Effects of Central Loop Length and Metal Ions on the Thermal Stability of G-Quadruplexes
Abstract
1. Introduction
2. Results
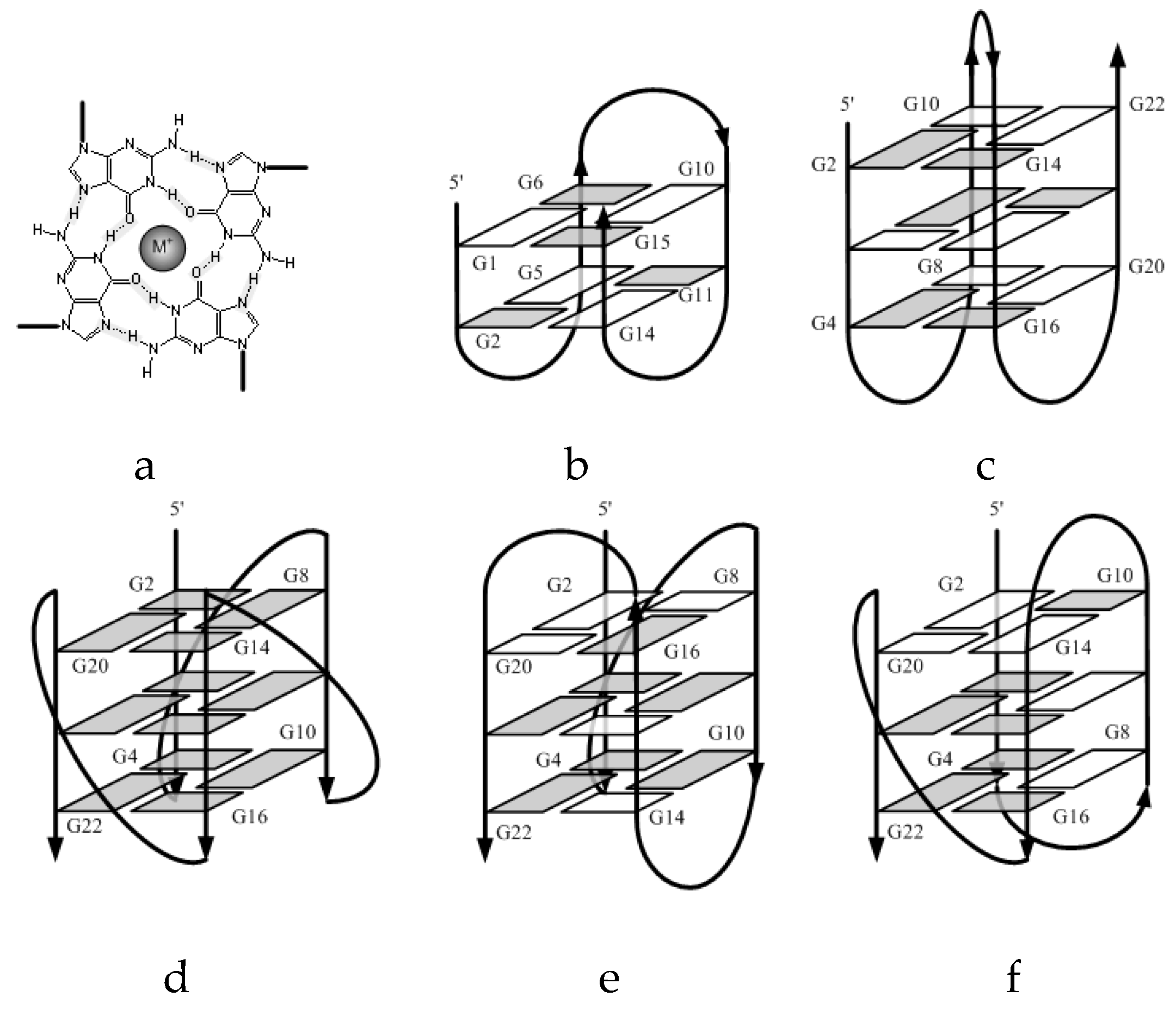
2.1. Formation of G-quadruplex
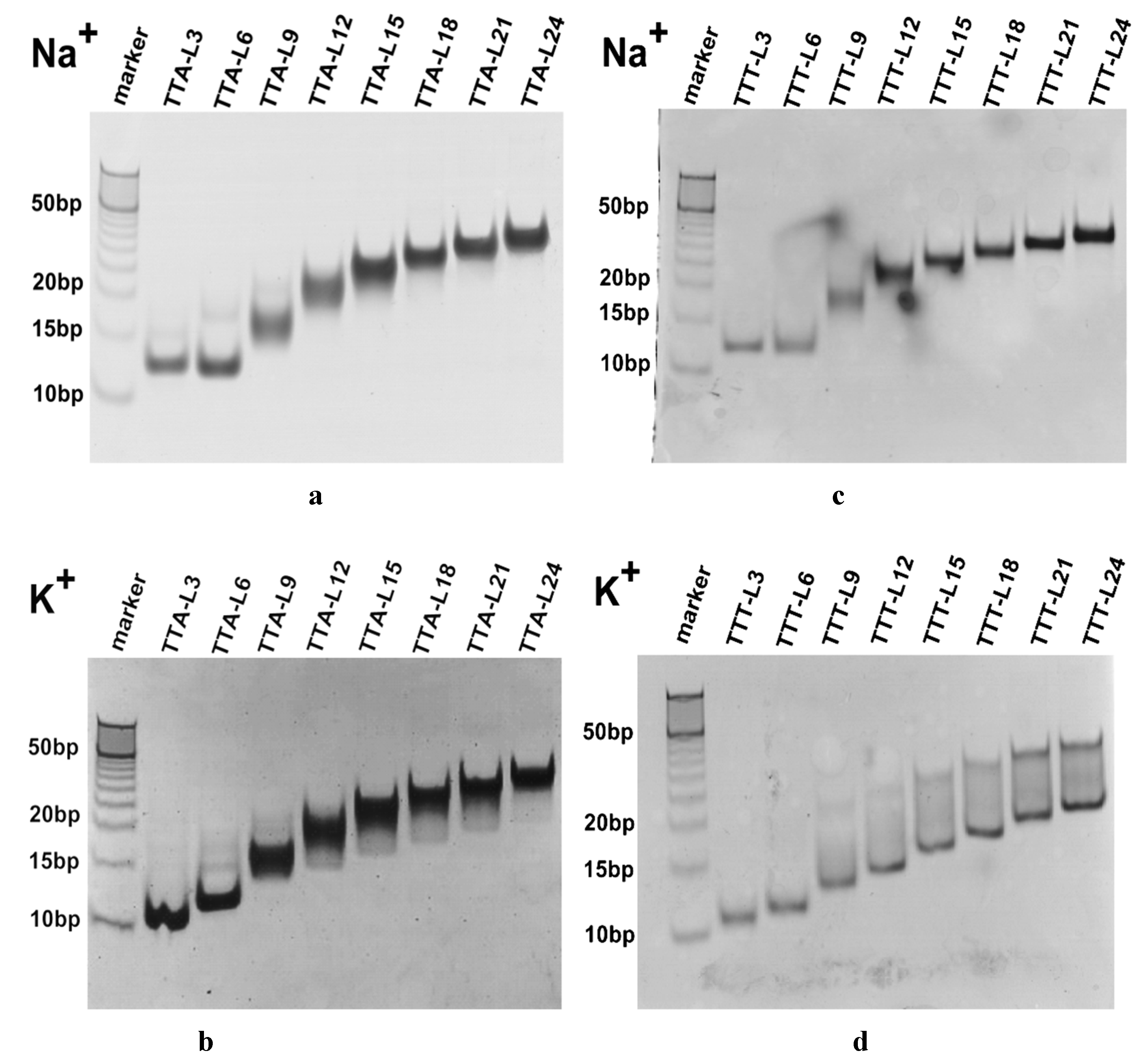
2.2. Molecularity of G4 Structures
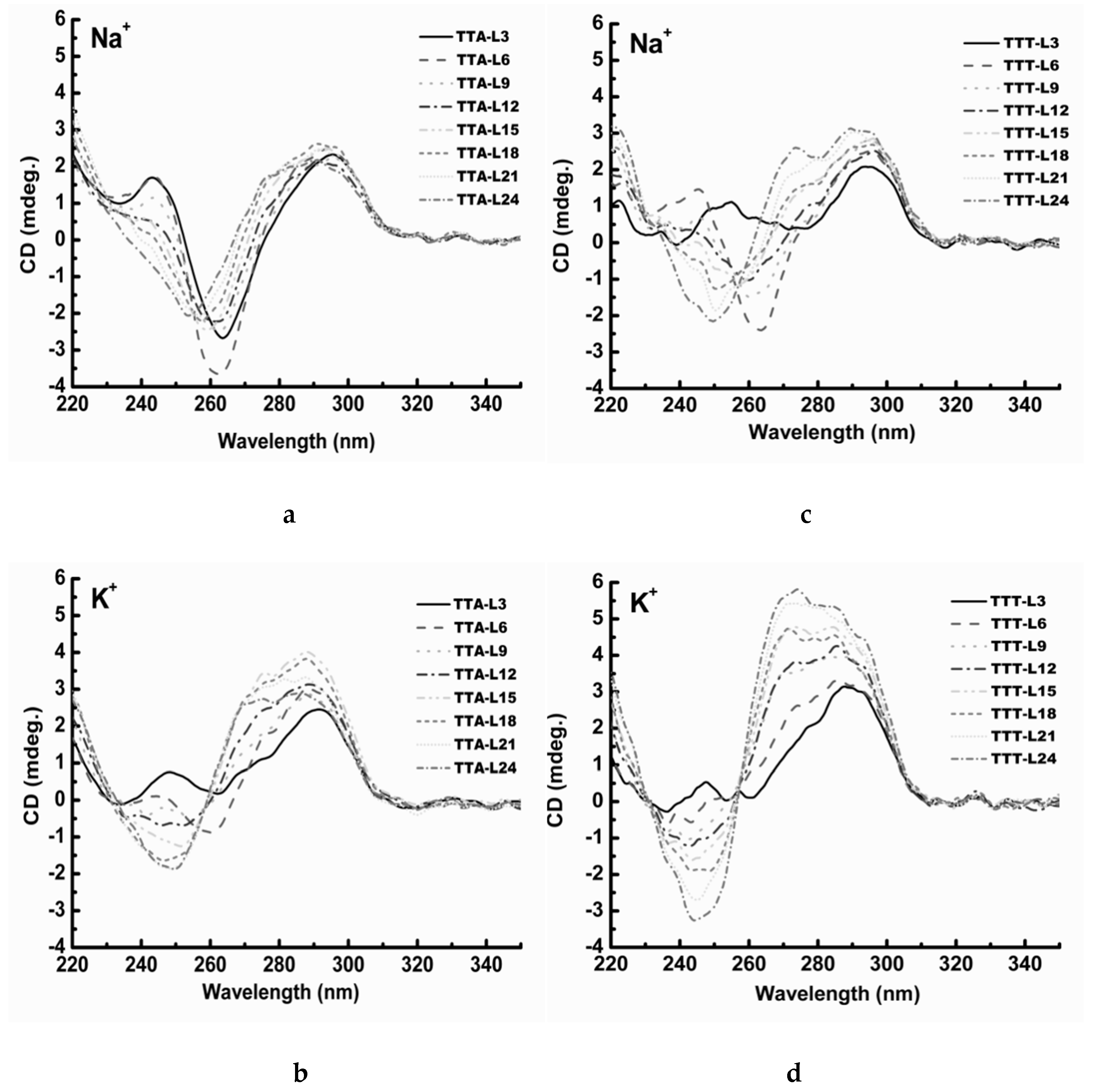
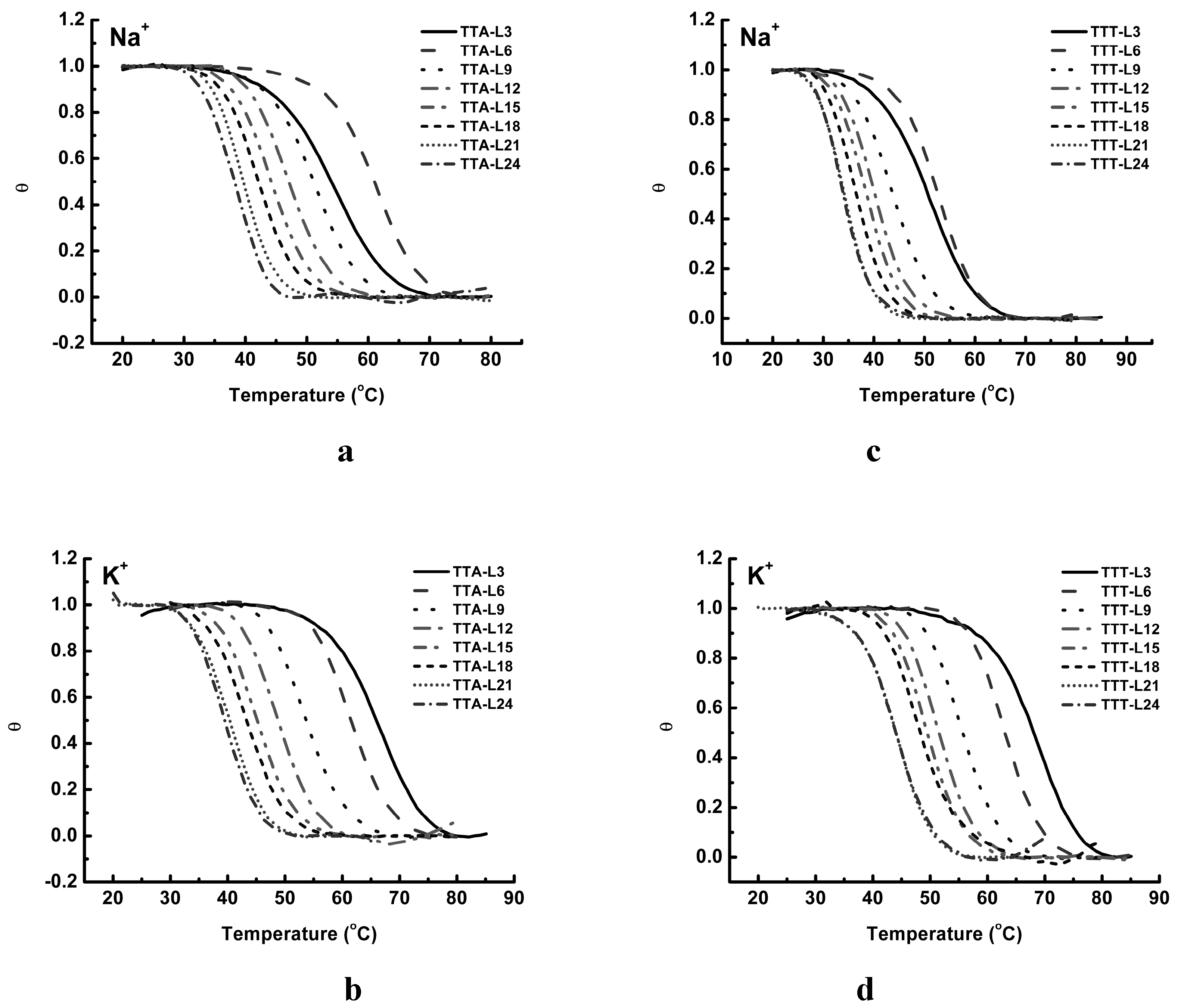
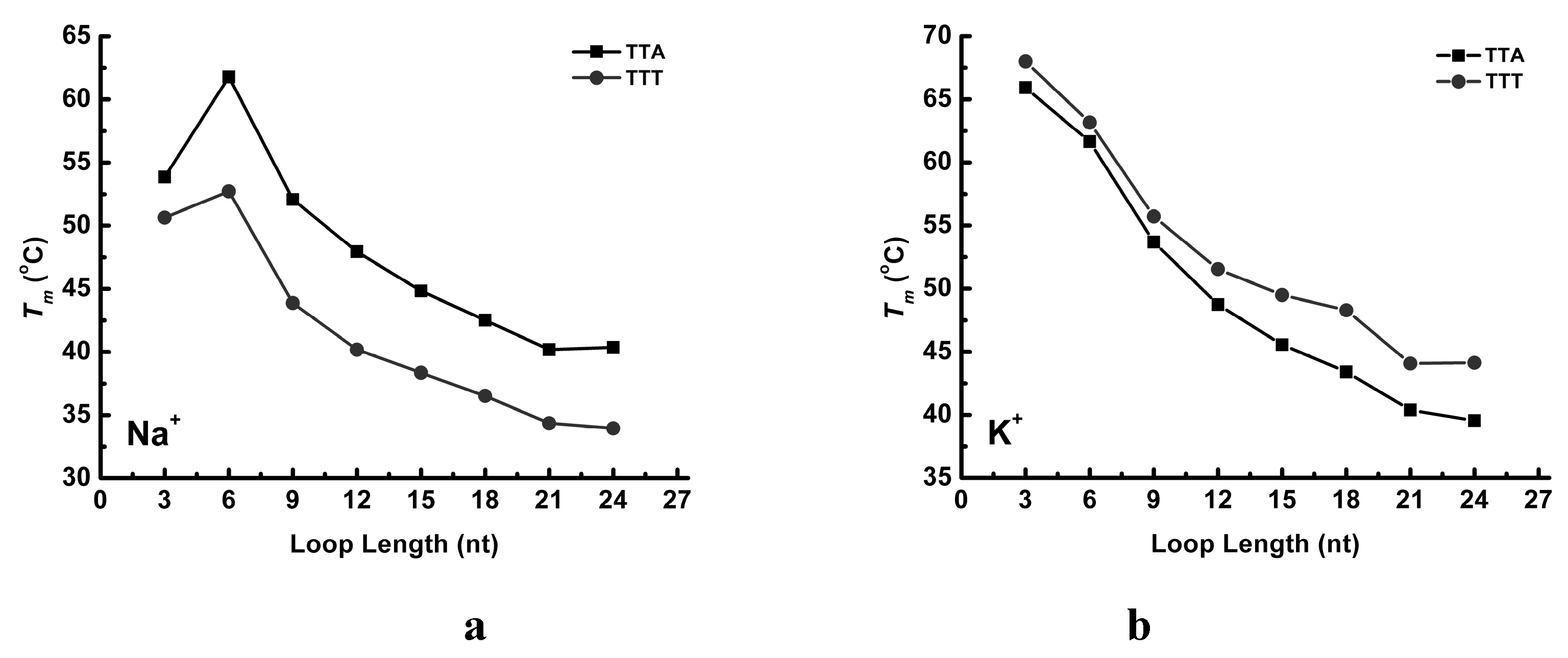
2.3. UV Melting Analysis of Structure Stability
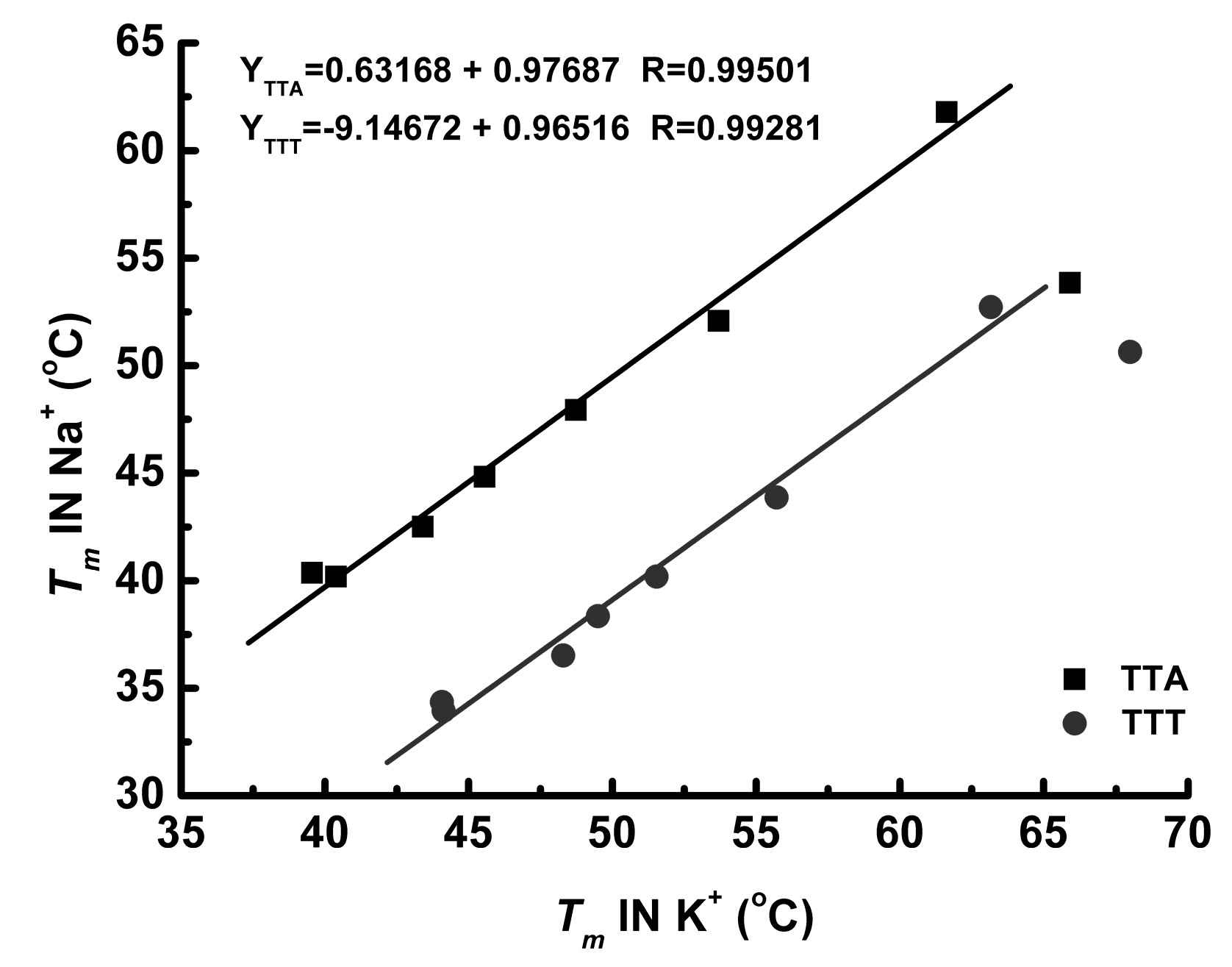
3. Discussion
3.1. Effect of the Central Loop on the G4 Structure
3.2. Effect of Cations
3.3. Thermal Properties of G4 Structures
4. Materials and Methods
4.1. Preparation of Oligonucleotides
4.2. Circular Dichroism Spectroscopy
4.3. UV Melting Measurement
4.4. Thermodynamic Parameters Calculation
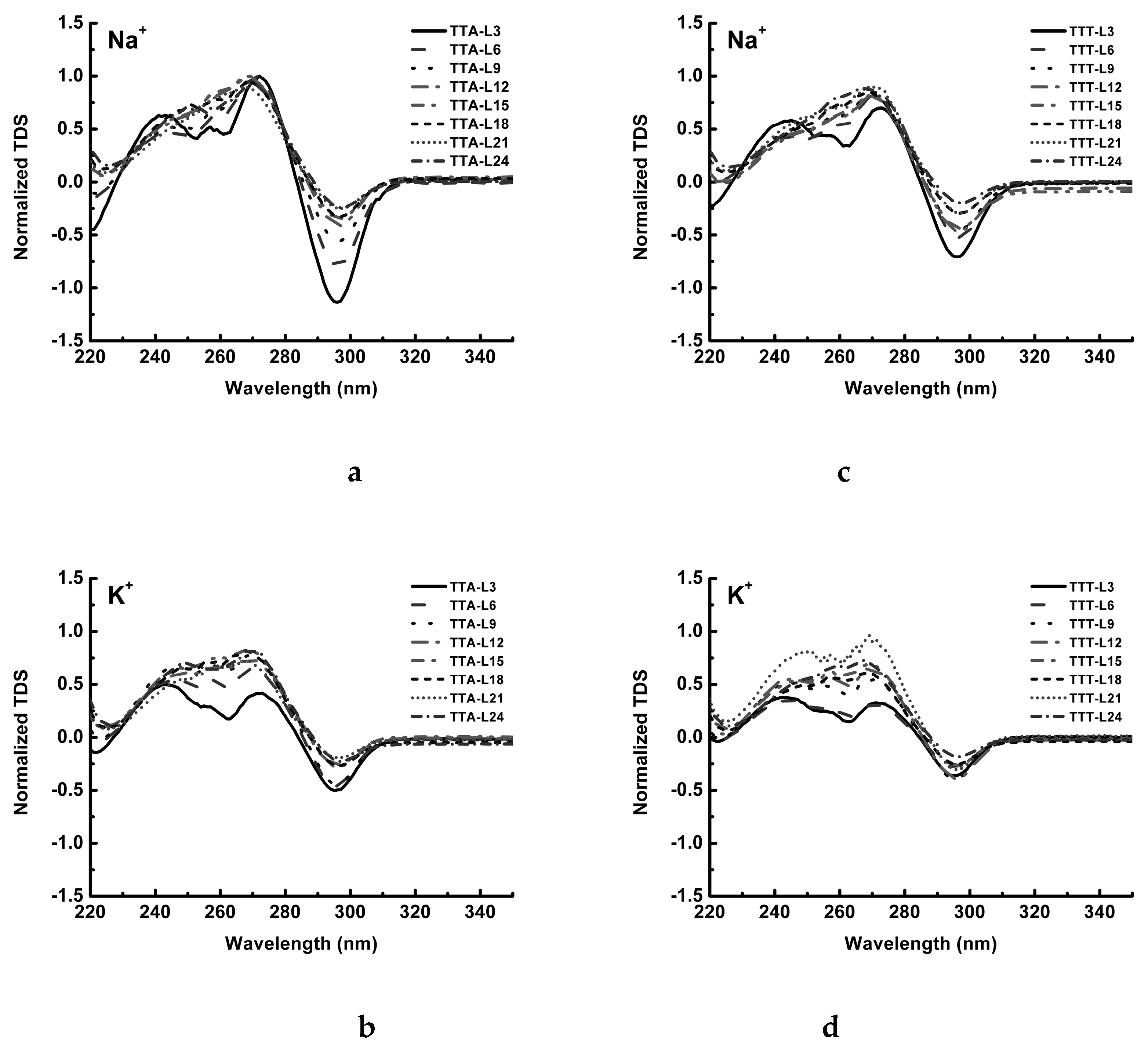
4.5. Thermal Difference Spectrum (TDS)
4.6. Non-Denatured Gel Electrophoresis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Blackburn, E.H. Structure and function of telomeres. Nature 1991, 350, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Okumus, B.; Kim, D.S.; Ha, T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 18938–18943. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sato, H.; Sannohe, Y.; Shinohara, K.; Sugiyama, H. Stable lariat formation based on a G-quadruplex scaffold. J. Am. Chem. Soc. 2008, 130, 16470–16471. [Google Scholar] [CrossRef]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef] [PubMed]
- Fernando, H.; Reszka, A.P.; Huppert, J.; Ladame, S.; Rankin, S.; Venkitaraman, A.R.; Neidle, S.; Balasubramanian, S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 2006, 45, 7854–7860. [Google Scholar] [CrossRef]
