Application of Natural Deep Eutectic Solvents in the Extraction of Quercetin from Vegetables
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Prepared NADESs
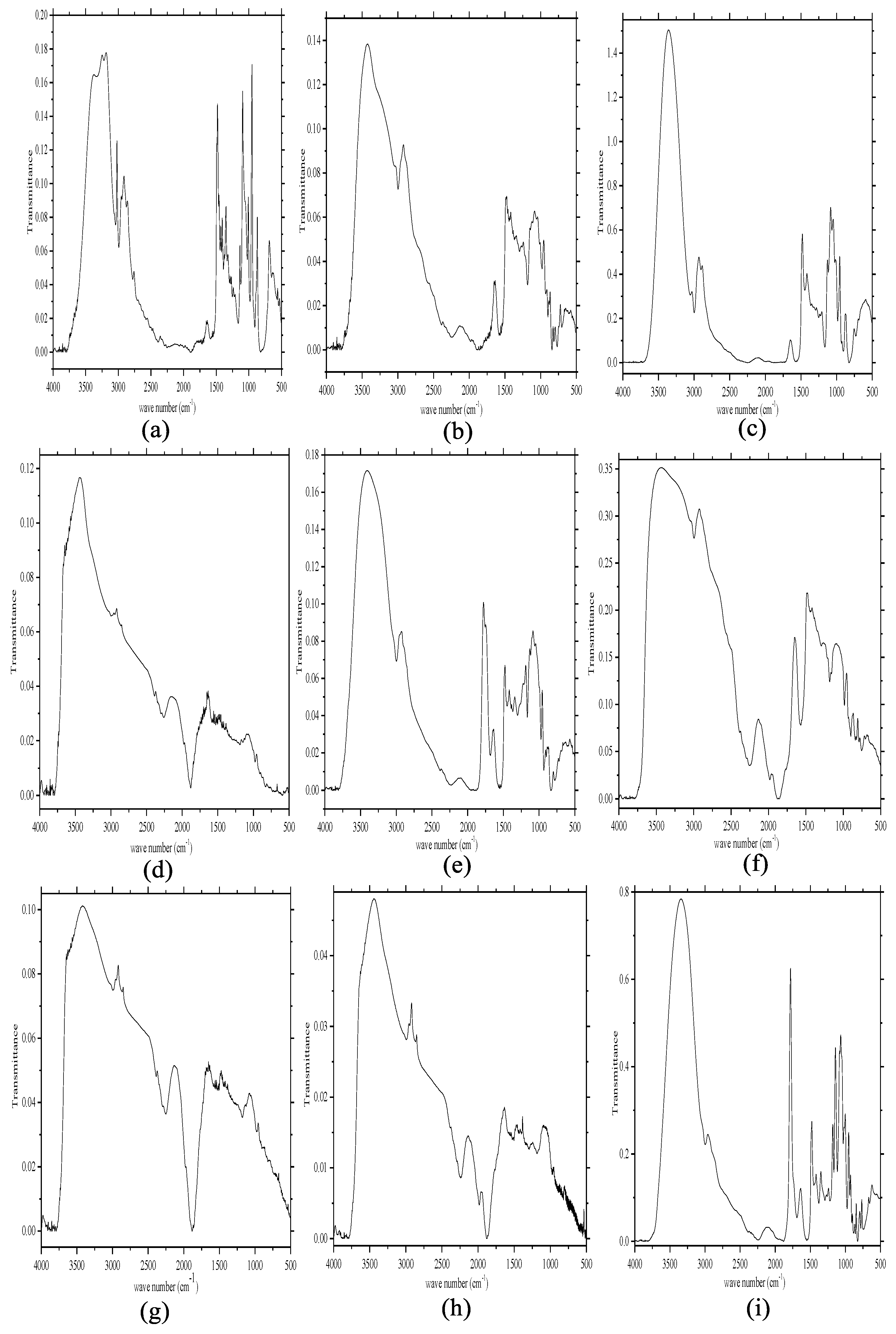
2.1.1. Fourier Transform-Infrared (FT-IR) Spectroscopy
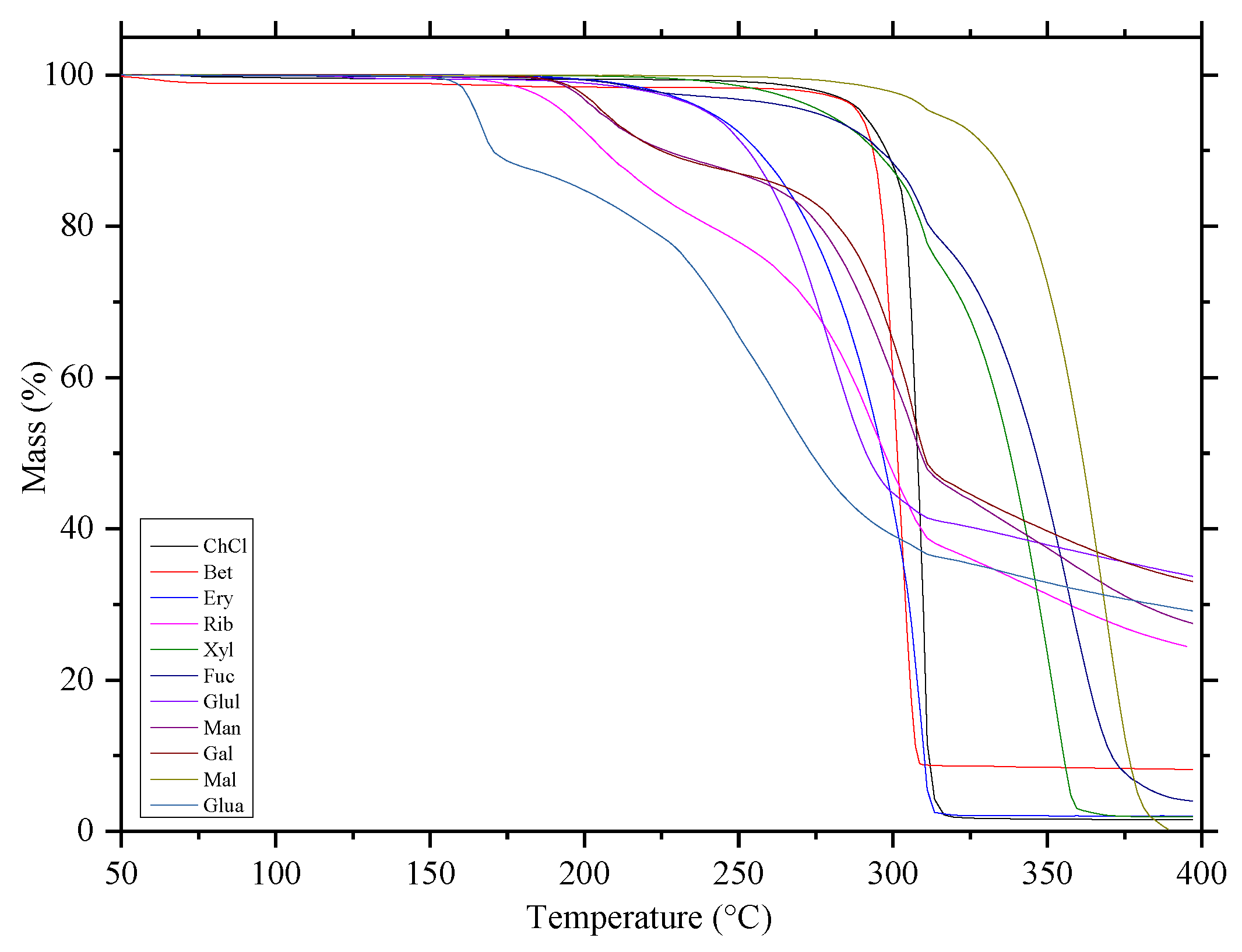
2.1.2. Thermogravimetric Analysis (TGA)
2.2. Selection Of The Extracting Method And Solvent
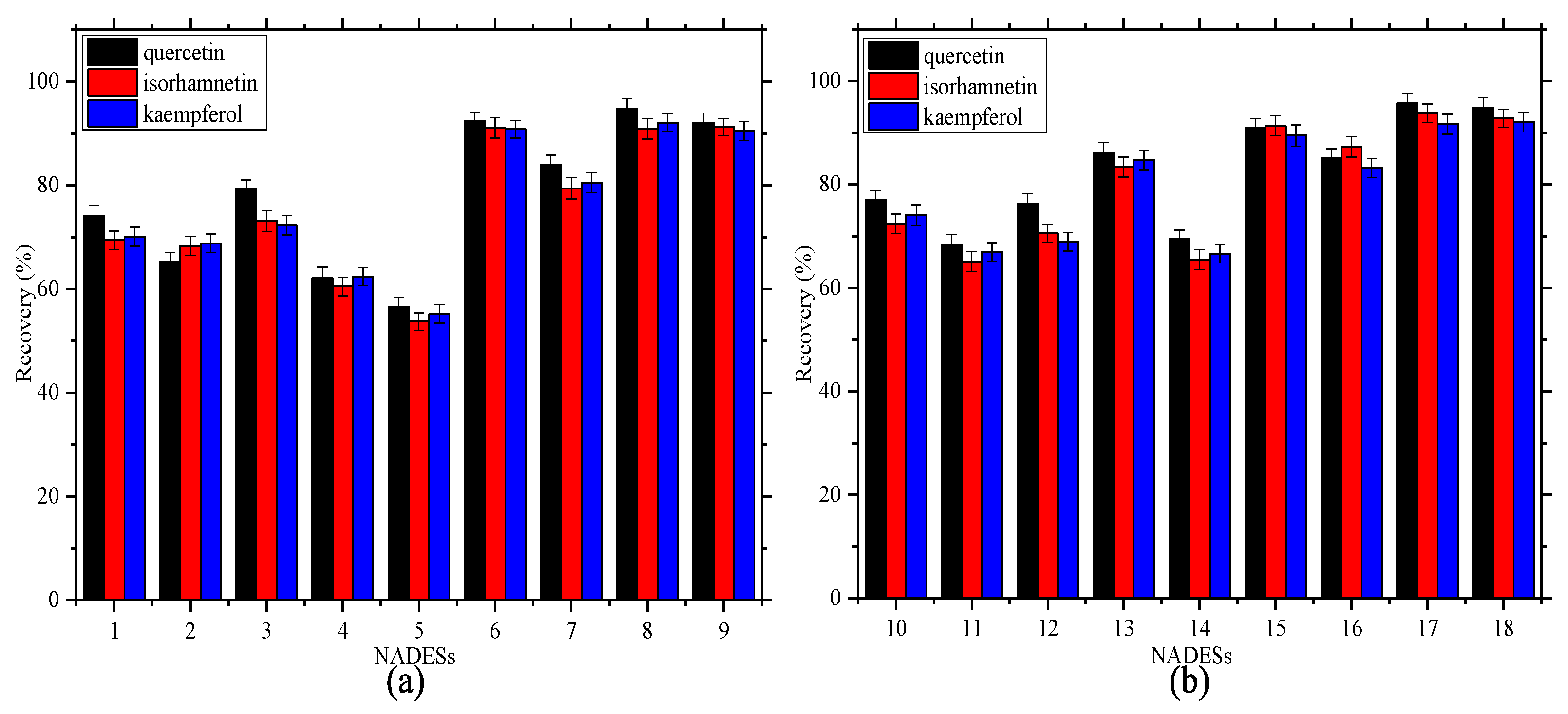
2.3. Selectivity of the NADESs
2.4. Optimization of Quercetin Extraction
2.5. Verification and Applications of the Method
2.6. Data Analyses
2.7. Comparison with Other Reported Methods
3. Experimental
3.1. Chemicals and Reagents
3.2. Instrumentation and Conditions
3.3. Preparation of Choline Chloride-Based NADESs
3.4. Preparation of Betaine-Based NADESs
3.5. Preparation of the Extraction Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lesjak, M.; Beara, I.; Simin, N.; Pintać, D.; Majkić, T.; Bekvalac, K.; Orčić, D.; Mimica-Dukić, N. Antioxidant and anti-inflammatory activities of quercetin and its derivatives. J. Funct. Foods 2018, 40, 68–75. [Google Scholar] [CrossRef]
- Ghoreishi, S.M.; Hedayati, A.; Mousavi, S.O. Quercetin extraction from Rosa damascena mill via supercritical CO2: Neural network and adaptive neuro fuzzy interface system modeling and response surface optimization. J. Supercrit. Fluid. 2016, 112, 57–66. [Google Scholar] [CrossRef]
- Iacopini, P.; Baldi, M.; Storchi, P.; Sebastiani, L. Catechin, epicatechin, quercetin, rutin and resveratrol in red grape: Content, in vitro antioxidant activity and interactions. J. Food Compos. Anal. 2008, 21, 589–598. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, B.; Shen, J.; Wan, L.; Zhu, Y.; Yi, T.; Xiao, Z. The beneficial effects of quercetin, curcumin, and resveratrol in obesity. Oxid. Med. Cell. Longev. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; He, H.; Chen, Q. Quercetin inhibits proliferation and invasion acts by up-regulating miR-146a in human breast cancer cells. Mol. Cell. Biochem. 