Development of Novel antimiRzymes for Targeted Inhibition of miR-21 Expression in Solid Cancer Cells
Abstract
:1. Introduction
2. Results
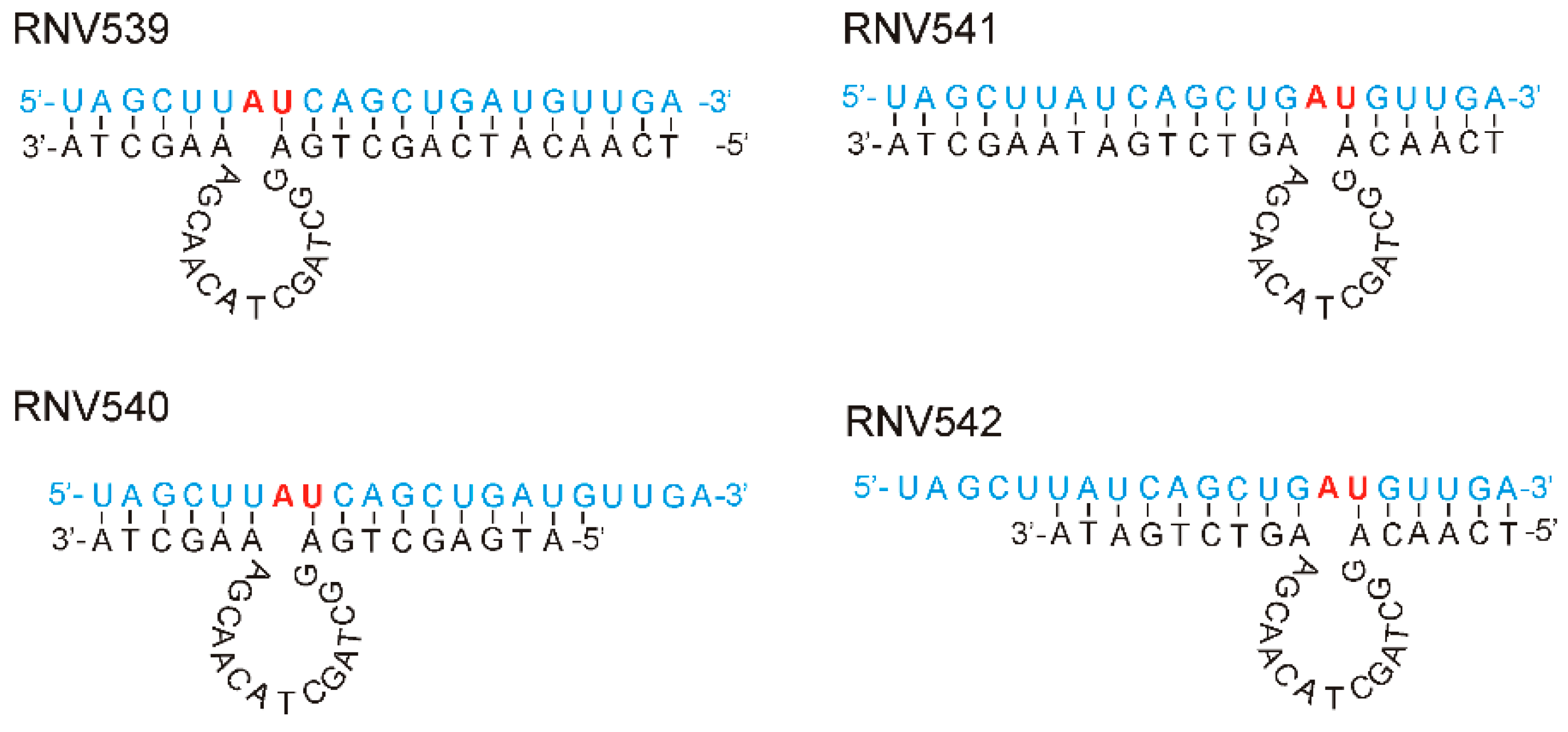
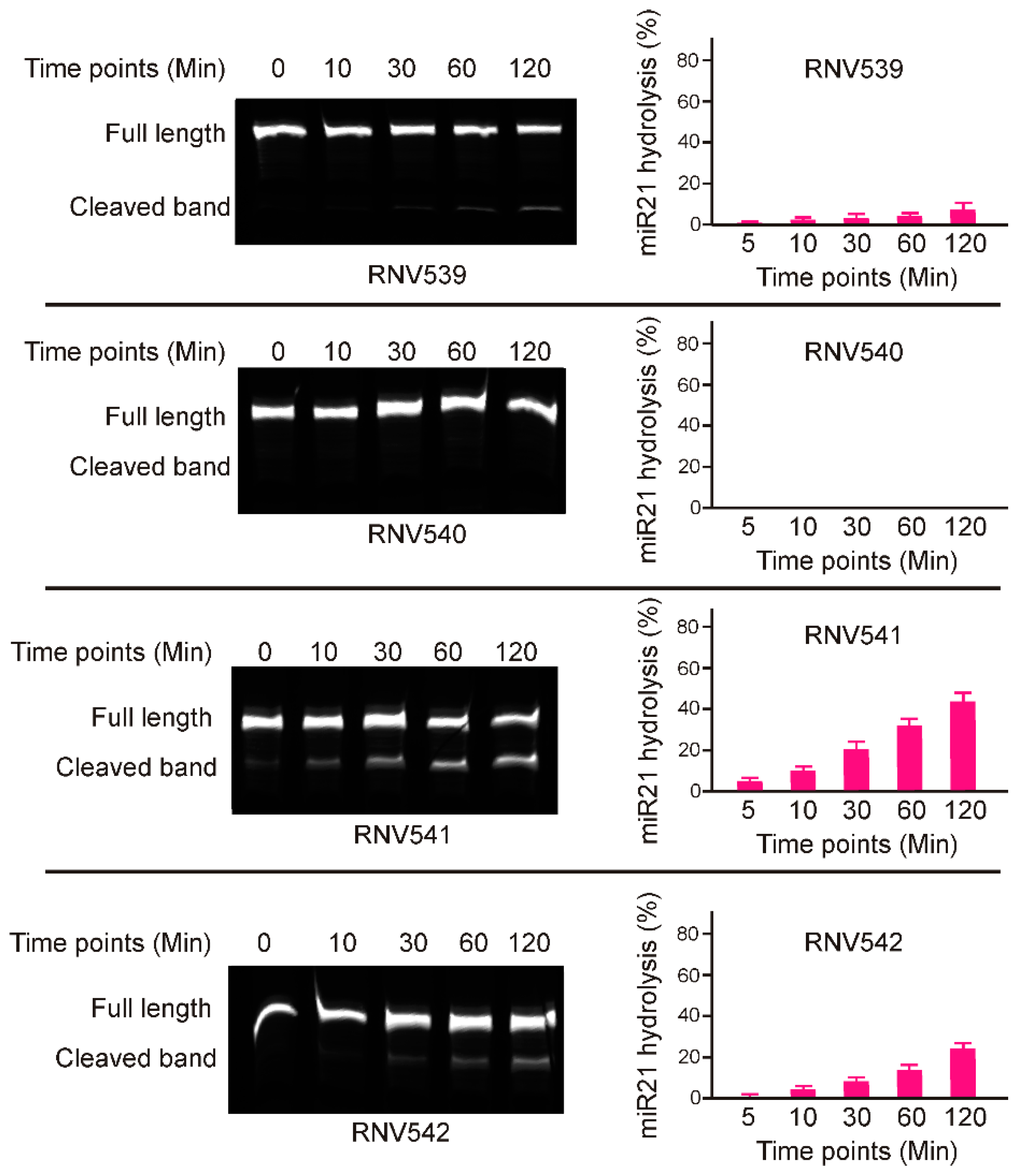
2.1. Rational Design and In Vitro Evaluation of antimiRzymes Targeting miR-21
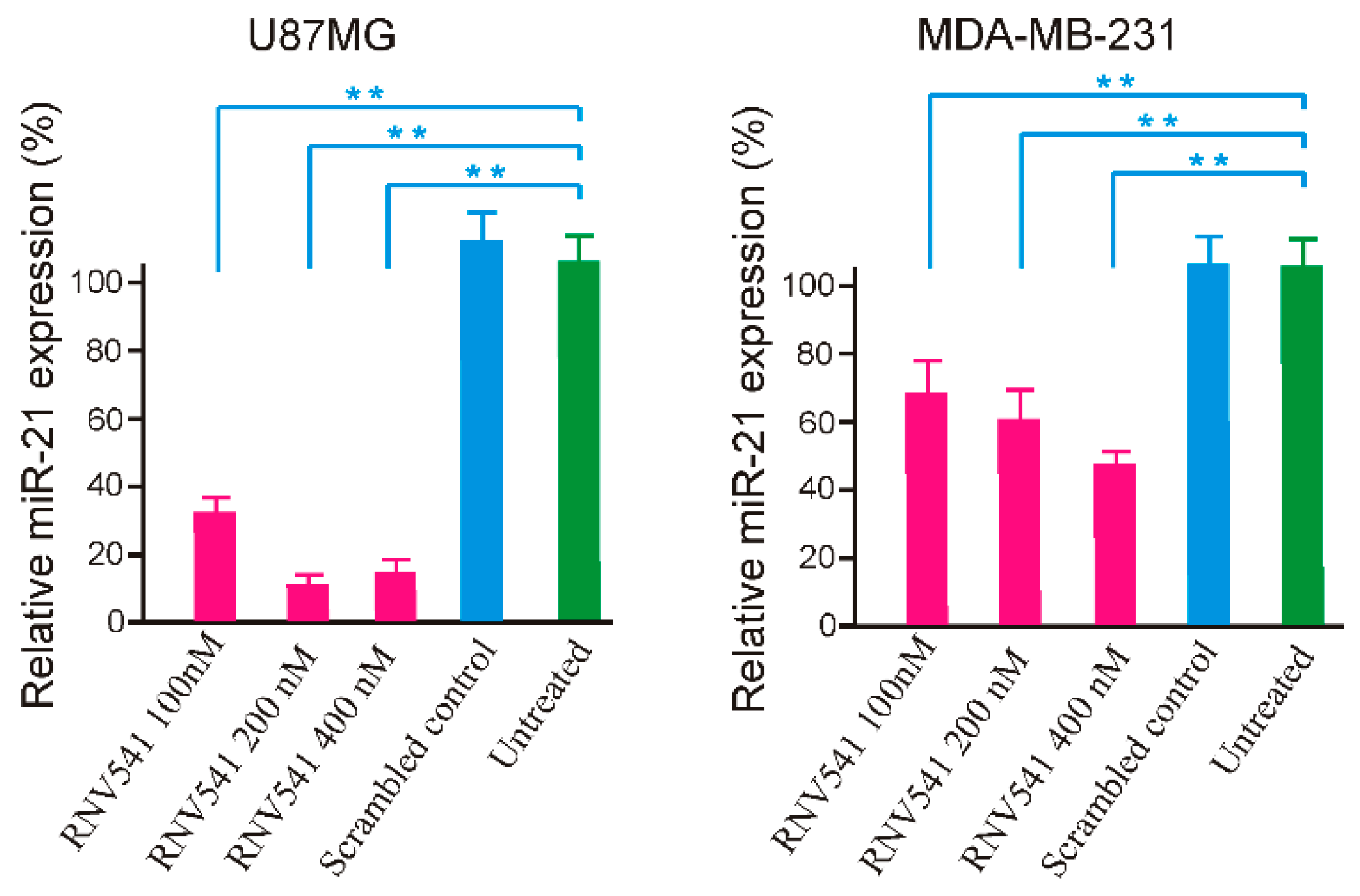
2.2. Evaluation of miR-21 Cleavage Activity of RNV541 antimiRzymes in Cancer Cell Lines
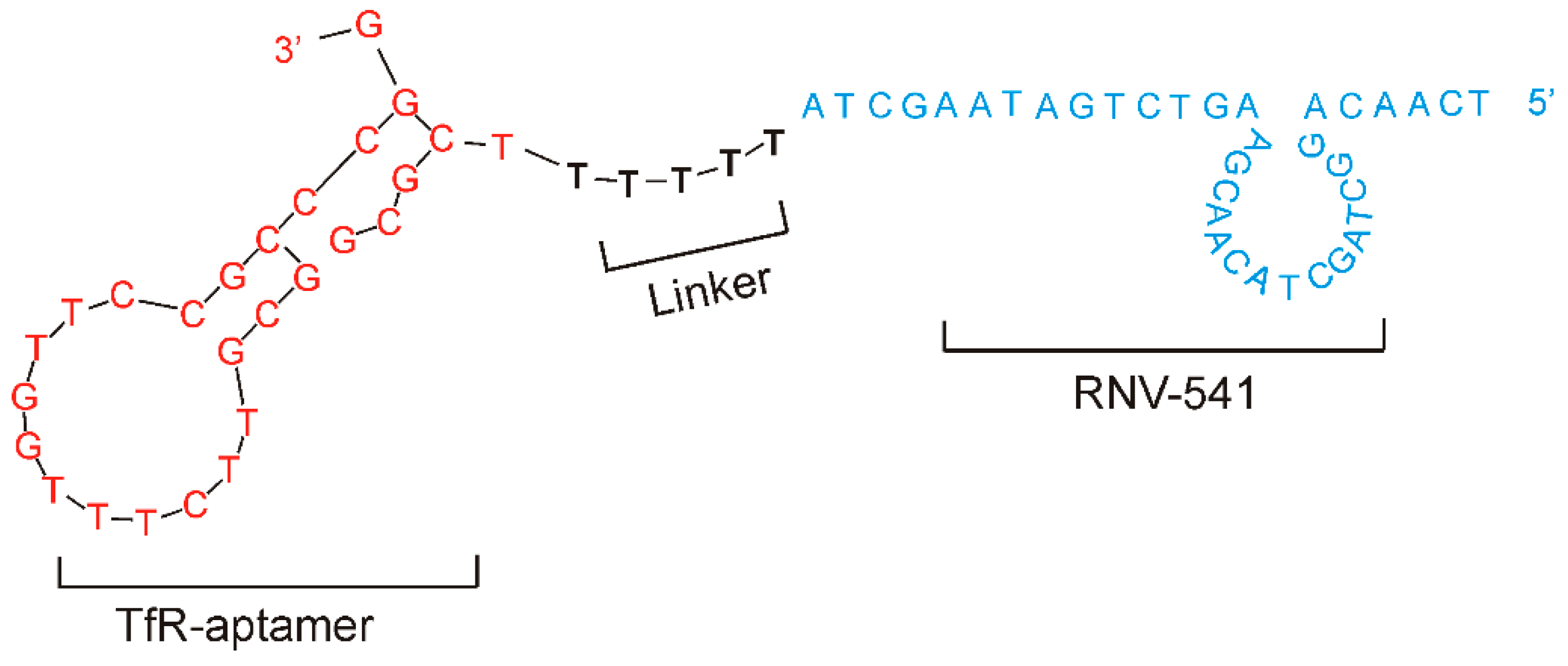
2.3. Evaluation of Transferrin Receptor Aptamer-RNV541 Chimera for Cancer Cell Targeted Inhibition of miR-21
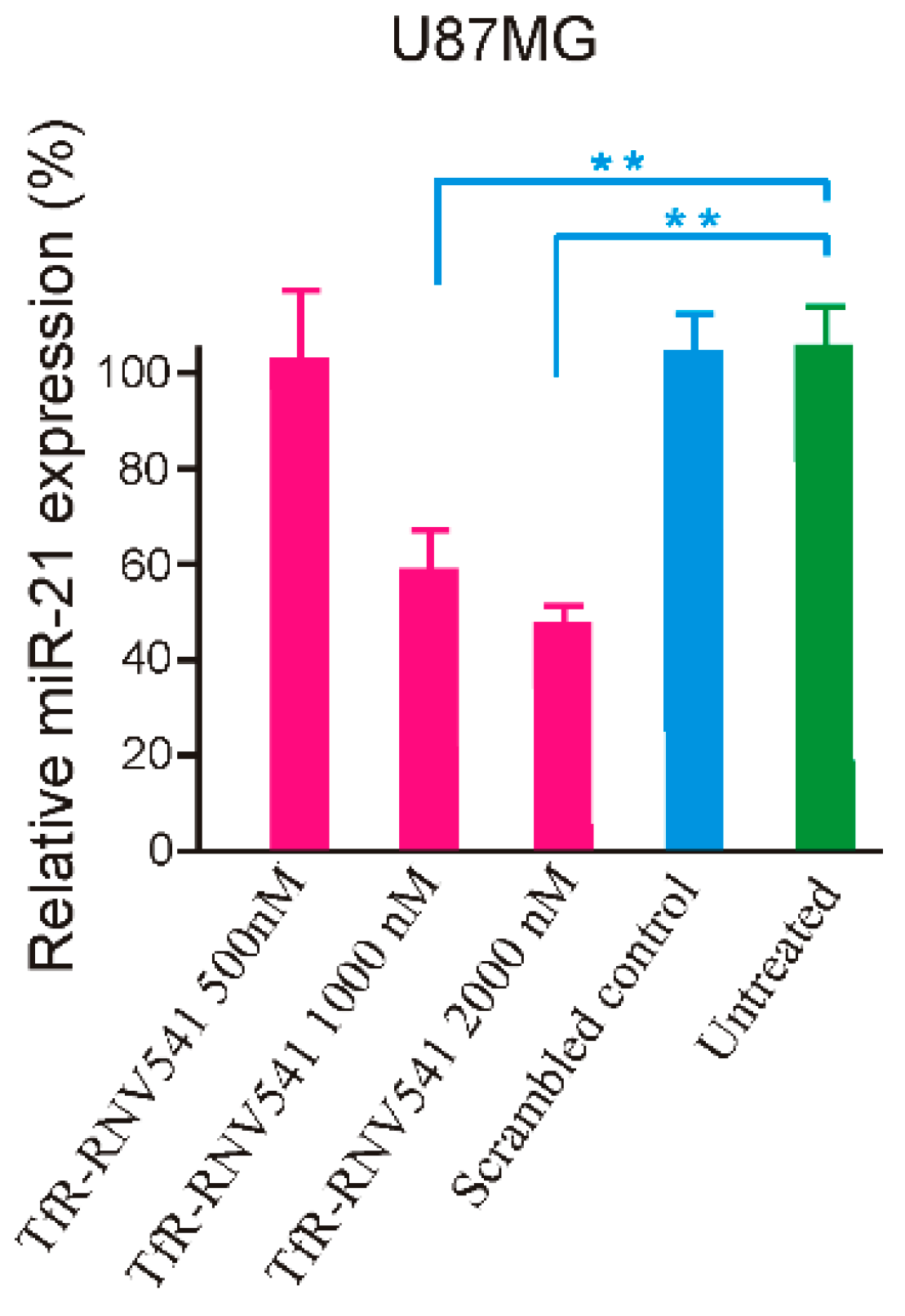
2.4. TfR Aptamer-RNV541 Chimera Effectively Suppressed miR-21 Expression in Glioblastoma Cells
3. Discussion
4. Materials and Methods
4.1. Oligonucleotide Sequences
4.2. Cell Culture and In Vitro Cleavage Assay
4.3. Transfection
4.4. Taqman qPCR to Measure the Expression of miR-21
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Li, W.D.; Lei, F.R.; Li, X.Q. The regulatory role of microRNAs in angiogenesis-related diseases. J. Cell. Mol. Med. 2018, 22, 4568–4587. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Masoudi, M.S.; Mehrabian, E.; Mirzaei, H. MiR-21: A key player in glioblastoma pathogenesis. J. Cell. Biochem. 2018, 119, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, K.; Li, T.; Fang, J.; Ding, Y.; Sun, L.; Tu, T.; Jiang, X.; Du, S.; Hu, J.; et al. miR-21: A gene of dual regulation in breast cancer. Int. J. Oncol. 