- Kumari, S.; Bugaut, A.; Balasubramanian, S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5’ UTR of the NRAS proto-oncogene. Biochemistry 2008, 47, 12664–12669. [Google Scholar] [CrossRef]
- Morris, M.J.; Basu, S. An unusually stable G-quadruplex within the 5’-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry 2009, 48, 5313–5319. [Google Scholar] [CrossRef]
- Rangan, A.; Fedoroff, O.Y.; Hurley, L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001, 276, 4640–4646. [Google Scholar] [CrossRef]
- Tornaletti, S. Transcriptional processing of G4 DNA. Mol. Carcinog. 2009, 48, 326–335. [Google Scholar] [CrossRef]
- Risitano, A.; Fox, K.R. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004, 32, 598–2606. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990, 344, 410–414. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Rachwal, P.A.; Brown, T.; Fox, K.R. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry 2007, 46, 3036–3044. [Google Scholar] [CrossRef] [PubMed]
- Viglasky, V.; Bauer, L.; Tluckova, K. Structural features of intra- and intermolecular G-quadruplexes derived from telomeric repeats. Biochemistry 2010, 49, 2110–2120. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.C.; Zhuang, J.; Ng, S.L.; New, L.L.; Hiew, S.; Guo, J.; Yang, Z.; Li, T. Conformational organizations of G-quadruplexes composed of d(G4Tn)3G4. Bioorg. Med. Chem. Lett. 2010, 20, 4689–4692. [Google Scholar] [CrossRef]
- Smirnov, I.; Shafer, R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry 2000, 39, 1462–1468. [Google Scholar] [CrossRef]
