [CrossRef]
- Galkin, A.; Kulakova, L.; Yoshimura, T.; Soda, K.; Esaki, N. Synthesis of optically active amino acids from alpha-keto acids with Escherichia coli cells expressing heterologous genes. Appl. Env. Microbiol. 1997, 63, 4651–4656. [Google Scholar]
- Batie, C.J.; LaHaie, E.; Ballou, D.P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J. Biol. Chem. 1987, 262, 1510–1518. [Google Scholar]
- Zheng, J.X.; Yang, T.W.; Zhou, J.P.; Xu, M.J.; Zhang, X.; Rao, Z.M. Elimination of a Free Cysteine by Creation of a Disulfide Bond Increases the Activity and Stability of Candida boidinii Formate Dehydrogenase. Appl. Environ. Microbiol. 2017, 83, 2. [Google Scholar] [CrossRef]
- Shao, M.; Zhang, X.; Zhiming, R.; Meijuan, X.; Yang, T.; Li, H.; Zhenghong, X.; Shangtian, Y. Efficient testosterone production by engineered Pichia pastoris co-expressing human 17β-hydroxysteroid dehydrogenase type 3 and Saccharomyces cerevisiae glucose 6-phosphate dehydrogenase with NADPH regeneration. Green Chem. 2016, 18, 1774–1784. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Z.; Liu, S.; Qian, K.; Xu, M.; Yang, T.; Xu, J.; Rao, Z. Improving the Production of Salt-Tolerant Glutaminase by Integrating Multiple Copies of Mglu into the Protease and 16S rDNA Genes of Bacillus subtilis 168. Molecules 2019, 24, 592. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.L.; Yang, T.W.; Zhou, J.P.; Zheng, J.X.; Xu, M.J.; Zhang, X.; Rao, Z.M.; Yang, S.T. Development of a multi-enzymatic desymmetrization and its application for the biosynthesis of L-norvaline from DL-norvaline. Process. Biochem. 2017, 55, 104–109. [Google Scholar] [CrossRef]
- Berrios-Rivera, S.J.; Sanchez, A.M.; Bennett, G.N.; San, K.Y. Effect of different levels of NADH availability on metabolite distribution in Escherichia coli fermentation in minimal and complex media. Appl. Microbiol. Biotechnol. 2004, 65, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.L.; Zhang, X.; Rao, Z.M.; Xu, M.J.; Yang, T.W.; Li, H.; Xu, Z.H. Enhanced Production of Androst-1,4-Diene-3,17-Dione by Mycobacterium neoaurum JC-12 Using Three-Stage Fermentation Strategy. PLoS ONE 2015, 10, 9. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
| Strains | Enzymes | Substrate | Activity (U/mL) | Total Protein (mg/mL) | Specific Activity (U/mg protein) |
|---|---|---|---|---|---|
| E. coli BL21 (DE3) | KshB | DCPIP | 0.64 ± 0.03 | 11.71 ± 0.15 | 0.05 ± 0.002 |
| E. coli BL21 (DE3) | FDH | NADH | n.d. | 12.21 ± 0.18 | n.d. |
| E. coli BL21 (DE3) | KSH | AD | n.d. | 2.095 ± 0.07 | n.d. |
| BL21 (DE3)/pET-28a(+)-kshB | KshB | DCPIP | 33.32 ± 0.12 | 11.25 ± 0.23 | 2.96 ± 0.12 |
| BL21 (DE3)/pET-28a(+)-fdh | FDH | NADH | 0.48 ± 0.02 | 12.13 ± 0.12 | 0.04 ± 0.002 |
| BL21(DE3)/pEtDuet-1-kshC | KSH | AD | 0.91 ± 0.06 | 2.07 ± 0.08 | 0.43 ± 0.02 |
| Strains/Plasmids/Primers | Characteristics | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| E. coli BL21(DE3) | F- dcm ompT hsdS (rB- mB-) gal λ(DE3) | Invitrogen |