2015, 402, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Ghayour, N.; Hosseini, S.M.H.; Eskandari, M.H.; Esteghlal, S.; Nekoei, A.; Gahruie, H.H.; Tatar, M.; Naghibalhossaini, F. Nanoencapsulation of quercetin and curcumin in casein-based delivery systems. Food Hydrocolloid. 2019, 87, 94–403. [Google Scholar] [CrossRef]
- Petersen, B.; Egert, S.; Bosy-Westphal, A.; Müller, M.J.; Wolffram, S.; Hubbermann, E.M.; Rimbach, G.; Schwarz, K. Bioavailability of quercetin in humans and the influence of food matrix comparing quercetin capsules and different apple sources. Food Res. Int. 2016, 88, 159–165. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, G. Quercetin: A flavonol with multifaceted therapeutic applications? Fitoterapia 2015, 106, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Russo, G.L.; Daglia, M.; Nabavi, S.M. Role of quercetin as an alternative for obesity treatment: You are what you eat! Food Chem. 2015, 179, 305–310. [Google Scholar] [CrossRef]
- Kim, K.H.; Dutta, T.; Sun, J.; Simmons, B.; Singh, S. Biomass pretreatment using deep eutectic solvents from lignin derived phenols. Green Chem. 2018, 20, 809–815. [Google Scholar] [CrossRef]
- Ma, C.; Laaksonen, A.; Liu, C.; Lu, X.; Ji, X. The peculiar effect of water on ionic liquids and deep eutectic solvents. Chem. Soc. Rev. 2018, 47, 8685–8720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capello, C.; Fischer, U.; Hungerbühler, K. What is a green solvent? A comprehensive framework for the environmental assessment of solvents. Green Chem. 2007, 9, 927–934. [Google Scholar] [CrossRef]
- Abbott, A.P.; Capper, G.; Davies, D.L.; Rasheed, R.K.; Tambyrajah, V. Novel solvent properties of choline chloride/urea mixtures. Chem. Commun. 2003, 1, 70–71. [Google Scholar] [CrossRef]
