Targeting Bacterial Biofilms by the Green Tea Polyphenol EGCG
Abstract
1. Introduction
2. Bacterial Biofilms
3. EGCG Can Interfere with Bacterial Biofilm Formation
4. Mechanisms of Action of EGCG on Bacterial Biofilms
5. Can EGCG Act Synergistically with Antibiotics on Bacterial Biofilms?
6. Why do Plants Produce Antibiofilm Agents such as EGCG?
7. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.G.; O’Toole, G.A. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 87–107. [Google Scholar]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Kjelleberg, S.; Normark, S.; Nyman, L.; Uhlin, B.E.; Åkerlund, B. Microbial biofilm formation: A need to act. J. Intern. Med. 2014, 276, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.J.; Melander, C. Controlling bacterial biofilms. Chembiochem 2009, 10, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Prospects for anti-biofilm pharmaceuticals. Pharmaceuticals 2015, 8, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Rabin, N.; Zheng, Y.; Opoku-Tememg, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Agents that inhibit bacterial biofilm formation. Future Med. Chem. 2015, 7, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Sintim, H.O.; Smith, J.A.; Wang, J.; Nakayama, S.; Yan, L. Pradigm shift in discovering next-generation anti-infective agents: Targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Future Med. Chem. 2010, 2, 1005–1035. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Givskov, M. Chemical biology strategies for biofilm control. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Peach, K.C.; Bray, W.M.; Shikuma, N.J.; Gassner, N.C.; Lokey, R.S.; Yildiz, F.H.; Linington, R.G. An image-based 384-well high-throughput screening method for the discovery of biofilm inhibitors in Vibrio cholerae. Mol. Biosyst. 2011, 7, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Budzynska, A.; Wieckowska-Szakiel, M.; Sadowska, B.; Kalemba, D.; Rozalska, B. Antibiofilm activity of selected plant essential oils and their major components. Polish. J. Microbiol. 2011, 60, 35–41. [Google Scholar]
- Stowe, S.D.; Richards, J.J.; Tucker, A.T.; Thompson, R.; Melander, C.; Cavanagh, J. Anti-biofilm compounds derived from marine sponges. Mar. Drugs 2011, 9, 2010–2035. [Google Scholar] [CrossRef]
- Khan, M.S.; Lee, J. Novel strategies for combating pathogenic biofilms using plant products and microbial antibiotics. Curr. Pharm. Biotechnol. 2015, 17, 126–140. [Google Scholar] [CrossRef]
- Nunes Silva, L.; Zimmer, K.R.; Macedo, A.J.; Silva Trentin, D. Plant natural products targeting bacterial virulence factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef]
- Mereles, D.; Hunstein, W. Epigallocatechin-3-gallate (EGCG) for clinical trials: More pitfalls than promises? Int. J. Mol. Sci. 2011, 12, 5592–5603. [Google Scholar] [CrossRef]
- Branda, S.S.; Vik, A.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Römling, U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 2005, 62, 1234–1246. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Richter, A.M.; Klauck, G.; Mika, F.; Hengge, R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio 2013, 4, e00103-13. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef] [PubMed]
- Okegbe, C.; Price-Whelan, A.; Dietrich, L.E. Redox-driven regulation of microbial community morphogenesis. Curr. Opin. Microbiol. 2014, 18, 39–45. [Google Scholar] [CrossRef]
- Vidakovic, L.; Singh, P.K.; Hartmann, R.; Nadell, C.D.; Drescher, K. Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat. Microbiol. 2018, 3, 26–31. [Google Scholar] [CrossRef]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Römling, U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef]
- Serra, D.O.; Hengge, R. Cellulose in bacterial biofilms. In Extracellular Sugar-Based Biopolymer Matrices; Cohen, E., Merzendorfer, H., Eds.; Springer International Publishing: Basel, Switzerland, 2019. [Google Scholar]
- Thongsomboon, W.; Serra, D.O.; Possling, A.; Hadjineophytou, C.; Hengge, R.; Cegelski, L. Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 2018, 359, 334–338. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef]
- Yildiz, F.H.; Schoolnick, G.K. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 1999, 96, 4028–4033. [Google Scholar] [CrossRef]