2016, 48, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Cheng, L.; Cao, D.K.; Zhang, H. Suppression of miR-21 Expression Inhibits Cell Proliferation and Migration of Liver Cancer Cells by Targeting Phosphatase and Tensin Homolog (PTEN). Med. Sci. Monit. 2018, 24, 3571–3577. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Scola, L.; Zanghi, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging 2018, 10, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Lupo, G.; Rosa, G.R.M.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Rapisarda, E.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancer 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCubrey, J.A.; Fitzgerald, T.L.; Yang, L.V.; Lertpiriyapong, K.; Steelman, L.S.; Abrams, S.L.; Montalto, G.; Cervello, M.; Neri, L.M.; Cocco, L.; et al. Roles of GSK-3 and microRNAs on epithelial mesenchymal transition and cancer stem cells. Oncotarget 2017, 8, 14221–14250. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Fan, M.; Paulus, E.; Hosni-Ahmed, A.; Sims, M.; Qayyum, S.; Davidoff, A.M.; Handorf, C.R.; et al. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J. Biol. Chem. 2014, 289, 25079–25087. [Google Scholar] [CrossRef] [PubMed]
- Lakomy, R.; Sana, J.; Hankeova, S.; Fadrus, P.; Kren, L.; Lzicarova, E.; Svoboda, M.; Dolezelova, H.; Smrcka, M.; Vyzula, R.; et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011, 102, 2186–2190. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucl. Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.; Yi, B.; Ma, R.X.; Zhang, X.G.; Zhao, H.Y.; Xi, Y.G. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 2016, 6, 22312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrayarthy, M.; Aung-Htut, M.T.; Le, B.T.; Veedu, R.N. Novel Chemically-modified DNAzyme targeting Integrin alpha-4 RNA transcript as a potential molecule to reduce inflammation in multiple sclerosis. Sci. Rep. 2017, 7, 1613. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Breaking up is easy to do (if you’re a DNA enzyme that cleaves RNA). Chem. Biol. 2004, 11, 7–8. [Google Scholar] [CrossRef]
- Zhang, J.J. RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and In Vivo Applications. Catalysts 2018, 8, 550. [Google Scholar] [CrossRef]
- Belter, A.; Rolle, K.; Piwecka, M.; Fedoruk-Wyszomirska, A.; Naskret-Barciszewska, M.Z.; Barciszewski, J. Inhibition of miR-21 in glioma cells using catalytic nucleic acids. Sci. Rep. 2016, 6, 24516. [Google Scholar] [CrossRef]
- Raz, M.H.; Hollenstein, M. Probing the effect of minor groove interactions on the catalytic efficiency of DNAzymes 8–17 and 10–23. Mol. Biosyst. 2015, 11, 1454–1461. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarthy, M.; AlShamaileh, H.; Huang, H.; Tannenberg, R.K.; Chen, S.; Worrall, S.; Dodd, P.R.; Veedu, R.N. Development of DNA aptamers targeting low-molecular-weight amyloid-beta peptide aggregates in vitro. Chem. Commun. 2018, 54, 4593–4596. [Google Scholar] [CrossRef] [PubMed]
- Hughes, Q.W.; Le, B.T.; Gilmore, G.; Baker, R.I.; Veedu, R.N. Construction of a Bivalent Thrombin Binding Aptamer and Its Antidote with Improved Properties. Molecules 2017, 22, 1770. [Google Scholar] [CrossRef] [PubMed]
- Lipi, F.; Chen, S.X.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.X.; Shigdar, S.; Bean, A.G.; Bruce, M.; Yang, W.R.; Mathesh, M.; Wang, T.; Yin, W.; Tran, P.H.L.; Al Shamaileh, H.; et al. Transforming doxorubicin into a cancer stem cell killer via EpCAM aptamer-mediated delivery. Theranostics 2017, 7, 4071–4086. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gantier, M.P.; Xiang, D.; Bean, A.G.; Bruce, M.; Zhou, S.F.; Khasraw, M.; Ward, A.; Wang, L.; Wei, M.Q.; et al. EpCAM Aptamer-mediated Survivin Silencing Sensitized Cancer Stem Cells to Doxorubicin in a Breast Cancer Model. Theranostics 2015, 5, 1456–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AlShamaileh, H.; Wang, T.; Xiang, D.X.; Yin, W.; Tran, P.H.L.; Barrero, R.A.; Zhang, P.Z.; Li, Y.; Kong, L.X.; Liu, K.; et al. Aptamer-mediated survivin RNAi enables 5-fluorouracil to eliminate colorectal cancer stem cells. Sci. Rep. 2017, 7, 5898. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Dellamaggiore, K.R.; Ouellette, C.P.; Sedano, C.D.; Lizadjohry, M.; Chernis, G.A.; Gonzales, M.; Baltasar, F.E.; Fan, A.L.; Myerowitz, R.; et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 15908–15913. [Google Scholar] [CrossRef] [Green Version]
- Trowbridge, I.S.; Omary, M.B. Human Cell-Surface Glycoprotein Related to Cell-Proliferation Is the Receptor for Transferrin. Proc. Natl. Acad. Sci. USA 1981, 78, 3039–3043. [Google Scholar] [CrossRef]
- Daniels, T.R.; Delgado, T.; Rodriguez, J.A.; Helguera, G.; Penichet, M.L. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin. Immunol. 2006, 121, 144–158. [Google Scholar] [CrossRef]
- Daniels, T.R.; Bernabeu, E.; Rodriguez, J.A.; Patel, S.; Kozman, M.; Chiappetta, D.A.; Holler, E.; Ljubimova, J.Y.; Helguera, G.; Penichet, M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. BBA Gen. Subj. 2012, 1820, 291–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pofahl, M.; Wengel, J.; Mayer, G. Multifunctional nucleic acids for tumor cell treatment. Nucleic Acid. Ther. 2014, 24, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front Pharm. 2018, 9, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Shigdar, S.; Gantier, M.P.; Hou, Y.C.; Wang, L.; Li, Y.; Al Shamaileh, H.; Yin, W.; Zhou, S.F.; Zhao, X.; et al. Cancer stem cell targeted therapy: Progress amid controversies. Oncotarget 2015, 6, 44191–44206. [Google Scholar] [CrossRef] [PubMed]