- Lee, C.B.T.L.; Wu, T.Y.; Ting, C.H.; Tan, J.K.; Siow, L.F.; Cheng, C.K.; Mohammad, A.W. One-pot furfural production using choline chloride-dicarboxylic acid based deep eutectic solvents under mild conditions. Bioresource Technol. 2019, 278, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.P.; Fernandez, L.; Conceição, J.H.; Martins, M.A.; Sosa, A.; Ortega, J.; Coutinho, J.A. Design and characterization of sugar-based deep eutectic solvents using conductor-like screening model for real solvents. ACS Sustain. Chem. Eng. 2018, 6, 10724–10734. [Google Scholar] [CrossRef]
- Cui, Y.; Kuroda, D.G. Evidence of molecular heterogeneities in amide-based deep eutectic solvents. J. Phys. Chem. A 2018, 122, 1185–1193. [Google Scholar] [CrossRef]
- Hossain, S.S.; Samanta, A. How do the hydrocarbon chain length and hydroxyl group position influence the solute dynamics in alcohol-based deep eutectic solvents? Phys. Chem. Chem. Phys. 2018, 20, 24613–24622. [Google Scholar] [CrossRef]
- García, A.; Rodríguez-Juan, E.; Rodríguez-Gutiérrez, G.; Rios, J.J.; Fernández-Bolaños, J. Extraction of phenolic compounds from virgin olive oil by deep eutectic solvents (DESs). Food Chem. 2016, 197, 554–561. [Google Scholar] [CrossRef]
- Florindo, C.; Oliveira, M.M.; Branco, L.C.; Marrucho, I.M. Carbohydrates-based deep eutectic solvents: Thermophysical properties and rice straw dissolution. J. Mol. Liq. 2017, 247, 441–447. [Google Scholar] [CrossRef]
- Alvarez-Vasco, C.; Ma, R.; Quintero, M.; Guo, M.; Geleynse, S.; Ramasamy, K.K.; Zhang, X. Unique low-molecular-weight lignin with high purity extracted from wood by deep eutectic solvents (DES): A source of lignin for valorization. Green Chem. 2016, 18, 5133–5141. [Google Scholar] [CrossRef]
- Mbous, Y.P.; Hayyan, M.; Hayyan, A.; Wong, W.F.; Hashim, M.A.; Looi, C.Y. Applications of deep eutectic solvents in biotechnology and bioengineering-promises and challenges. Biotechnol. Adv. 2017, 35, 105–134. [Google Scholar] [CrossRef] [PubMed]