- Sutherland, I.W. Structural studies on colanic acid, the common exopolysaccharide found in the Enterobacteriaceae, by partial acid hydrolysis. Biochem. J. 1970, 115, 935–945. [Google Scholar] [CrossRef]
- Mann, E.E.; Wozniak, C.E. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012, 36, 893–916. [Google Scholar] [CrossRef] [PubMed]
- Whitney, J.C.; Howell, P.L. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013, 21, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.; Nielsen, J.L.; Dueholm, M.S.; Wetzel, R.; Otzen, D.; Nielsen, P.H. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007, 9, 3077–3090. [Google Scholar] [CrossRef] [PubMed]
- Blanco, L.P.; Evans, M.L.; Smith, D.R.; Badtke, M.P.; Chapman, M.R. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012, 20, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid structures as biofilm matrix scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Erskine, E.; MacPhee, C.E.; Stanley-Wall, N.R. Functional amyloid and other protein fibers in the biofilm matrix. J. Mol. Biol. 2018, 430, 3642–3656. [Google Scholar] [CrossRef]
- Van Gerven, N.; Van der Verren, S.E.; Reiter, D.M.; Remaut, H. The role of functional amyloids in bacterial virulence. J. Mol. Biol. 2018, 430, 3657–3684. [Google Scholar] [CrossRef]
- Evans, M.L.; Chapman, M. Curli biogenesis: Order out of disorder. Biochim. Biophys. Acta 2014, 1843, 1551–1558. [Google Scholar] [CrossRef]
- Fändrich, M. Oligomeric intermediates in amyloid formation: Structure determination and mechanisms of toxicity. J. Mol. Biol. 2012, 421, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Nielsen, S.B.; Hein, K.L.; Nissen, P.; Chapman, M.; Christiansen, G.; Nielsen, P.H.; Otzen, D.E. Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 2011, 50, 8281–8290. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, M.; Evans, M.L.; Chapman, M.R. Amyloid by design: Intrinsic regulation of microbial amyloid assembly. J. Mol. Biol. 2018, 430, 3631–3641. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Rohde, M.; Olsén, A.; Normark, S.; Reinköster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000, 36, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Brombacher, E.; Dorel, C.; Zehnder, A.J.B.; Landini, P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 2003, 149, 2847–2857. [Google Scholar] [CrossRef] [PubMed]
- Bokranz, W.; Wang, X.; Tschape, H.; Römling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.M.; Povolotsky, T.L.; Wieler, L.H.; Hengge, R. C-di-GMP signaling and biofilm-related properties of the Shiga toxin-producing German outbreak Escherichia coli O104:H4. EMBO Mol. Med. 2014, 6, 1622–1637. [Google Scholar] [CrossRef] [PubMed]
- Tükel, C.; Nishimori, J.H.; Wilson, R.P.; Winter, M.G.; Keestra, A.M.; van Putten, J.P.; Bäumler, A.J. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol. 2010, 12, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Gallo, P.M.; Rapsinski, G.J.; Wilson, R.P.; Oppong, G.O.; Sriram, U.; Goulian, M.; Buttaro, B.; Caricchio, R.; Gallucci, S.; Tükel, C. Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 2015, 42, 1171–1184. [Google Scholar] [CrossRef]
- Spaulding, C.N.; Dodson, K.W.; Chapman, M.R.; Hultgren, S.J. Fueling the fire with fibres: Bacterial amyloids promote inflammatory disorders. Cell Host Microbe 2015, 18, 1–2. [Google Scholar] [CrossRef][Green Version]
- Rouse, S.L.; Matthews, S.J.; Dueholm, M.S. Ecology and biogenesis of functional amyloids in Pseudomonas. J. Mol. Biol. 2018, 430, 3685–3695. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid fibres provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef] [PubMed]
- Diehl, A.; Roske, Y.; Ball, L.; Chowdhury, A.; Hiller, M.; Molière, N.; Kramer, R.; Stöppler, D.; Worth, C.L.; Schlegel, B.; et al. Structural changes of TasA in biofilms formation of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2018, 115, 3237–3242. [Google Scholar] [CrossRef] [PubMed]
- Costalonga, M.; Herzberg, M.C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 2014, 162, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Hengge, R. Stress responses go three-dimensional—The spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 2014, 16, 1455–1471. [Google Scholar] [CrossRef] [PubMed]
