Metabolic Brain Network Analysis of FDG-PET in Alzheimer’s Disease Using Kernel-Based Persistent Features
Abstract
1. Introduction
2. Results
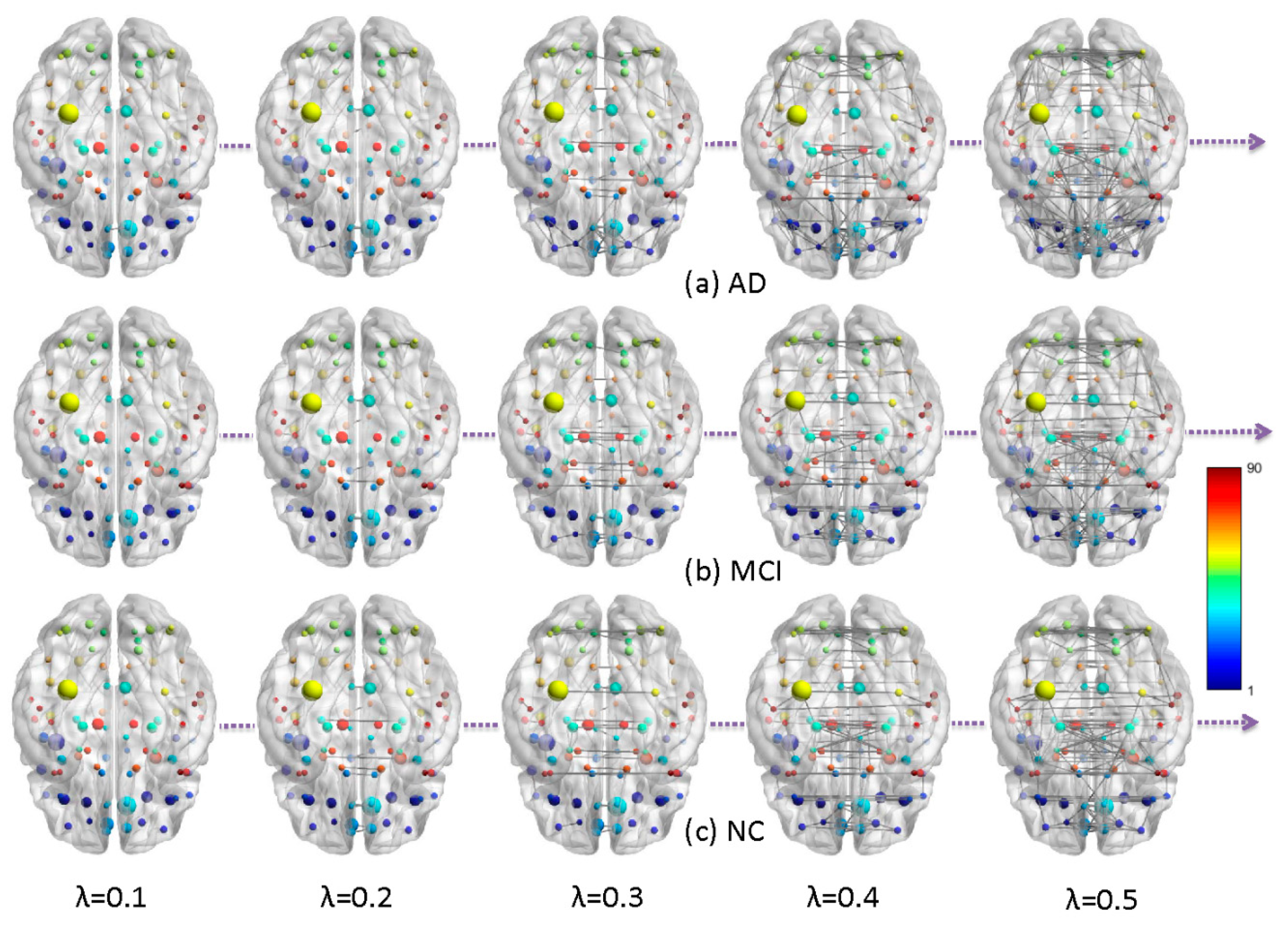
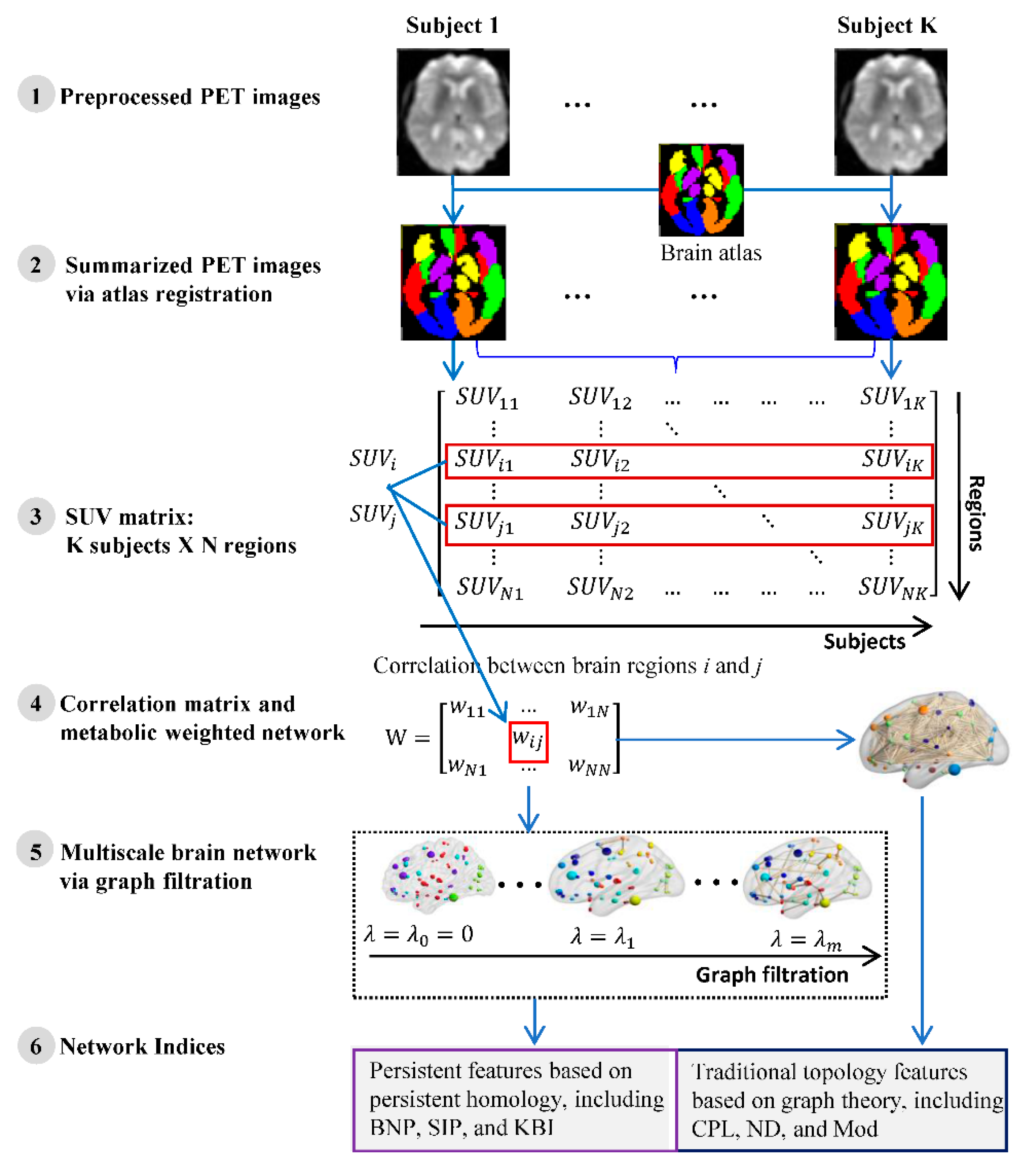
2.1. Metabolic Brain Networks
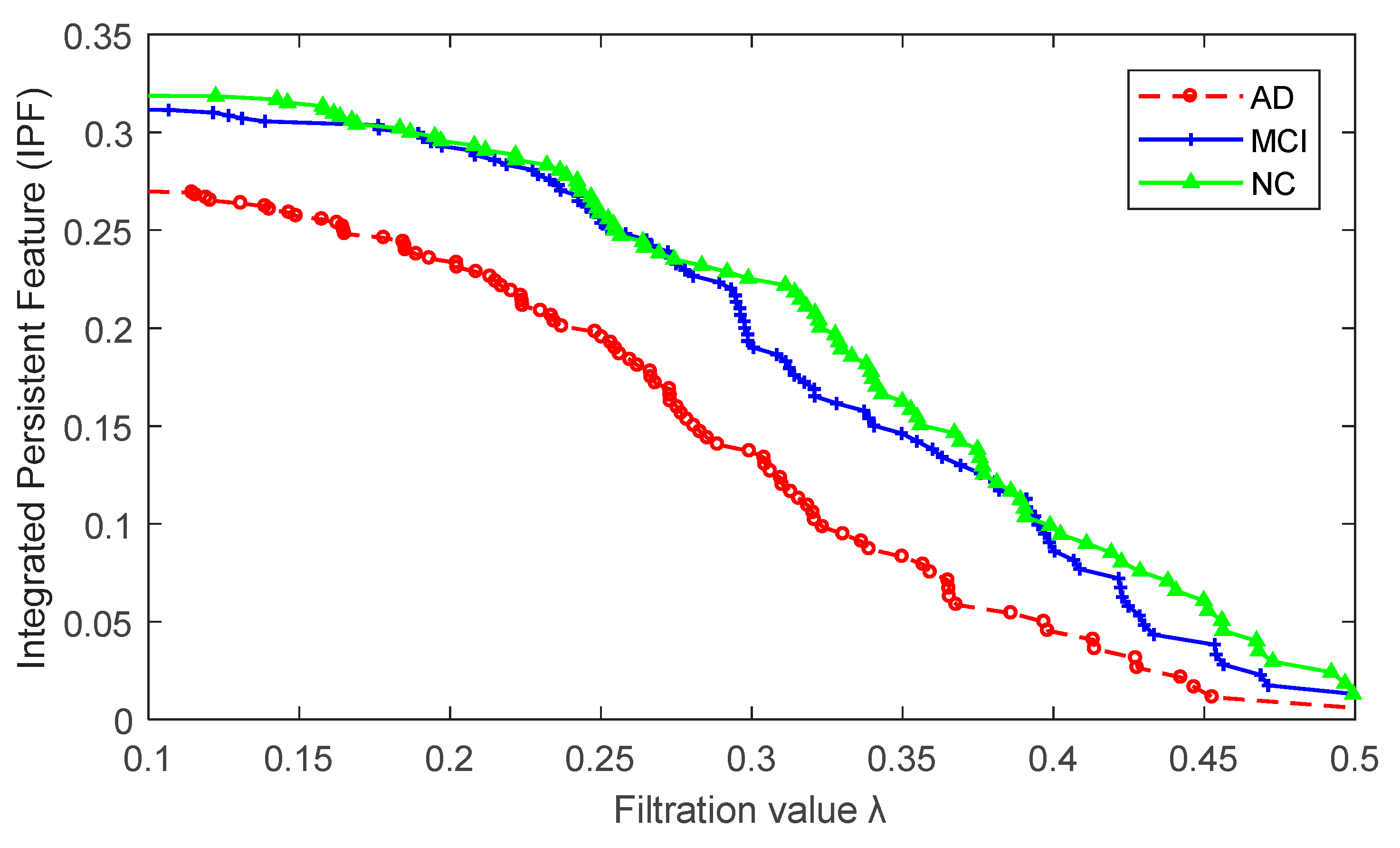
2.2. Brain Network Features
2.3. Statistical Group Difference Performance
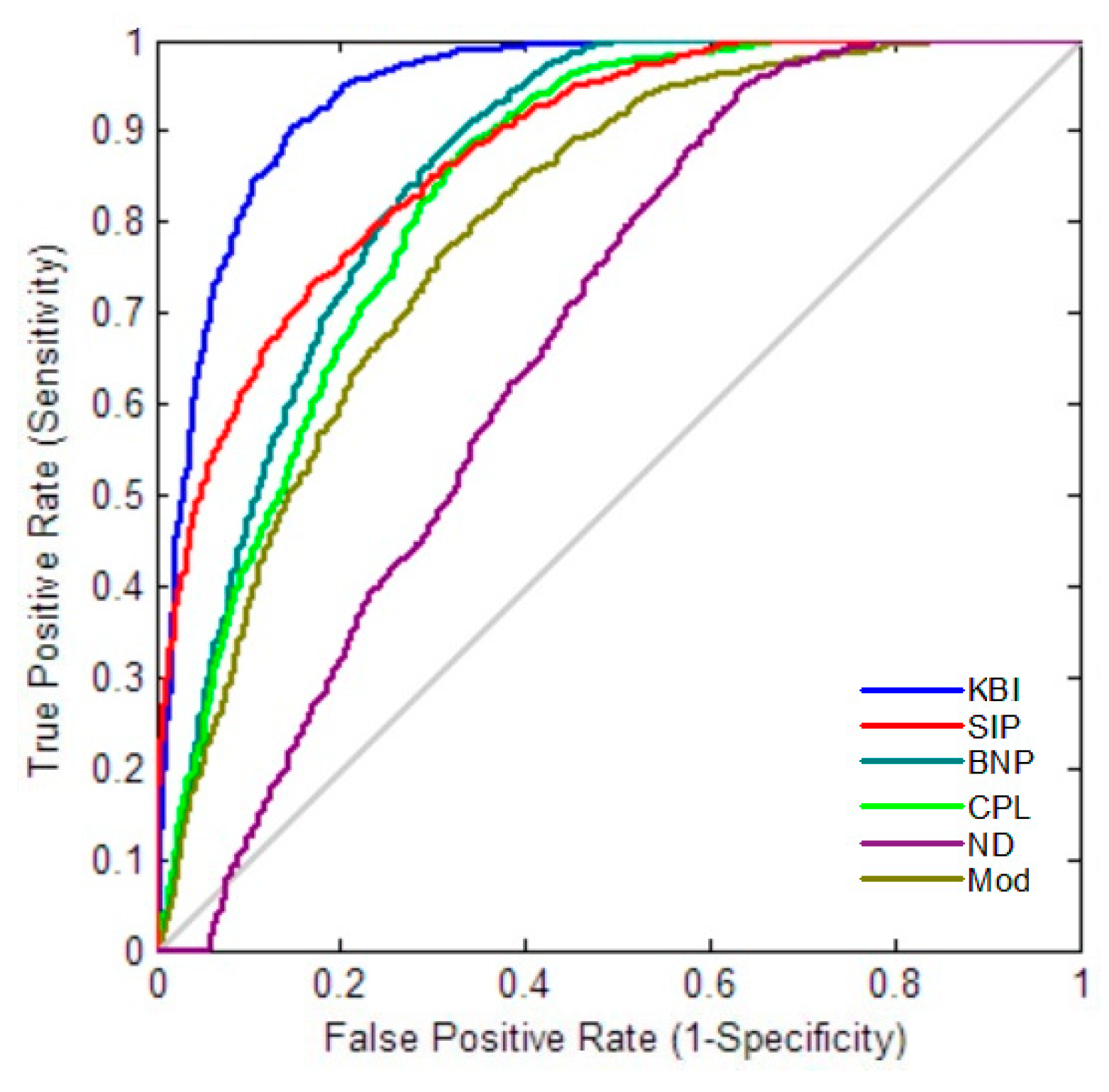
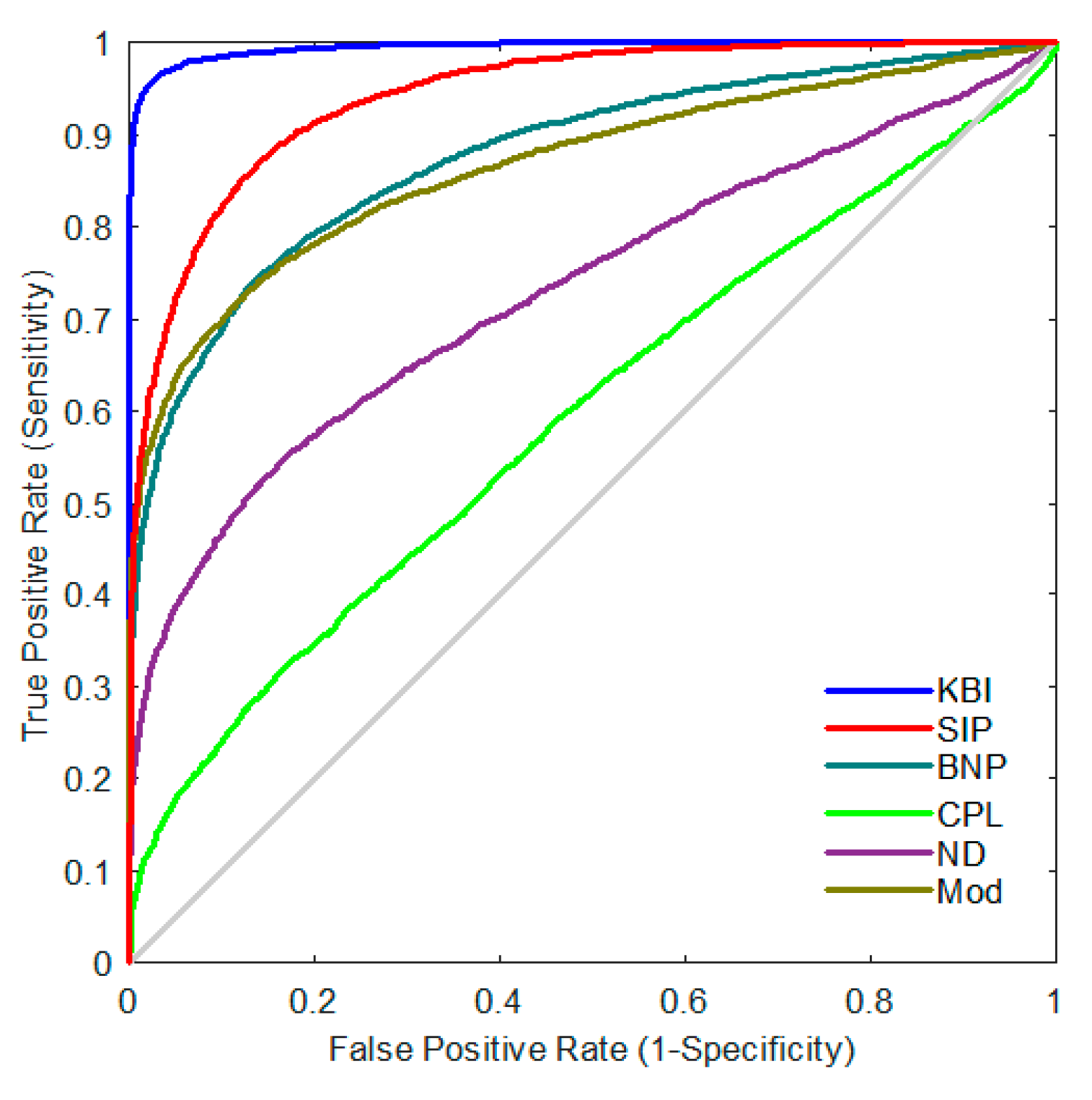
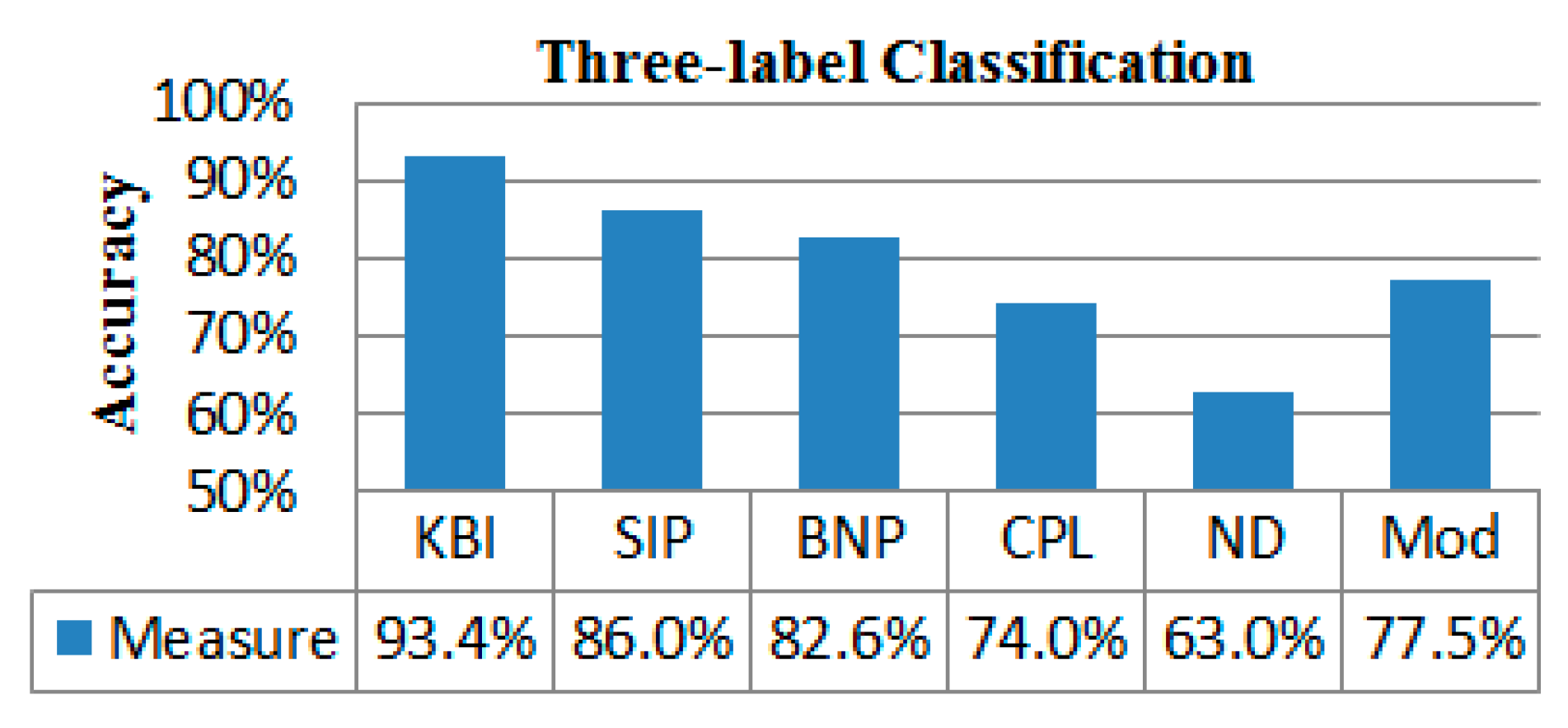
2.4. Classification Performance
3. Discussion
3.1. Present Findings
3.2. Exploring Other Connectivity Definitions
3.3. Ways of Network Construction
3.4. Limitations and Future Work
4. Materials and Methods
4.1. Participants
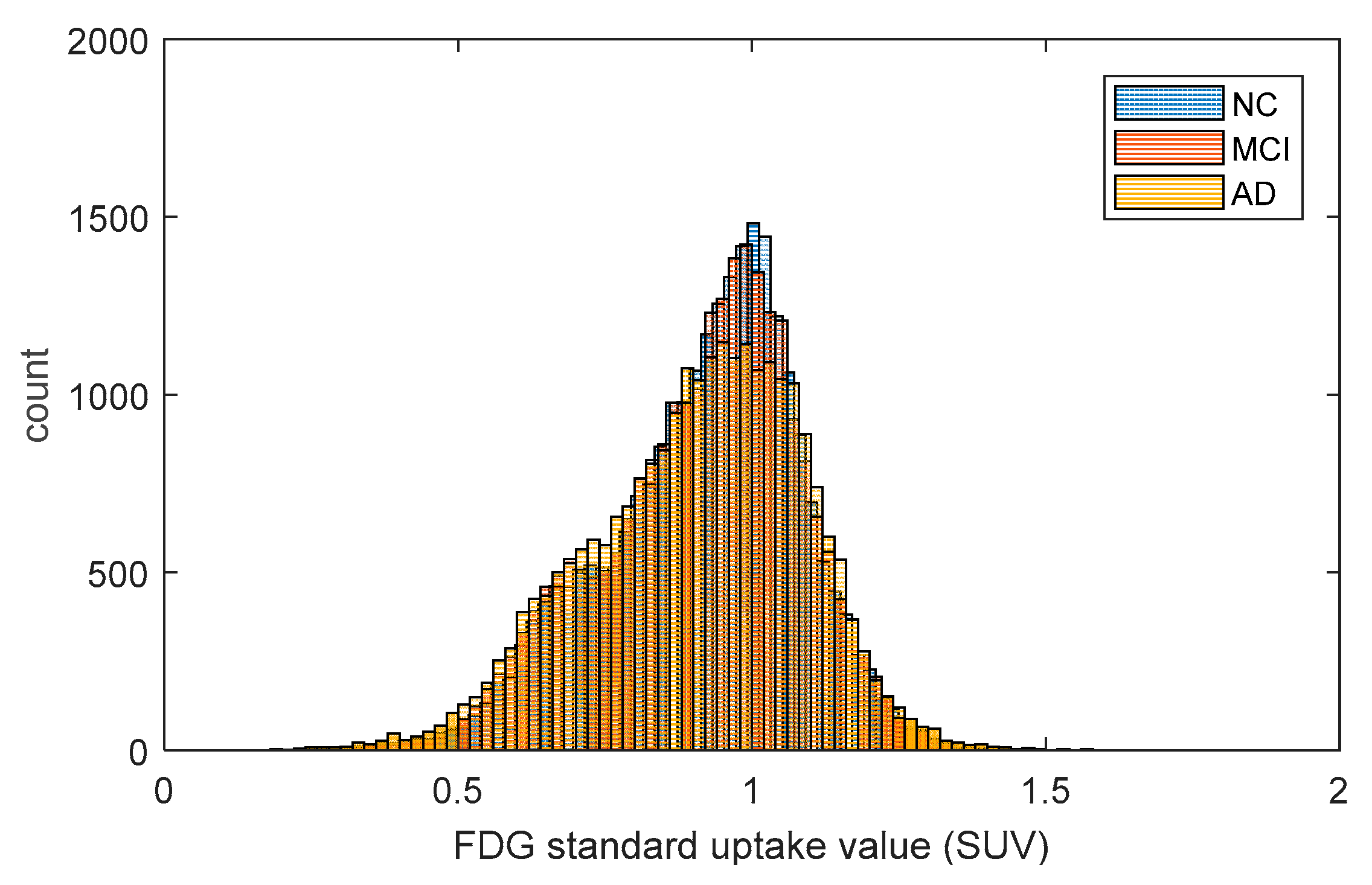
4.2. FDG-PET Data Acquisition and Preprocessing
4.3. Network Construction
4.4. Network Indices
4.4.1. Traditional Graph Theory Indices
4.4.2. Persistent Features Based on Persistent Homology
4.4.3. The Kernel-Based IPF (KBI) Index
4.5. Statistical Analysis
4.6. Classification
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Fukuyama, H.; Senda, M.; Ishii, K.; Maeda, K.; Yamamoto, Y.; Ouchi, Y.; Ishii, K.; Okumura, A.; Fujiwara, K. Prediction of outcomes in mild cognitive impairment by using 18F-FDG-PET: A multicenter study. J. Alzheimers Dis. 2015, 45, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O. Graph theory methods: Applications in brain networks. Dialogues Clin. Neuro. 2018, 20, 111–121. [Google Scholar]