- Procopio, A.; Alcaro, S.; Nardi, M.; Oliverio, M.; Ortuso, F.; Sacchetta, P.; Sindona, G. Synthesis, biological evaluation, and molecular modeling of oleuropein and its semisynthetic derivatives as cyclooxygenase inhibitors. J. Agric. Food Chem. 2009, 57, 11161–11167. [Google Scholar] [CrossRef] [PubMed]
- Nardi, M.; Bonacci, S.; De Luca, G.; Maiuolo, J.; Oliverio, M.; Sindona, G.; Procopio, A. Biomimetic synthesis and antioxidant evaluation of 3, 4-DHPEA-EDA [2-(3, 4-hydroxyphenyl) ethyl (3S, 4E)-4-formyl-3-(2-oxoethyl) hex-4-enoate]. Food Chem. 2014, 162, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Abo-Hamad, A.; Hayyan, M.; AlSaadi, M.A.; Hashim, M.A. Potential applications of deep eutectic solvents in nanotechnology. Chem. Eng. J. 2015, 273, 551–567. [Google Scholar] [CrossRef]
- Tan, T.; Zhang, M.; Wan, Y.; Qiu, H. Utilization of deep eutectic solvents as novel mobile phase additives for improving the separation of bioactive quaternary alkaloids. Talanta 2016, 149, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.C.; Chen, J.; Zhang, H.; Li, Z.; Zhao, L.; Qiu, H. Effective extraction of flavonoids from Lycium barbarum L. fruits by deep eutectic solvents-based ultrasound-assisted extraction. Talanta 2019, 203, 16–22. [Google Scholar] [CrossRef]
- Ali, M.C.; Liu, R.; Chen, J.; Cai, T.; Zhang, H.; Li, Z.; Qiu, H. New deep eutectic solvents composed of crown ether, hydroxide and polyethylene glycol for extraction of non-basic N-compounds. Chinese Chem. Lett. 2019, 30, 871–874. [Google Scholar] [CrossRef]
- Tome, L.I.; Baiao, V.; da Silva, W.; Brett, C.M. Deep eutectic solvents for the production and application of new materials. Appl. Mater. Today 2018, 10, 30–50. [Google Scholar] [CrossRef]
- Nardi, M.; Bonacci, S.; Cariati, L.; Costanzo, P.; Oliverio, M.; Sindona, G.; Procopio, A. Synthesis and antioxidant evaluation of lipophilic oleuropein aglycone derivatives. Food Funct. 2017, 8, 4684–4692. [Google Scholar] [CrossRef]