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell Physiol. 2019, 234, 12369–12384. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef]
- Javanmardi, S.; Aghamaali, M.R.; Abolmaali, S.S.; Mohammadi, S.; Tamaddon, A.M. miR-21, An Oncogenic Target miRNA for Cancer Therapy: Molecular Mechanisms and Recent Advancements in Chemo and Radio-resistance. Curr. Gene 2016, 16, 375–389. [Google Scholar] [CrossRef]
- Zhou, W.H.; Ding, J.S.; Liu, J.W. Theranostic DNAzymes. Theranostics 2017, 7, 1010–1025. [Google Scholar] [CrossRef]
- Wang, T.; Yin, W.; AlShamaileh, H.; Zhang, Y.M.; Tran, P.H.L.; Nguyen, T.N.G.; Li, Y.; Chen, K.S.; Sun, M.M.; Hou, Y.C.; et al. A Detailed Protein-SELEX Protocol Allowing Visual Assessments of Individual Steps for a High Success Rate. Hum. Gene Ther. Methods 2019, 30, 1–16. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.Y.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Henri, J.; Goodman, L.; Xiang, D.; Duan, W.; Shigdar, S. Development of a Bifunctional Aptamer Targeting the Transferrin Receptor and Epithelial Cell Adhesion Molecule (EpCAM) for the Treatment of Brain Cancer Metastases. Acs Chem. Neurosci. 