- Bullmore, E.; Sporns, O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O. The human connectome: A complex network. Ann. N. Y. Acad. Sci. 2011, 1224, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Brier, M.R.; Thomas, J.B.; Fagan, A.M.; Hassenstab, J.; Holtzman, D.M.; Benzinger, T.L.; Morris, J.C.; Ances, B.M. Functional connectivity and graph theory in preclinical alzheimer’s disease. Neurobiol. Aging 2014, 35, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Sporns, O.; Betzel, R.F. Modular brain networks. Annu. Rev. Psychol. 2016, 67, 613–640. [Google Scholar] [CrossRef]
- Fagerholm, E.D.; Hellyer, P.J.; Scott, G.; Leech, R.; Sharp, D.J. Disconnection of network hubs and cognitive impairment after traumatic brain injury. Brain 2015, 138, 1696–1709. [Google Scholar] [CrossRef]
- Stam, C.; Jones, B.; Nolte, G.; Breakspear, M.; Scheltens, P. Small-world networks and functional connectivity in alzheimer’s disease. Cereb. Cortex 2006, 17, 92–99. [Google Scholar] [CrossRef]
- Qiu, T.; Luo, X.; Shen, Z.; Huang, P.; Xu, X.; Zhou, J.; Zhang, M. Disrupted brain network in progressive mild cognitive impairment measured by eigenvector centrality mapping is linked to cognition and cerebrospinal fluid biomarkers. J. Alzheimers Dis. 2016, 54, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- De Haan, W.; Van Der Flier, W.M.; Koene, T.; Smits, L.L.; Scheltens, P.; Stam, C.J. Disrupted modular brain dynamics reflect cognitive dysfunction in alzheimer’s disease. NeuroImage 2012, 59, 3085–3093. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Aganj, I.; Ge, T.; Polimeni, J.R.; Fischl, B. Functional density and edge maps: Characterizing functional architecture in individuals and improving cross-subject registration. NeuroImage 2017, 158, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Daianu, M.; Jahanshad, N.; Nir, T.M.; Jack, C.R.; Weiner, M.W.; Bernstein, M.A.; Thompson, P.M. Rich club analysis in the alzheimer’s disease connectome reveals a relatively undisturbed structural core network. Hum. Brain Mapp. 2015, 36, 3087–3103. [Google Scholar] [CrossRef] [PubMed]
- McKenna, F.; Koo, B.B.; Killiany, R. Comparison of apoe-related brain connectivity differences in early mci and normal aging populations: An fmri study. Brain Imaging Behav. 2016, 10, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.-W.; Krishnan, A.; Wager, T.D. Cluster-extent based thresholding in fmri analyses: Pitfalls and recommendations. NeuroImage 2014, 91, 412–419. [Google Scholar] [CrossRef]
- Zalesky, A.; Cocchi, L.; Fornito, A.; Murray, M.M.; Bullmore, E. Connectivity differences in brain networks. NeuroImage 2012, 60, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Edelsbrunner, H.; Harer, J. Computational Topology: An Introduction; American Mathematical Society: Heidelberg, Germany, 2010. [Google Scholar]
- Lee, H.; Kang, H.; Chung, M.K.; Kim, B.-N.; Lee, D.S. Persistent brain network homology from the perspective of dendrogram. IEEE Trans. Med. Imaging 2012, 31, 2267–2277. [Google Scholar]