- Bravo, L. Polyphenols: Chemistry, dietary sources, metabolism, and nutritional significance. Nutr. Rev. 1998, 56, 317–333. [Google Scholar] [CrossRef]
- Steinmann, J.; Pietschmann, T.; Steinmann, E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 2013, 168, 1059–1073. [Google Scholar] [CrossRef]
- Reygaert, W.C. Green tea catechins: Their use in treating and preventing infectious diseases. BioMed Res. Int. 2018, 9105261. [Google Scholar] [CrossRef]
- Hamilton-Miller, J.M.T. Anti-cariogenic properties of tea (Camellia sinensis). J. Med. Microbiol. 2001, 50, 299–302. [Google Scholar] [CrossRef]
- Taylor, P.W.; Hamilton-Miller, J.M.T.; Stapleton, P.D. Antimicrobial properties of green tea catechins. Food Sci. Technol. Bull. 2005, 2, 71–81. [Google Scholar] [CrossRef]
- Huber, B.; Eberl, L.; Feucht, W.; Polster, J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z. Naturforsch. 2003, 58, 879–884. [Google Scholar] [CrossRef]
- Sudano Roccaro, A.; Blanco, A.R.; Giuliano, F.; Rusciano, D.; Enea, V. Epigallocatechin-gallate enhances the activity of tetracycline in staphylococci by inhibiting its efflux from bacterial cells. Antimicrob. Agents Chemother. 2004, 48, 1968–1973. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.R.; Sudano-Roccaro, A.; Spoto, G.C.; Nostro, A.; Rusciano, D. Epigallocatechin gallate inhibits biofilm formation by ocular staphylococcal isolates. Antimicrob. Agents Chemother. 2005, 49, 4339–4343. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Kim, W.S.; Lim, J.; Nam, S.; Youn, M.; Nam, S.W.; Kim, Y.; Kim, S.H.; Park, W.; Park, S. Antipathogenic properties of green tea polyphenol epigallocatechin gallate at concentration below the MIC against enterohemorrhagic Escherichia coli O157:H7. J. Food Prot. 2009, 72, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Y.; Sternberg, C.; Molin, S. Evaluation of enoyl-acyl carrier protein reductase inhibitors as Pseudomonas aeruginosa quorum-quenching reagents. Molecules 2010, 15, 780–792. [Google Scholar] [CrossRef]
- Asahi, Y.; Noiri, Y.; Miura, J.; Maezono, H.; Yamaguchi, M.; Yamamoto, R.; Azakami, H.; Hayashi, M.; Ebisu, S. Effects of the tea catechin epigallocatechin gallate on Porphyromonas gingivalis biofilms. J. Appl. Microbiol. 2014, 116, 1164–1171. [Google Scholar] [CrossRef]
- Fournier-Larente, J.; Morin, M.P.; Grenier, D. Green tea catechins potentiate the effect of antibiotics and modulate adherence and gene expression in Porphyromonas gingivalis. Arch. Oral Biol. 2016, 65, 35–43. [Google Scholar] [CrossRef]
- Vidigal, P.G.; Müsken, M.; Becker, K.A.; Häussler, S.; Wingender, J.; Steinmann, E.; Kehrmann, J.; Gulbins, E.; Buer, J.; Rath, P.M.; et al. Effects of green tea compound epigallocatechin-3-gallate against Stenotrophomonas maltophilia infection and biofilm. PLoS ONE 2014, 9, e92876. [Google Scholar] [CrossRef]
- Lee, P.; Tan, K.S. Effects of epigallocatechin gallate against Enterococcus faecalis biofilm and virulence. Arch. Oral Biol. 2015, 60, 393–399. [Google Scholar] [CrossRef]
- Castillo, S.; Heredia, N.; García, S. 2(5H)-Furanone, epigallocatechin gallate, and a citric-based disinfectant disturb quorum-sensing activity and reduce motility and biofilm formation of Campylobacter jejuni. Folia Microbiol. 2015, 60, 89–95. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, X.; Zhang, F.; Feng, L.; Li, J. Inhibition of quorum sensing, biofilm, and spoilage potential in Shewanella baltica by green tea polyphenols. J. Microbiol. 2015, 53, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Teng, Z.; Li, M.; Niu, X.; Wang, J.; Deng, X. Epigallocatechin gallate inhibits Streptococcus pneumoniae virulence by simultaneously targeting pneumolysin and sortase A. J. Cell. Mol. Med. 2017, 21, 2586–2598. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Takagi, S.; Ando, T.; Yoneyama, H.; Ito, K.; Mizugai, H.; Isogai, E. Antimicrobial activity of tea catechin against canine oral bacteria and the functional mechanisms. J. Vet. Med. Sci. 2016, 78, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Ben Lagha, A.; Haas, B.J.; Grenier, D. Tea polyphenols inhibit the growth and virulence properties of Fusobacterium nucleatum. Sci. Rep. 2017, 7, 44815. [Google Scholar] [CrossRef] [PubMed]