2017, 8, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Zhang, Y.; Kenrick, M.; Hoyte, K.; Luk, W.; Lu, Y.M.; Atwal, J.; Elliott, J.M.; Prabhu, S.; Watts, R.J.; et al. Boosting Brain Uptake of a Therapeutic Antibody by Reducing Its Affinity for a Transcytosis Target. Sci. Transl. Med. 2011, 3, 84ra44. [Google Scholar] [CrossRef] [PubMed]
- Aaldering, L.J.; Tayeb, H.; Krishnan, S.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Smart functional nucleic acid chimeras: Enabling tissue specific RNA targeting therapy. RNA Biol. 2015, 12, 412–425. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larcher, L.M.; Wang, T.; Veedu, R.N. Development of Novel antimiRzymes for Targeted Inhibition of miR-21 Expression in Solid Cancer Cells. Molecules 2019, 24, 2489. https://doi.org/10.3390/molecules24132489
Larcher LM, Wang T, Veedu RN. Development of Novel antimiRzymes for Targeted Inhibition of miR-21 Expression in Solid Cancer Cells. Molecules. 2019; 24(13):2489. https://doi.org/10.3390/molecules24132489
Chicago/Turabian StyleLarcher, Leon M., Tao Wang, and Rakesh N. Veedu. 2019. "Development of Novel antimiRzymes for Targeted Inhibition of miR-21 Expression in Solid Cancer Cells" Molecules 24, no. 13: 2489. https://doi.org/10.3390/molecules24132489
APA StyleLarcher, L. M., Wang, T., & Veedu, R. N. (2019). Development of Novel antimiRzymes for Targeted Inhibition of miR-21 Expression in Solid Cancer Cells. Molecules, 24(13), 2489. https://doi.org/10.3390/molecules24132489