- Guedin, A.; De Cian, A.; Gros, J.; Lacroix, L.; Mergny, J.L. Sequence effects in single-base loops for quadruplexes. Biochimie 2008, 90, 686–696. [Google Scholar] [CrossRef]
- Jaumot, J.; Gargallo, R. Using principal component analysis to find correlations between loop-related and thermodyn amic variable s for G-quadruplex-forming sequences. Biochimie 2010, 92, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Findlow, I.S.; Werner, J.M.; Brown, T.; Fox, K.R. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007, 35, 4214–4222. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Maiti, S. A thermodynamic overview of naturally occurring intramolecular DNA quadruplexes. Nucleic Acids Res. 2008, 36, 5610–5622. [Google Scholar] [CrossRef]
- Guedin, A.; Alberti, P.; Mergny, J.L. Stability of intramolecular quadruplexes: Sequence effects in the central loop. Nucleic Acids Res. 2009, 37, 5559–5567. [Google Scholar] [CrossRef]
- Bugaut, A.; Balasubramanian, S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 2008, 47, 689–697. [Google Scholar] [CrossRef]
- Zhang, A.Y.; Bugaut, A.; Balasubramanian, S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry 2011, 50, 7251–7258. [Google Scholar] [CrossRef]
- Kumar, N.; Sahoo, B.; Varun, K.A.; Maiti, S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008, 36, 4433–4442. [Google Scholar] [CrossRef]
- Cang, X.; Sponer, J.; Cheatham, T.E. Explaining the varied glycosidic conformational, G-tract length and sequence preferences for anti-parallel G-quadruplexes. Nucleic Acids Res. 2011, 39, 4499–4512. [Google