- Sugita-Konishi, Y.; Hara-Kudo, Y.; Amano, F.; Okubo, T.; Aoi, N.; Iwaki, M.; Kumagai, S. Epigallocatechin gallate and gallocatechin gallate in green tea catechins inhibit extracellular release of Vero toxin from enterohemorrhagic Escherichia coli O157:H7. Biochem. Biophys. Acta 1999, 1472, 42–50. [Google Scholar] [CrossRef]
- Hisano, M.; Yamaguchi, K.; Inoue, Y.; Ikeda, Y.; Iijima, M.; Adachi, M.; Shimamura, T. Inhibitory effect of catechin against the superantigen staphylococcal enterotoxin B (SEB). Arch. Dermatol. Res. 2003, 295, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhou, X.D.; Wu, C.D. The tea catechin epigallocatechin gallate suppresses cariogenic virulence factors of Streptococcus mutans. Antimicrob. Agents Chemother. 2011, 55, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhou, X.D.; Wu, C.D. Tea catechin epigallocatechin gallate inhibits Streptococcus mutans biofilm formation by suppressing gtf genes. Arch. Oral Biol. 2012, 57, 678–683. [Google Scholar] [CrossRef]
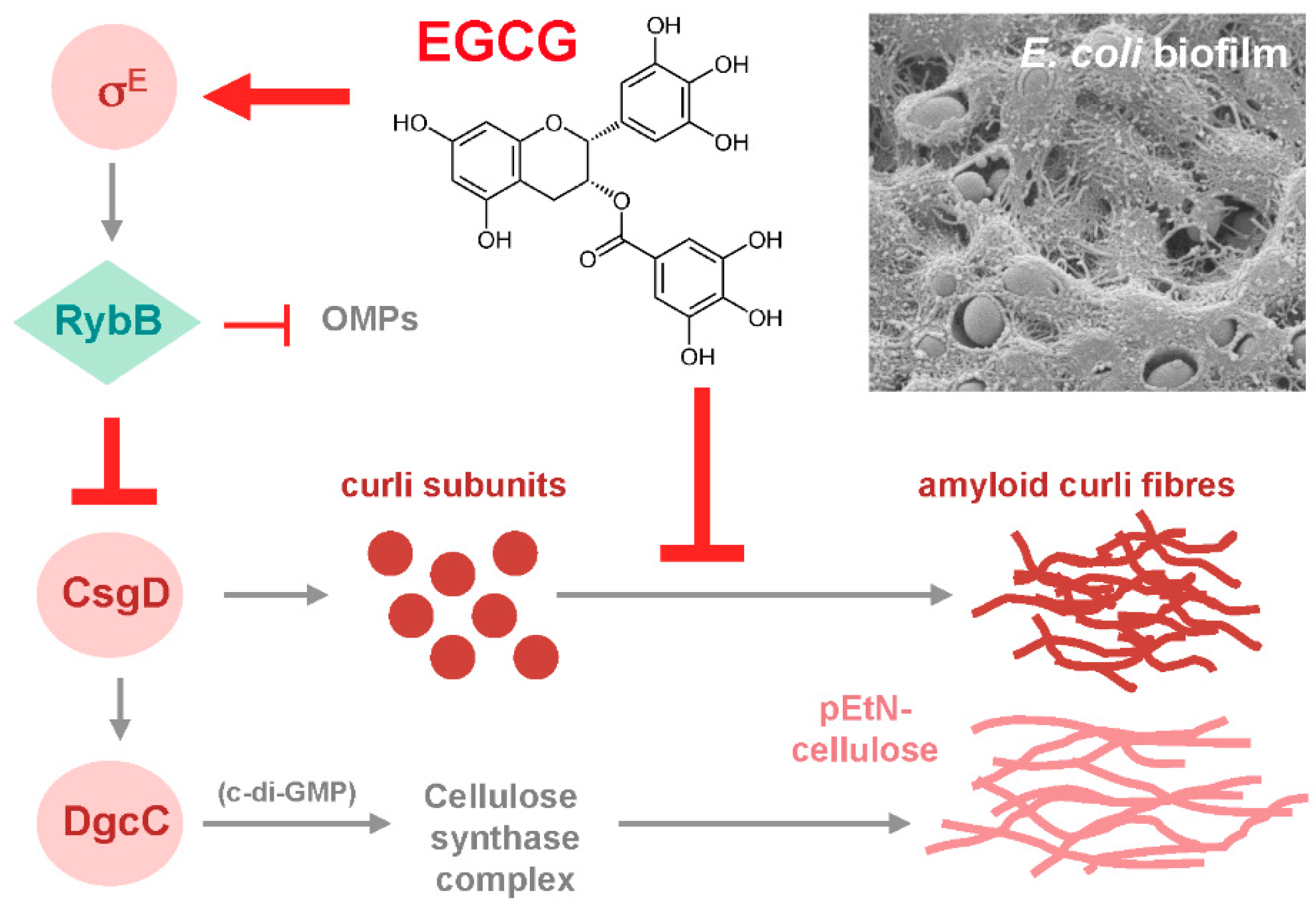
- Serra, D.O.; Mika, F.; Richter, A.M.; Hengge, R. The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and down-regulating the biofilm regulator CsgD via the σE-dependent sRNA RybB. Mol. Microbiol. 2016, 101, 136–151. [Google Scholar] [CrossRef]
- Serra, D.O.; Hengge, R. Experimental detection and visualization of the extracellular matrix in macrocolony biofilms. In c-di-GMP Signaling: Methods & Protocols—Methods in Molecular Biology; Sauer, K., Ed.; Humana Press, Springer Nature: New York, NY, USA, 2017; pp. 133–145. [Google Scholar]
- Römling, U.; Amikam, D. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 2006, 9, 218–228. [Google Scholar] [CrossRef]
- Jenal, U.; Malone, J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006, 40, 385–407. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. Principles of cyclic-di-GMP signaling. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic-di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, T. C-di-GMP synthesis: Structural aspects of evolution, catalysis and regulation. J. Mol. Biol. 2016, 428, 3683–3701. [Google Scholar] [CrossRef] [PubMed]
- Ehrnhoefer, D.E.; Bieschke, J.; Boeddrich, A.; Herbst, M.; Masino, L.; Lurz, R.; Engemann, S.; Pastore, A.; Wanker, E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Bieschke, J.; Russ, J.; Friedrich, R.P.; Ehrnhoefer, D.E.; Wobst, H.; Neugebauer, K.; Wanker, E.E. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. USA 2010, 107, 7710–7715. [Google Scholar] [CrossRef] [PubMed]
- Lopez del Amo, J.M.; Fink, U.; Dasari, M.; Grelle, G.; Wanker, E.E.; Bieschke, J.; Reif, B. Structural properties of EGCG-induced, nontoxic Alzheimer’s Disease Aß oligomers. J. Mol. Biol. 2012, 421, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Epigallocatechin-3-gallate as a potential therapeutic drug for TTR-related amyloidosis: “in vivo” evidence from FAP mice models. PLoS ONE 2012, 7, e29933. [Google Scholar] [CrossRef] [PubMed]
- Sirk, T.W.; Brown, E.F.; Sum, A.K.; Friedman, M. Molecular dynamics study on the biophysical interactions of seven green tea catechins with lipid bilayers of cell membranes. J. Agric. Food Chem. 2008, 56, 7750–7758. [Google Scholar] [CrossRef]
- Kajiya, K.; Kumazawa, S.; Naito, A.; Nakayama, T. Solid-state NMR analysis of the orientation and dynamics of epigallocatechin gallate, a green tea polyphenol, incorporated into lipid bilayers. Magn. Reson. Chem. 2008, 46, 174–177. [Google Scholar] [CrossRef]
- Cui, Y.; Oh, Y.J.; Lim, J.; Youn, M.; Lee, I.; Pak, H.K.; Park, W.; Jo, W.; Park, S. AFM study of the differential inhibitory effects of the green tea polyphenol (-)-epigallocatechin-3-gallate (EGCG) against Gram-positive and Gram-negative bacteri. Food Microbiol. 2012, 29, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Papenfort, K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006, 9, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Grabowicz, M.; Silhavy, T.J. Envelope stress responses: An interconnected safety net. Trends Biochem. Sci. 2017, 42, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Wagner, E.G.H. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, K.S.; Gottesman, S. Small regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.H.; Hu, Z.Q.; Hara, Y.; Shimamura, T. Inhibition of epigallocatechin gallate (EGCg) of conjugative R plasmid transfer in Escherichia coli. J. Infect. Chemother. 2001, 7, 195–197. [Google Scholar] [CrossRef]