- Choi, H.; Kim, Y.K.; Kang, H.; Lee, H.; Im, H.-J.; Kim, E.E.; Chung, J.-K.; Lee, D.S. Abnormal metabolic connectivity in the pilocarpine-induced epilepsy rat model: A multiscale network analysis based on persistent homology. NeuroImage 2014, 99, 226–236. [Google Scholar] [CrossRef]
- Chung, M.K.; Hanson, J.L.; Ye, J.; Davidson, R.J.; Pollak, S.D. Persistent homology in sparse regression and its application to brain morphometry. IEEE Trans. Med. Imaging 2015, 34, 1928–1939. [Google Scholar] [CrossRef]
- Yoo, K.; Lee, P.; Chung, M.K.; Sohn, W.S.; Chung, S.J.; Na, D.L.; Ju, D.; Jeong, Y. Degree-based statistic and center persistency for brain connectivity analysis. Hum. Brain Mapp. 2017, 38, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kang, H.; Chung, M.K.; Lim, S.; Kim, B.N.; Lee, D.S. Integrated multimodal network approach to pet and mri based on multidimensional persistent homology. Hum. Brain Mapp. 2017, 38, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Giusti, C.; Ghrist, R.; Bassett, D.S. Two’s company, three (or more) is a simplex: Algebraic-topological tools for understanding higher-order structure in neural data. J. Comput. Neurosci. 2016, 41, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Han, X.; Chen, K. A concise and persistent feature to study brain resting-state network dynamics: Findings from the alzheimer’s disease neuroimaging initiative. Hum. Brain Mapp. 2019, 40, 1062–1081. [Google Scholar] [CrossRef] [PubMed]
- Kusano, G.; Hiraoka, Y.; Fukumizu, K. Persistence Weighted Gaussian Kernel for Topological Data Analysis; International Conference on Machine Learning: New York, NY, USA, 2016; pp. 2004–2013. [Google Scholar]