Scholar] [CrossRef]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- Liu, H.; Kugimiya, A.; Sakurai, T.; Katahira, M.; Uesugi, S. A comparison of the properties and the solution structure for RNA and DNA quadruplexes which contain two GGAGG sequences joined with a tetranucleotide linker. Nucleosides Nucleotides Nucleic Acids 2002, 21, 785–801. [Google Scholar] [CrossRef]
- Bardin, C.; Leroy, J.L. The formation pathway of tetramolecular G-quadruplexes. Nucleic Acids Res. 2008, 36, 477–488. [Google Scholar] [CrossRef]
- Gray, R.D.; Chaires, J.B. Isothermal folding of G-quadruplexes. Methods 2012, 57, 47–55. [Google Scholar] [CrossRef]
- Kankia, B.I.; Marky, L.A. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: Stability, heat, and hydration. J. Am. Chem. Soc. 2001, 123, 10799–10804. [Google Scholar] [CrossRef]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef]
- Qi, H.; Lin, C.P.; Fu, X.; Wood, L.M.; Liu, A.A.; Tsai, Y.C.; Chen, Y.; Barbieri, C.M.; Pilch, D.S.; Liu, L.F. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006, 66, 11808–11816. [Google Scholar] [CrossRef]
- Jing, N.; Li, Y.; Xiong, W.; Sha, W.; Jing, L.; Tweardy, D.J. G-quartet oligonucleotides: A new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004, 64, 6603–6609. [Google Scholar] [CrossRef]
- Pavlov, V.; Xiao, Y.; Gill, R.; Dishon, A.; Kotler, M.; Willner, I. Amplified chemiluminescence surface detection of DNA and telomerase activity using catalytic nucleic acid labels. Analyt. Chem. 2004, 7, 2152–2156. [Google Scholar] [CrossRef]
- Kong, D.M.; Yang, W.; Wu, J.; Li, C.X.; Shen, H.X. Structure-function study of peroxidase-like G-quadruplex-hemin complexes. Analyst 2010, 135, 321–326. [Google Scholar] [CrossRef]