- Stenvang, M.; Dueholm, M.S.; Vad, B.S.; Sevlour, T.; Zeng, G.; Gelfman-Shochat, S.; Søndergaard, M.T.; Christiansen, G.; Meyer, R.L.; Kjelleberg, S.; et al. Epigallocatechin gallate remodels overexpressed functional amyloids in Pseudomonas aeruginosa and increases biofilm susceptibility to antibiotic treatment. J. Biol. Chem. 2016, 291, 26540–26553. [Google Scholar] [CrossRef]
- Turner, K.H.; Everett, J.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014, 10, e1004518. [Google Scholar] [CrossRef]
- Marinelli, P.; Pallares, I.; Navarro, S.; Ventura, S. Dissecting the contribution of Staphylococcus aureus a-phenol-soluble modulins to biofilm amyloid structure. Sci. Rep. 2016, 6, 34552. [Google Scholar] [CrossRef] [PubMed]
- Kirketerp-Møller, K.; Jensen, P.Ø.; Fazli, M.; Madsen, K.G.; Pedersen, J.; Moser, C.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M.; Bjarnsholt, T. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 2008, 46, 2717–2722. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Grande, R.; Butrico, L.; Rossi, A.; Settimio, U.F.; Caroleo, B.; Amato, B.; Gallelli, L.; de Franciscis, S. Chronic wound infections: The role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev. Anti Infect. Ther. 2015, 13, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 2009, 75, 4093–4100. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Rojo-Molinero, E.; Macià, M.D.; Oliver, A. Antibiotic treatment of biofilm infections. Acta Pathol. Microbiol. Immunol. Scand. 2017, 125, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Mah, T.F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef]
- Kai-Larsen, Y.; Lüthje, P.; Chromek, M.; Peters, V.; Wang, X.; Holm, A.; Kádas, L.; Hedlund, K.O.; Johansson, J.; Chapman, M.R.; et al. Coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010, 6, e1001010. [Google Scholar] [CrossRef]
- Hobley, L.; Harkins, C.; MacPhee, C.E.; Stanley-Wall, N.R. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 2015, 39, 649–669. [Google Scholar] [CrossRef]
- Peterson, B.W.; He, Y.-Q.; Ren, Y.; Zerdoum, A.; Libera, M.R.; Sharma, P.K.; van Winkelhoff, A.-J.; Neut, D.; Stoodley, P.; van der Mei, H.C.; et al. Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol. Rev. 2015, 39, 234–245. [Google Scholar] [CrossRef]
- Walters, M.C.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef]
- Hengge, R. The general stress response in Gram-negative bacteria. In Bacterial Stress Responses; Storz, G., Hengge, R., Eds.; ASM Press: Washington, DC, USA, 2011; pp. 251–289. [Google Scholar]
- Nguyen, D.; Joshi-Datar, A.; Lepine, F.; Bauerle, E.; Olakanmi, O.; Beer, K.; McKay, G.; Siehnel, R.; Schafhauser, J.; Wang, Y.; et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011, 334, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mah, T.F. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 2008, 190, 4447–4452. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, B.; Sauer, K. The ABC of biofilm drug tolerance: The MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob. Agents Chemother. 2018, 62, e01981-17. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 2008, 322, 107–131. [Google Scholar]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Monds, R.D.; O’Toole, G.A. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol. 2009, 17, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Klauck, G.; Serra, D.O.; Possling, A.; Hengge, R. Spatial organization of different sigma factor activities and c-di-GMP signalling within the three-dimensional landscape of a bacterial biofilm. Open Biol. 2018, 8, 180066. [Google Scholar] [CrossRef]
- Shiota, S.; Shimizu, M.; Mizushima, T.; Ito, H.; Hatano, T.; Yoshida, T.; Tsuchiya, T. Marked reduction in the minimum inhibitory concentration (MIC) of beta-lactams in methicillin-resistent Staphylococcus aureus produced by epicatechine gallate, an ingredient of green tea (Camellia sinensis). Biol. Pharm. Bull. 1999, 22, 1388–1390. [Google Scholar] [CrossRef]