- Carrière, M.; Cuturi, M.; Oudot, S. Sliced wasserstein kernel for persistence diagrams. Working Papers 2017. [Google Scholar]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. NeuroImage 2002, 15, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.J. Functional and effective connectivity: A review. Brain Connect 2011, 1, 13–36. [Google Scholar] [CrossRef]
- Valdes-Sosa, P.A.; Roebroeck, A.; Daunizeau, J.; Friston, K. Effective connectivity: Influence, causality and biophysical modeling. NeuroImage 2011, 58, 339–361. [Google Scholar] [CrossRef]
- Kendall, M.G. A new measure of rank correlation. Biometrika 1938, 30, 81–93. [Google Scholar] [CrossRef]
- Best, D.; Roberts, D. Algorithm as 89: The upper tail probabilities of spearman’s rho. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1975, 24, 377–379. [Google Scholar] [CrossRef]
- McIntosh, A.R.; Chau, W.K.; Protzner, A.B. Spatiotemporal analysis of event-related fmri data using partial least squares. NeuroImage 2004, 23, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Roebroeck, A.; Formisano, E.; Goebel, R. The identification of interacting networks in the brain using fmri: Model selection, causality and deconvolution. NeuroImage 2011, 58, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.; Lange, N.; Makris, N.; Bates, J.; Meyer, J.; Caviness Jr, V. Gyri of the human neocortex: An mri-based analysis of volume and variance. Cereb. Cortex 