- Dai, N.; Kool, E.T. Fluorescent DNA-based enzyme sensors. Chem. Soc. Rev. 2011, 40, 5756–5770. [Google Scholar] [CrossRef]
- Bourdoncle, B.; Este’ vez Torres, A.; Gosse, C.; Lacroix, L.; Vekhoff, P.; Le Saux, T.; Jullien, L.; Mergny, J.-L. Quadruplex-Based Molecular Beacons as Tunable DNA Probes. J. Am. Chem. Soc. 2006, 128, 11094–11105. [Google Scholar] [CrossRef]
- Zheng, Y.T.; Zhu, J.H.; Ma, G.; Zhu, Q.; Yang, P.; Tan, B.; Zhang, J.L.; Shen, H.X.; Xu, J.L.; Zhu, Y.Z.; et al. Preclinical assessment of the distribution, metabolism, and excretion of S-propargyl-cysteine, a novel H2S donor, in Sprague-Dawley rats. Acta Pharmacol. Sinica 2012, 33, 839–844. [Google Scholar] [CrossRef][Green Version]
- Mergny, J.L.; Phan, A.T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. UV Melting of G-Quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Xu, Y.; Noguchi, Y.; Sugiyama, H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006, 14, 5584–5591. [Google Scholar] [CrossRef]
- Dai, J.; Dexheimer, T.S.; Chen, D.; Carver, M.; Ambrus, A.; Jones, R.A.; Yang, D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006, 128, 1096–1098. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-quartet layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef]
- Phan, A.T.; Patel, D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Ford, K.G.; Neidle, S. Perturbations in DNA structure upon interaction with porphyrins revealed by chemical probes, DNA footprinting and molecular modelling. Bioorg. Med. Chem. 1995, 3, 671–677. [Google Scholar] [CrossRef]