- Hu, Z.-Q.; Zhao, W.-H.; Hara, Y.; Shimamura, T. Epigallocatechin gallate synergy with ampicillin/sulbactam against 28 clinical isolates of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2001, 48, 361–364. [Google Scholar] [CrossRef]
- Hu, Z.-Q.; Zhao, W.-H.; Asano, N.; Yoda, Y.; Hara, Y.; Shimamura, T. Epigallocatechin gallate synergistically enhances the activity of carbapenems against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 558–560. [Google Scholar] [CrossRef]
- Zhao, W.H.; Hu, Z.Q.; Hara, Y.; Shimamura, T. Inhibition of penicillinase by epigallocatechin gallate resulting in restoration of antibacterial activity of penicillin against penicillinase-producing Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2266–2268. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Ghosh, S.; Ray, R.; Hazra, B. Polyphenolic secondary metabolites synergize the activity of commercial antibiotics against clinical isolates of b-lactamase-producing Klebsiella pneumoniae. Phytother. Res. 2016, 30, 272–282. [Google Scholar] [CrossRef] [PubMed]
- O’May, C.; Ciobanu, A.; Lam, H.; Tufenkji, N. Tannin derived materials can block swarming motility and enhance biofilm formation in Pseudomonas aeruginosa. Biofouling 2012, 28, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Bikels-Goshen, T.; Landau, E.; Saguy, S.; Shapira, R. Staphylococcal strains adapted to epigallocathechin gallate (EGCG) show reduce susceptibility to vancomycin, oxacillin and ampicillin, increased heat tolerance, and altered cell morphology. Int. J. Food Microbiol. 2010, 138, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Ide, K.; Matsuoka, N.; Yamada, H.; Furushima, D.; Kawakami, K. Effects of tea catechins on Alzheimer’s Disease: Recent updates and perspectives. Molecules 2018, 23, 2357. [Google Scholar] [CrossRef] [PubMed]
- Pervin, M.; Unno, K.; Ohishi, T.; Tanabe, H.; Miyoshi, N.; Nakamura, Y. Beneficial effects of green tea catechins on neurodegenerative diseases. Molecules 2018, 23, 1297. [Google Scholar] [CrossRef] [PubMed]
- Shirakami, Y.; Shimizu, M. Possible mechanisms of green tea and its constituents against cancer. Molecules 2018, 23, 2284. [Google Scholar] [CrossRef] [PubMed]
- Reto, M.; Figueira, M.E.; Filipe, H.M.; Almeida, C.M. Chemical composition of green tea (Camellia sinensis) infusions commercialized in Portugal. Plant Foods Hum. Nutr. 2007, 62, 139–144. [Google Scholar] [CrossRef]
- Solovchenko, A.; Schmitz-Eiberger, M. Significance of skin flavonoids for UV-B-protection in apple fruits. J. Exp. Bot. 2003, 54, 1977–1984. [Google Scholar] [CrossRef]
- Matern, U.; Kneusel, R.E. Phenolic compounds in plant disease resistance. Phytoparasitica 1988, 16, 153–170. [Google Scholar] [CrossRef]
- Field, B.; Jordán, F.; Osbourn, A. First encounters—Deployment of defence-related natrual products by plants. New. Phytol. 2006, 172, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Ullah, C.; Unsicker, S.B.; Fellenberg, C.; Constabel, C.P.; Schmidt, A.; Gershenzon, J.; Hammerbacher, A. Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol. 2017, 175, 1560–1578. [Google Scholar] [CrossRef] [PubMed]
- Teplitski, M.; de Moraes, M. Of mice and men... and plants: Comparative genomics of the dual lifestyles of enteric pathogens. Trends Microbiol. 2018, 26, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Hengge, R. Bacterial Stress Responses; ASM Press: Washington, DC, USA, 2011. [Google Scholar]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Yaron, S.; Römling, U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechn. 2014, 7, 496–516. [Google Scholar] [CrossRef] [PubMed]
- Barak, J.D.; Gorski, L.; Naraghi-Arani, P.; Charkowski, A.O. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 2005, 71, 5685–5691. [Google Scholar] [CrossRef]
- Jeter, C.; Matthysse, A.G. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol. Plant Microbe Interact. 2005, 18, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Matthysse, A.G.; Deora, R.; Mishra, M.; Torres, A.G. Polysaccharides cellulose, poly-beta-1,6-n-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl. Environ. Microbiol. 2008, 74, 2384–2390. [Google Scholar] [CrossRef]