1998, 8, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Makris, N.; Meyer, J.W.; Bates, J.F.; Yeterian, E.H.; Kennedy, D.N.; Caviness, V.S. Mri-based topographic parcellation of human cerebral white matter and nuclei: Ii. Rationale and applications with systematics of cerebral connectivity. NeuroImage 1999, 9, 18–45. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Yao, Z.; Zheng, W.; Jing, Y.; Ding, Z.; Mi, L.; Lu, S. Predicting mci Progression with Individual Metabolic Network Based on Longitudinal FDG-PET; IEEE International Conference on Bioinformatics & Biomedicine: Kansas City, MO, USA, 2017. [Google Scholar]
- Jagust, W.J.; Bandy, D.; Chen, K.; Foster, N.L.; Landau, S.M.; Mathis, C.A.; Price, J.C.; Reiman, E.M.; Skovronsky, D.; Koeppe, R.A. The alzheimer’s disease neuroimaging initiative positron emission tomography core. Alzheimer’s Dement. 2010, 6, 221–229. [Google Scholar] [CrossRef]
- Jack, C.R.; Bernstein, M.A.; Fox, N.C.; Thompson, P.; Alexander, G.; Harvey, D.; Borowski, B.; Britson, P.J.L.; Whitwell, J.; Ward, C. The alzheimer’s disease neuroimaging initiative (adni): Mri methods. J. Magn. Reson. Imaging 2008, 27, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Zhang, W.; Zhang, J.; Fan, Y.; Goradia, D.; Chen, K.; Reiman, E.M.; Gu, X.; Wang, Y. An optimal transportation based univariate neuroimaging index. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 2017; pp. 182–191. [Google Scholar]