- Hsu, S.T.; Varnai, P.; Bugaut, A.; Reszka, A.P.; Neidle, S.; Balasubramanian, S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-quartet dynamics. J. Am. Chem. Soc. 2009, 131, 13399–13409. [Google Scholar] [CrossRef]
- Tong, X.; Lan, W.; Zhang, X.; Wu, H.; Liu, M.; Cao, C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753–6763. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Ma, J.B.; Faure, A.; Andreola, M.L.; Patel, D.J. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 2005, 102, 634–639. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Gao, X.; Rando, R.F.; Hogan, M.E. Potassium-induced loop conformational transition of a potent anti-HIV oligonucleotide. J. Biomol. Struct. Dyn. 1997, 15, 573–585. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Species | Na+ | K+ | ||||||
|---|---|---|---|---|---|---|---|---|
| ΔH (kJ/mol) | −TΔS (kJ/mol) | ΔG (kJ/mol) | Tm (°C) | ΔH (kJ/mol) | −TΔS (kJ/mol) | ΔG (kJ/mol) | Tm (°C) | |
| TTA-L3 | −198.0 (±0.7) | 180.7 (±0.6) | −17.3 (±0.1) | 53.8 (±0.1) | −221.3 (±6.6) | 194.7 (±5.8) | −26.6 (±0.7) | 65.9 (±0.0) |
| TTA-L6 | −212.2 (±5.4) | 188.9 (±4.9) | −23.4 (±0.5) | 61.8 (±0.0) | −300.3 (±4.5) | 267.4 (±3.9) | −32.9 (±0.6) | 61.6 (±0.2) |
| TTA-L9 | −243.9 (±2.9) | 223.8 (±2.7) | −20.2 (±0.2) | 52.1 (±0.0) | −265.4 (±2.9) | 242.0 (±2.7) | −23.3 (±0.2) | 53.7 (±0.0) |
| TTA-L12 | −261.8 (±1.1) | 243.2 (±1.1) | −18.7 (±0.1) | 47.9 (±0.0) | −299.6 (±3.7) | 277.4 (±3.4) | −22.2 (±0.3) | 48.7 (±0.0) |
| TTA-L15 | −283.9 (±0.8) | 266.3 (±0.8) | −17.7 (±0.0) | 44.8 (±0.0) | −285.2 (±2.8) | 266.8 (±2.6) | −18.4 (±0.2) | 45.6 (±0.1) |