| BL21 (DE3)/pET-28a(+) | BL21 (DE3) containing pET-28a(+) (KmR) | This study |
| BL21 (DE3)/pET-28a(+)-kshA | BL21 (DE3) containing pET-28a(+)-kshA (KmR) | This study |
| BL21 (DE3)/pET-28a(+)-kshB | BL21 (DE3) containing pET-28a(+)-kshB (KmR) | This study |
| BL21 (DE3)/pET-28a(+)-kshC | BL21 (DE3) containing pET-28a(+)-kshC (KmR) | This study |
| BL21 (DE3)/pET-28a(+)-fdh | BL21 (DE3) containing pET-28a(+)-fdh (KmR) | This study |
| BL21 (DE3)/pEtDuet-1 | BL21 (DE3) containing pEtDuet-1 (AmpR) | This study |
| BL21 (DE3)/pEtDuet-1-kshC | BL21 (DE3) containing pEtDuet-1-kshC (AmpR) | This study |
| Plasmids | ||
| pET-28a(+) | Expression plasmid, KmR, T7 promoter | This lab |
| pET-28a(+)-kshA | pET-28a(+) containing kshA | This study |
| pET-28a(+)-kshB | pET-28a(+) containing ksbB-His | This study |
| pET-28a(+)-kshC | pET-28a(+) containing kshC | This study |
| pET-28a(+)-fdh | pET-28a(+) containing fdh | This lab |
| pEtDuet-1 | Expression plasmid, AmpR, T7 promoter | This lab |
| pEtDuet-1-kshC | pEtDuet-1 containing kshC-His | This study |
| Primers (5′-3′) | ||
| P1 | CGGGATCCATGACGACTGAGCACGCCGG | |
| P2 | CCCAAGCTTTCAGCTTGATTGAGCGGTTTC | |
| P3 | CGGGATCCATGACTGATGAACCGTTAGGTAG | |
| P4 | CCCAAGCTTTCACTCGTCGTAGGTCACCTC | |
| P5 | CGGGATCCGATGGCCGGTCTGAACAACGATAG | |
| P6 | CCCAAGCTTTCAGCCGCTGGCCGGGGCGGCC | |
| P7 | CGGGATCCGATGGCCGGTCTGAACAACGATAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhu, M.; Han, R.; Zhao, Y.; Chen, K.; Qian, K.; Shao, M.; Yang, T.; Xu, M.; Xu, J.; et al. A Novel 3-Phytosterone-9α-Hydroxylase Oxygenation Component and Its Application in Bioconversion of 4-Androstene-3,17-Dione to 9α-Hydroxy-4-Androstene-3,17-Dione Coupling with A NADH Regeneration Formate Dehydrogenase. Molecules 2019, 24, 2534. https://doi.org/10.3390/molecules24142534
Zhang X, Zhu M, Han R, Zhao Y, Chen K, Qian K, Shao M, Yang T, Xu M, Xu J, et al. A Novel 3-Phytosterone-9α-Hydroxylase Oxygenation Component and Its Application in Bioconversion of 4-Androstene-3,17-Dione to 9α-Hydroxy-4-Androstene-3,17-Dione Coupling with A NADH Regeneration Formate Dehydrogenase. Molecules. 2019; 24(14):2534. https://doi.org/10.3390/molecules24142534
Chicago/Turabian StyleZhang, Xian, Manchi Zhu, Rumeng Han, Youxi Zhao, Kewei Chen, Kai Qian, Minglong Shao, Taowei Yang, Meijuan Xu, Jianzhong Xu, and et al. 2019. "A Novel 3-Phytosterone-9α-Hydroxylase Oxygenation Component and Its Application in Bioconversion of 4-Androstene-3,17-Dione to 9α-Hydroxy-4-Androstene-3,17-Dione Coupling with A NADH Regeneration Formate Dehydrogenase" Molecules 24, no. 14: 2534. https://doi.org/10.3390/molecules24142534
APA StyleZhang, X., Zhu, M., Han, R., Zhao, Y., Chen, K., Qian, K., Shao, M., Yang, T., Xu, M., Xu, J., & Rao, Z. (2019). A Novel 3-Phytosterone-9α-Hydroxylase Oxygenation Component and Its Application in Bioconversion of 4-Androstene-3,17-Dione to 9α-Hydroxy-4-Androstene-3,17-Dione Coupling with A NADH Regeneration Formate Dehydrogenase. Molecules, 24(14), 2534. https://doi.org/10.3390/molecules24142534