- Penny, W.D.; Friston, K.J.; Ashburner, J.T.; Kiebel, S.J.; Nichols, T.E. Statistical Parametric Mapping: The Analysis of Functional Brain Images; Academic press: Pittsburgh, PA, USA, 2011. [Google Scholar]
- De Floriani, L.; Magillo, P. Multiresolution mesh representation: Models and data structures. In Tutorials on Multiresolution in Geometric Modelling; Springer: Berlin/Heidelberg, Germany, 2002; pp. 363–417. [Google Scholar]
- Chen, S.; Tian, D.; Feng, C.; Vetro, A.; Kovačević, J. Fast resampling of 3d point clouds via graphs. arXiv 2017, arXiv:1702.06397. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
| Cohort | KBI | SIP | BNP | CPL | ND | Mod |
|---|---|---|---|---|---|---|
| AD | 0.577 | 0.790 | 268.7 | 1.20 | 1.601 | 1.289 |
| MCI | 0.826 | 0.842 | 265.2 | 1.17 | 1.457 | 0.770 |
| NC | 0.988 | 0.882 | 245.5 | 1.15 | 1.436 | 1.004 |
| Cohort | KBI | SIP | BNP | CPL | ND | Mod |
|---|---|---|---|---|---|---|
| AD vs. MCI | 0.017 | 0.036 | 0.048 | 0.058 | 0.397 | 0.074 |
| AD vs. NC | 0.002 | 0.042 | 0.054 | 0.285 | 0.266 | 0.063 |
| MCI vs. NC | 0.036 | 0.076 | 0.339 | 0.062 | 0.031 | 0.458 |
| Between-Group | Definitions of Distance Function | |||
|---|---|---|---|---|
| Kendall Correlation | Spearman Correlation | Partial Least Squares | Granger Causality Modeling | |
| AD vs. MCI | 0.003 | 0.007 | 0.159 | 0.399 |
| AD vs. NC | 0.005 | 0.003 | 0.047 | 0.375 |
| MCI vs. NC | 0.109 | 0.224 | 0.198 | 0.238 |
| Between-Group | KBI | SIP | BNP | CPL | ND | Mod |
|---|---|---|---|---|---|---|
| AD vs. MCI | 0.022 | 0.032 | 0.063 | 0.092 | 0.424 | 0.035 |