| TTA-L18 | −294.1 (±2.3) | 277.8 (±2.1) | −16.3 (±0.2) | 42.5 (±0.0) | −275.8 (±1.7) | 259.6 (±1.7) | −16.2 (±0.1) | 43.4 (±0.1) |
| TTA-L21 | −304.5 (±2.1) | 289.7 (±1.9) | −14.8 (±0.2) | 40.2 (±0.0) | −301.2 (±3.7) | 286.4 (±3.5) | −14.8 (±0.2) | 40.4 (±0.0) |
| TTA-L24 | −329.2 (±0.7) | 313.0 (±0.8) | −16.1 (±0.0) | 40.4 (±0.0) | −310.0 (±6.7) | 295.4 (±6.3) | −14.5 (±0.3) | 39.6 (±0.0) |
| Notation | Na+ | K+ | ||||||
|---|---|---|---|---|---|---|---|---|
| ΔH (kJ/mol) | −TΔS (kJ/mol) | ΔG (kJ/mol) | Tm (°C) | ΔH (kJ/mol) | −TΔS (kJ/mol) | ΔG (kJ/mol) | Tm (°C) | |
| TTT-L3 | −194.4 (±1.3) | 179.2 (±1.2) | −15.3 (±0.1) | 50.6 (±0.0) | −217.9 (±2.7) | 190.5 (±2.4) | −27.4 (±0.3) | 67.9 (±0.1) |
| TTT-L6 | −244.8 (±3.4) | 224.1 (±3.1) | −20.7 (±0.3) | 52.7 (±0.0) | −309.7 (±2.1) | 274.5 (±1.9) | −35.2 (±0.2) | 63.2 (±0.0) |
| TTT-L9 | −235.2 (±2.0) | 221.1 (±1.8) | −14.0 (±0.2) | 43.9 (±0.0) | −295.8 (±2.5) | 268.0 (±2.3) | −27.7 (±0.3) | 55.7 (±0.0) |
| TTT-L12 | −251.2 (±0.8) | 238.9 (±0.8) | −12.2 (±0.1) | 40.2 (±0.0) | −290.9 (±1.6) | 267.1 (±1.5) | −23.8 (±0.1) | 51.5 (±0.0) |
| TTT-L15 | −268.5 (±1.9) | 256.9 (±1.7) | −11.5 (±0.2) | 38.4 (±0.1) | −290.5 (±1.1) | 268.4 (±1.1) | −22.1 (±0.1) | 49.5 (±0.0) |
| TTT-L18 | −287.2 (±4.3) | 276.4 (±4.2) | −10.7 (±0.2) | 36.5 (±0.1) | −229.0 (±2.3) | 212.5 (±2.2) | −16.5 (±0.2) | 48.3 (±0.0) |
| TTT-L21 | −303.8 (±2.9) | 294.4 (±2.8) | −9.3 (±0.2) | 34.4 (±0.1) | −276.9 (±1.1) | 260.5 (±1.0) | −16.4 (±0.0) | 44.1 (±0.0) |
| TTT-L24 | −252.1 (±1.8) | 245.1 (±1.7) | −7.0 (±0.1) | 33.9 (±0.0) | −265.2 (±3.0) | 249.2 (±2.8) | −15.9 (±0.1) | 44.1 (±0.1) |
| Species | Sequence | Central Loop Length |