- Saldana, Z.; Sánchez, E.; Xicohtencatl-Cortes, J.; Puente, J.L.; Girón, J.A. Surface structure involved in plant stomata and leaf colonization by Shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2011, 2, 119. [Google Scholar] [CrossRef]
- Fink, R.C.; Black, E.P.; Hou, Z.; Sugawara, M.; Sadowsky, M.J.; Diez-Gonzalez, F. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl. Environ. Microbiol. 2012, 78, 1752–1764. [Google Scholar] [CrossRef]
- Macarisin, D.; Patel, J.; Bauchan, G.; Giron, J.A.; Sharma, V.K. Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog. Dis. 2012, 9, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Osbourn, A. Plant-microbe interactions: Chemical diversity in plant defense. Science 2009, 324, 746–748. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Jedrzeyczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef]
- Radulovic, N.S.; Blagojevic, P.D.; Stojanovic-Radic, Z.Z.; Stojanovic, N.M. Antimicrobial plant metabolites: Structural diversity and mechanism of action. Curr. Med. Chem. 2013, 20, 932–952. [Google Scholar]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Thermaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Rosier, A.; Bishnoi, U.; Lakshmanan, V.; Sherrier, D.J.; Bais, H.P. A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 2016, 90, 537–548. [Google Scholar] [CrossRef]
- Chen, D.; Cao, Y.; Yu, L.; Tao, Y.; Zhou, Y.; Zhi, Q.; Lin, H. Characteristics and influencing factors of amyloid fibers in S. mutans biofilm. AMB Expr. 2019, 9, 31. [Google Scholar] [CrossRef]
- Du, X.; Huang, X.; Huang, C.; Wang, Y.; Zhang, Y. Epigallocatechin-3-gallate (EGCG) enhances the therapeutic activity of a dental adhesive. J. Dent. 2012, 40, 485–492. [Google Scholar] [CrossRef]
- Hu, J.; Du, X.; Huang, C.; Fu, D.; Ouyang, X.; Wang, Y. Antibacterial and physical properties of EGCG-containing glass ionomer cements. J. Dent. 2013, 41, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.H.; Zhang, L.; Yu, F.; Liu, Z.Y.; Chen, J.H. Epigallocatechin-3-gallate and epigallocatechin-3-O-(3-O-methyl)-gallate enhance the bonding stability of an etch-and-rinse adhesive to dentin. Materials 2017, 10, 183. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, H.; Li, K.; Ren, H.; Lei, J.; Huang, C. Development of epigallocatechin-3-gallate-encapsulated nanohydroxyappatite/mesoprous silica for therapeutic management of dentin surface. ACS Appl. Mater. Interfaces 2017, 9, 25796–25807. [Google Scholar] [CrossRef] [PubMed]
- Melok, A.L.; Lee, L.H.; Yussof, S.A.M.; Chu, T. Green tea polyphenol epigallocatechin-3-gallate-stearate inhibits the growth of Streptococcus mutans. A promising new approach in caries prevention. Dent. J. 2018, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Francesko, A.; Soares da Costa, D.; Reis, R.L.; Pashkuleva, I.; Tzanov, T. Functional biopolymer-based matrices for modulation of chronic wound enzyme activities. Acta Biomater. 2013, 9, 5216–5225. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.L.; Lee, J.H.; Kwon, B.H.; Lee, M.H.; Han, D.W.; Hyon, S.H.; Park, J.C. Promotion of full-thickness wound healing using epigallocatechin-3-O-gallate/poly (lactic-co-glycolic acid) membrane as temporary wound dressing. Artif. Organs 2014, 38, 411–417. [Google Scholar] [CrossRef]
- Li, M.; Xu, J.; Shi, T.; Yu, H.; Bi, J.; Chen, G. Epigallocatechin-3-gallate augments therapeutic effects of mesenchymal stem cells in skin wound healing. Clin. Exp. Pharmacol. Physiol. 2016, 43, 1115–1124. [Google Scholar] [CrossRef]
- Huang, Y.W.; Zhu, Q.Q.; Yang, X.Y.; Xu, H.H.; Sun, B.; Wang, X.J.; Sheng, J. Wound healing can be improved by (-)-epigallocatechin gallate through targeting Notch in streptozotocin-induced diabetic mice. FASEB J. 2019, 33, 952–964. [Google Scholar] [CrossRef]
- Ullmann, U. Plasma-kinetic characteristics of purified and isolated green tea catechin epigallocatechin gallate (EGCG) after 10 days repeated dosing in healthy volunteers. Int. J. Vitam. Nutr. Res. 2004, 74, 269–278. [Google Scholar] [CrossRef]
- Schantz, M.; Erk, T.; Richling, E. Metabolism of green tea catechins by the human small intestine. Biotech. J. 2010, 5, 1050–1059. [Google Scholar] [CrossRef]