- Nardi, M.; Costanzo, P.; De Nino, A.; Di Gioia, M.L.; Olivito, F.; Sindona, G.; Procopio, A. Water excellent solvent for the synthesis of bifunctionalized cyclopentenones from furfural. Green Chem. 2017, 19, 5403–5411. [Google Scholar] [CrossRef]
- Aroso, I.M.; Paiva, A.; Reis, R.L.; Duarte, A.R.C. Natural deep eutectic solvents from choline chloride and betaine–Physicochemical properties. J. Mol. Liq. 2017, 241, 654–661. [Google Scholar] [CrossRef]
- Al-Rifai, A.A.; Aqel, A.; Awaad, A.; ALOthman, Z.A. Analysis of quercetin and kaempferol in an alcoholic extract of convolvulus pilosellifolius using HPLC. Commun. Soil Sci. Plan. 2015, 46, 1411–1418. [Google Scholar] [CrossRef]
- Rasoolzadeh, F.; Hashemi, P.; Nazari Serenjeh, F. Ionic liquid-based cloud-point extraction of quercetin for its sensitive HPLC–UV determination in juice samples. Acta Chromatogr. 2017, 29, 493–496. [Google Scholar] [CrossRef]
- Abdelkawy, K.S.; Balyshev, M.E.; Elbarbry, F. A new validated HPLC method for the determination of quercetin: Application to study pharmacokinetics in rats. Biomed. Chromatogr. 2017, 31, e3819. [Google Scholar] [CrossRef]
- Fan, L.; Lin, C.; Duan, W.; Wang, X.; Liu, J.; Liu, F. Rapid and quantitative determination of 10 major active components in Lonicera japonica Thunb. by ultrahigh pressure extraction-HPLC/DAD. High Pressure Res. 2015, 35, 57–68. [Google Scholar] [CrossRef]
- Bi, W.; Tian, M.; Row, K.H. Evaluation of alcohol-based deep eutectic solvent in extraction and determination of flavonoids with response surface methodology optimization. J. Chromatogr. A 2013, 1285, 22–30. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Analyte | Range (μg/mL) | Equation | R2 | LOD (μg/mL) | LOQ (μg/mL) |
|---|---|---|---|---|---|
| quercetin | 1–200 | Y = 4.2756X − 20.2 | 0.99926 | 0.16 | 0.51 |
| isorhamnetin | Y = 4.2444X − 36.884 | 0.99853 | 0.14 | 0.43 | |
| kaempferol | Y = 4.0205X − 18.683 | 0.99931 | 0.17 | 0.52 |
| Analyte | Spike Concentration (μg/mL) | Inter-Day | Intra-Day | ||
|---|---|---|---|---|---|