| AD vs. NC | 0.013 | 0.053 | 0.033 | 0.458 | 0.290 | 0.081 |
| MCI vs. NC | 0.042 | 0.051 | 0.242 | 0.088 | 0.076 | 0.404 |
| Between-Group | CPL | ND | Mod |
|---|---|---|---|
| AD vs. MCI | 0.214 | 0.040 | 0.081 |
| AD vs. NC | 0.063 | 0.083 | 0.173 |
| MCI vs. NC | 0.004 | 0.281 | 0.005 |
| AD (n = 140) | MCI (n = 280) | NC (n = 280) | p-value c | |
|---|---|---|---|---|
| Age a | 74.27.8 | 73.98.0 | 75.06.6 | 0.20 |
| Gender b | 70/70 | 140/140 | 140/140 | 1 |
| CDR | 0.5 | 0 | -- |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, L.; Zhao, D.; Xing, J.; Chen, Z.; Xiong, F.; Han, X. Metabolic Brain Network Analysis of FDG-PET in Alzheimer’s Disease Using Kernel-Based Persistent Features. Molecules 2019, 24, 2301. https://doi.org/10.3390/molecules24122301
Kuang L, Zhao D, Xing J, Chen Z, Xiong F, Han X. Metabolic Brain Network Analysis of FDG-PET in Alzheimer’s Disease Using Kernel-Based Persistent Features. Molecules. 2019; 24(12):2301. https://doi.org/10.3390/molecules24122301
Chicago/Turabian StyleKuang, Liqun, Deyu Zhao, Jiacheng Xing, Zhongyu Chen, Fengguang Xiong, and Xie Han. 2019. "Metabolic Brain Network Analysis of FDG-PET in Alzheimer’s Disease Using Kernel-Based Persistent Features" Molecules 24, no. 12: 2301. https://doi.org/10.3390/molecules24122301
APA StyleKuang, L., Zhao, D., Xing, J., Chen, Z., Xiong, F., & Han, X. (2019). Metabolic Brain Network Analysis of FDG-PET in Alzheimer’s Disease Using Kernel-Based Persistent Features. Molecules, 24(12), 2301. https://doi.org/10.3390/molecules24122301