- Gan, R.Y.; Li, H.B.; Sui, Z.Q.; Corke, H. Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCG): An updated review. Crit. Rev. Food Sci. Nutr. 2018, 58, 924–941. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bruins, M.E.; Ni, L.; Vincken, J.P. Green and black tea phenolics: Bioavailability, transformation by colonic microbiota, and modulation of colonic microbiota. J. Agric. Food Chem. 2018, 66, 8469–8477. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Brauner, A.; Li, Y.; Normark, S. Expression of and cytokine activation by Escherichia coli curli fibres in human sepsis. J. Infect. Dis. 2000, 181, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Tükel, C.; Wilson, R.P.; Nishimori, J.H.; Pezeshki, M.; Chromy, B.A.; Bäumler, A.J. Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe 2009, 6, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Tükel, C.; Raffatellu, M.; Humphries, A.D.; Wilson, R.P.; Andrews-Polymenis, H.L.; Gull, T.; Figueiredo, J.F.; Wong, M.H.; Michelsen, K.S.; Akcelik, M.; et al. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 2005, 58, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Bäumler, A.J. Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes 2014, 5, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Oz, H.S.; Barve, S.; De Villiers, W.J.; McClain, C.J.; Varilek, G.W. The green tea polyphenol (-)-epigallocatechin-3-gallate blocks nuclear factor-kappa B activation by inhibiting I kappa B kinase activity in the intestinal epithelial cell line IEC-6. Mol. Pharmacol. 2001, 60, 528–533. [Google Scholar]
- Oz, H.S.; Chen, T.; De Villiers, W.J. Green tea polyphenols and sulfasalazine have parallel anti-inflammatory properties in colitis models. Front. Immunol. 2013, 4, 132. [Google Scholar] [CrossRef]
- Barbalho, S.M.; Bosso, H.; Salzedas-Pescinini, L.M.; de Alvares Goulart, R. Green tea: A possibility in the therapeutic approach of inflammatory bowel diseases? Green tea and inflammatory bowel diseases. Complement. Ther. Med. 2019, 43, 148–153. [Google Scholar] [CrossRef]
- Lecumberri, E.; Dupertuis, Y.M.; Miralbell, R.; Pichard, C. Green tea polyphenol epigallocatechin-3-gallate (EGCG) as adjuvant in cancer therapy. Clin. Nutr. 2013, 32, 894–903. [Google Scholar] [CrossRef]
- Hara-Terawaki, A.; Takagaki, A.; Kobayashi, H.; Nanjo, F. Inhibitory activity of catechin metabolites produced by intestinal microbiota on proliferation of HeLa cells. Biol. Pharm. Bull. 2017, 40, 1331–1335. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, Y.; Ohland, C.; Jobin, C.; Sang, S. Microbiota facilitates the formation of the aminated metabolite of green tea polyphenol (-) epigallocatechin-3-gallate which tap deleterious reactive endogeneous metabolites. Free Radic. Biol. Med. 2019, 131, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Gil-Cardoso, K.; Ginés, I.; Pinent, M.; Ardévol, A.; Blay, M.; Terra, X. Effects of flavonoids on intestinal inflammation, barrier integrity and changes in gut microbiota during diet-induced obesity. Nutr. Res. Rev. 2016, 29, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Remely, M.; Ferk, F.; Sterneder, S.; Setayesh, T.; Roth, S.; Kepcija, T.; Noorizadeh, R.; Rebhan, I.; Greunz, M.; Beckmann, J.; et al. EGCG prevents high fat diet-induced changes in gut microbiota, decreases of DNA strand breaks, and changes in expression and DNA methylation of Dnmt1 and MLH1 in C57BL/6J male mice. Oxid. Med. Cell. Longev. 2017, 3079148. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Jena, P.K.; Liu, H.X.; Hu, Y.; Nagar, N.; Bronner, D.N.; Settles, M.L.; Bäumler, A.J.; Wan, Y.Y. Obesity treatment by epigallocatechin-3-gallate-regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J. 2018, 32, 6371–6384. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hengge, R. Targeting Bacterial Biofilms by the Green Tea Polyphenol EGCG. Molecules 2019, 24, 2403. https://doi.org/10.3390/molecules24132403
Hengge R. Targeting Bacterial Biofilms by the Green Tea Polyphenol EGCG. Molecules. 2019; 24(13):2403. https://doi.org/10.3390/molecules24132403
Chicago/Turabian StyleHengge, Regine. 2019. "Targeting Bacterial Biofilms by the Green Tea Polyphenol EGCG" Molecules 24, no. 13: 2403. https://doi.org/10.3390/molecules24132403
APA StyleHengge, R. (2019). Targeting Bacterial Biofilms by the Green Tea Polyphenol EGCG. Molecules, 24(13), 2403. https://doi.org/10.3390/molecules24132403