|---|---|---|
| TTA-L3 | AG3-TTA-G3-(TTA)1-G3-TTA-G3T | 3 |
| TTA-L6 | AG3-TTA-G3-(TTA)2-G3-TTA-G3T | 6 |
| TTA-L9 | AG3-TTA-G3-(TTA)3-G3-TTA-G3T | 9 |
| TTA-L12 | AG3-TTA-G3-(TTA)4-G3-TTA-G3T | 12 |
| TTA-L15 | AG3-TTA-G3-(TTA)5-G3-TTA-G3T | 15 |
| TTA-L18 | AG3-TTA-G3-(TTA)6-G3-TTA-G3T | 18 |
| TTA-L21 | AG3-TTA-G3-(TTA)7-G3-TTA-G3T | 21 |
| TTA-L24 | AG3-TTA-G3-(TTA)8-G3-TTA-G3T | 24 |
| TTT-L3 | AG3-TTT-G3-(TTA)1-G3-TTT-G3T | 3 |
| TTT-L6 | AG3-TTT-G3-(TTA)2-G3-TTT-G3T | 6 |
| TTT-L9 | AG3-TTT-G3-(TTA)3-G3-TTT-G3T | 9 |
| TTT-L12 | AG3-TTT-G3-(TTA)4-G3-TTT-G3T | 12 |
| TTT-L15 | AG3-TTT-G3-(TTA)5-G3-TTT-G3T | 15 |
| TTT-L18 | AG3-TTT-G3-(TTA)6-G3-TTT-G3T | 18 |
| TTT-L21 | AG3-TTT-G3-(TTA)7-G3-TTT-G3T | 21 |
| TTT-L24 | AG3-TTT-G3-(TTA)8-G3-TTT-G3T | 24 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, F.; Ma, Y.; Guan, Y. Effects of Central Loop Length and Metal Ions on the Thermal Stability of G-Quadruplexes. Molecules 2019, 24, 1863. https://doi.org/10.3390/molecules24101863
Hao F, Ma Y, Guan Y. Effects of Central Loop Length and Metal Ions on the Thermal Stability of G-Quadruplexes. Molecules. 2019; 24(10):1863. https://doi.org/10.3390/molecules24101863
Chicago/Turabian StyleHao, Fengjin, Yushu Ma, and Yifu Guan. 2019. "Effects of Central Loop Length and Metal Ions on the Thermal Stability of G-Quadruplexes" Molecules 24, no. 10: 1863. https://doi.org/10.3390/molecules24101863
APA StyleHao, F., Ma, Y., & Guan, Y. (2019). Effects of Central Loop Length and Metal Ions on the Thermal Stability of G-Quadruplexes. Molecules, 24(10), 1863. https://doi.org/10.3390/molecules24101863