| Recovery (%) | Mean ± Standard Deviation (%, n = 3) | Recovery (%) | Mean ± Standard Deviation (%, n = 3) | ||
| quercetin | 10 | 85.23 | 85.23 ± 2.41 | 88.91 | 88.91 ± 5.01 |
| 50 | 90.54 | 90.54 ± 4.03 | 90.02 | 90.02 ± 4.68 | |
| 100 | 98.76 | 98.76 ± 3.75 | 98.99 | 98.99 ± 3.78 | |
| isorhamnetin | 10 | 84.17 | 84.17 ± 3.48 | 88.45 | 88.45 ± 4.32 |
| 50 | 90.35 | 90.35 ± 2.15 | 92.13 | 92.13 ± 4.02 | |
| 100 | 97.92 | 97.92 ± 3.82 | 99.01 | 99.01 ± 4.33 | |
| kaempferol | 10 | 84.52 | 84.52 ± 3.95 | 89.56 | 89.56 ± 3.95 |
| 50 | 89.73 | 89.73 ± 2.51 | 91.05 | 91.05 ± 3.89 | |
| 100 | 98.02 | 98.02 ± 3.78 | 98.74 | 98.74 ± 4.06 | |
| Method | Detection System | Extraction Solvent | Linear Range (μg/mL) | R2 | LOD (μg/mL) | LOQ (μg/mL) | Average Recovery (%, n = 3) | References |
|---|---|---|---|---|---|---|---|---|
| ultrasonic- assisted solid liquid extraction | HPLC | methanol | 1–200 | 0.99926 | 0.18 | 0.51 | 92.64 | This study |
| solid liquid extraction | Methanol + H2O (0.1% v/v formic acid) | 1–300 | 0.9993 | 0.33 | 1.06 | 99.2 | [32] | |
| ionic liquid-based cloud-point extraction | 25% HCl | 1–20 | 0.998 | 0.002 | - | 92.5 | [33] | |
| rat plasma extraction | rat plasma | 0.1–25 | 0.9941 | 0.05 | 0.1 | - | [34] | |
| ultrahigh pressure extraction | 60% methanol | 0.005–0.05 | 0.9994 | 0.039 | 0.13 | 98.8 | [35] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Y.; Row, K.H. Application of Natural Deep Eutectic Solvents in the Extraction of Quercetin from Vegetables. Molecules 2019, 24, 2300. https://doi.org/10.3390/molecules24122300
Dai Y, Row KH. Application of Natural Deep Eutectic Solvents in the Extraction of Quercetin from Vegetables. Molecules. 2019; 24(12):2300. https://doi.org/10.3390/molecules24122300
Chicago/Turabian StyleDai, Yunliang, and Kyung Ho Row. 2019. "Application of Natural Deep Eutectic Solvents in the Extraction of Quercetin from Vegetables" Molecules 24, no. 12: 2300. https://doi.org/10.3390/molecules24122300