Binding of Small Molecules to G-quadruplex DNA in Cells Revealed by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci
Abstract
1. Introduction
2. Results
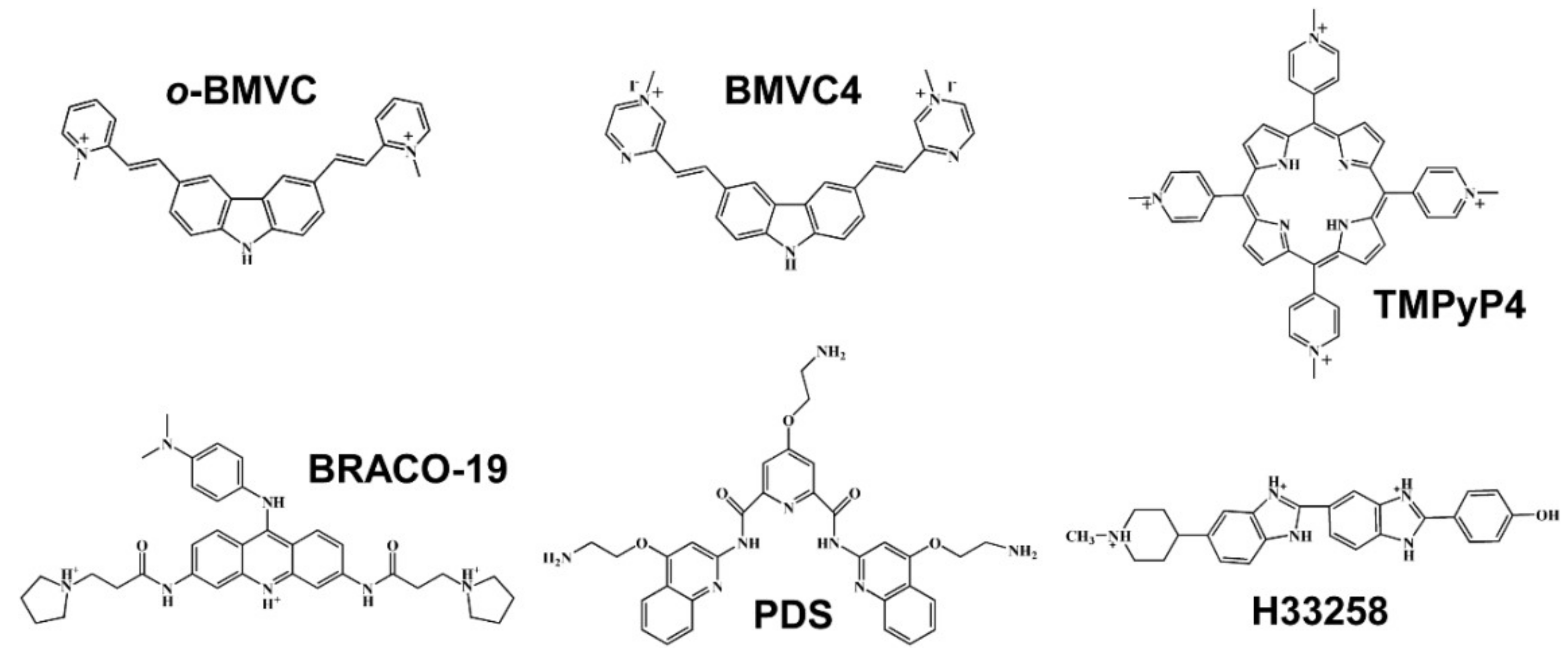
2.1. Study of G4 Ligands In Vitro
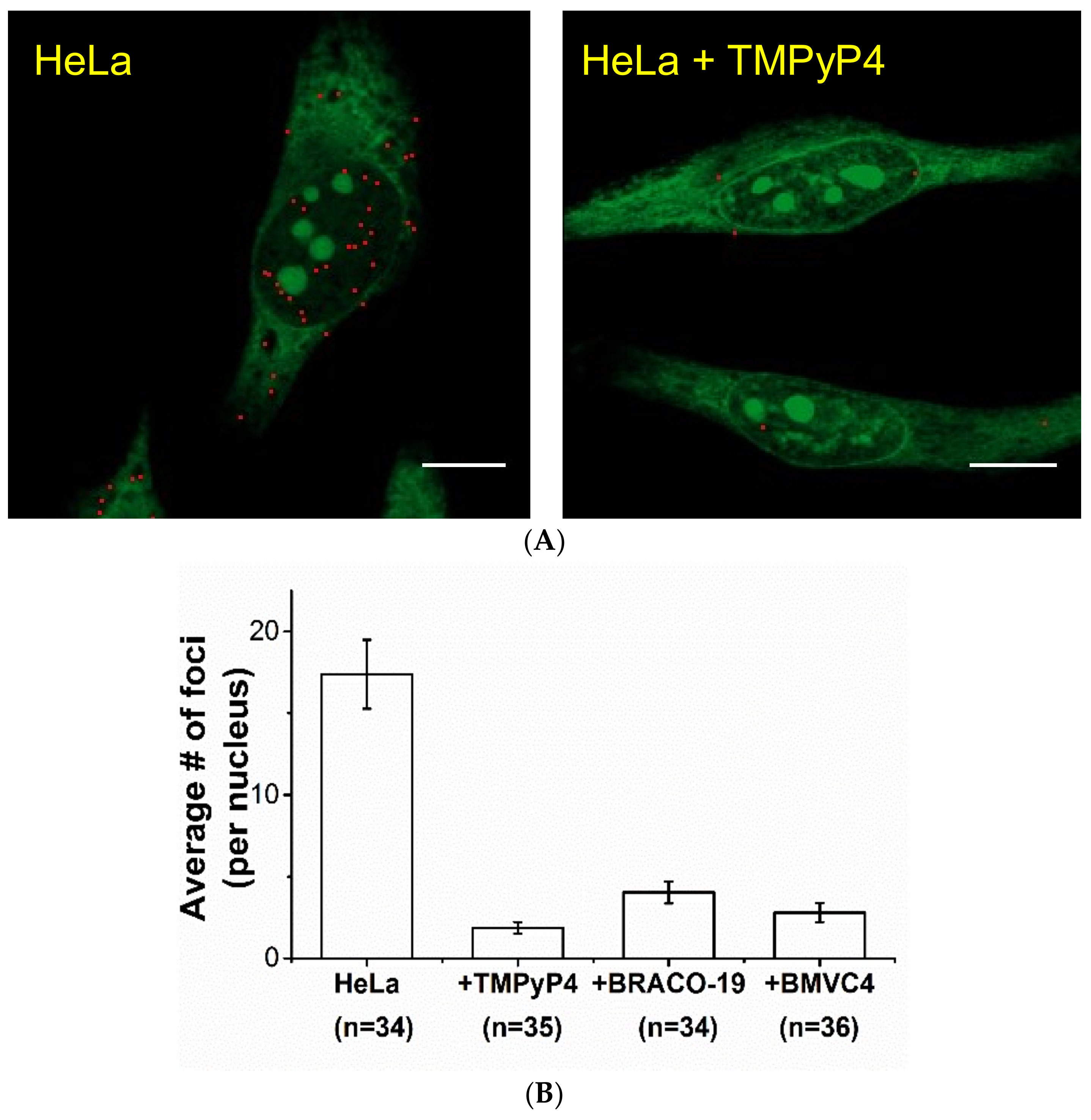
2.2. Study of G4 Ligands in Cells
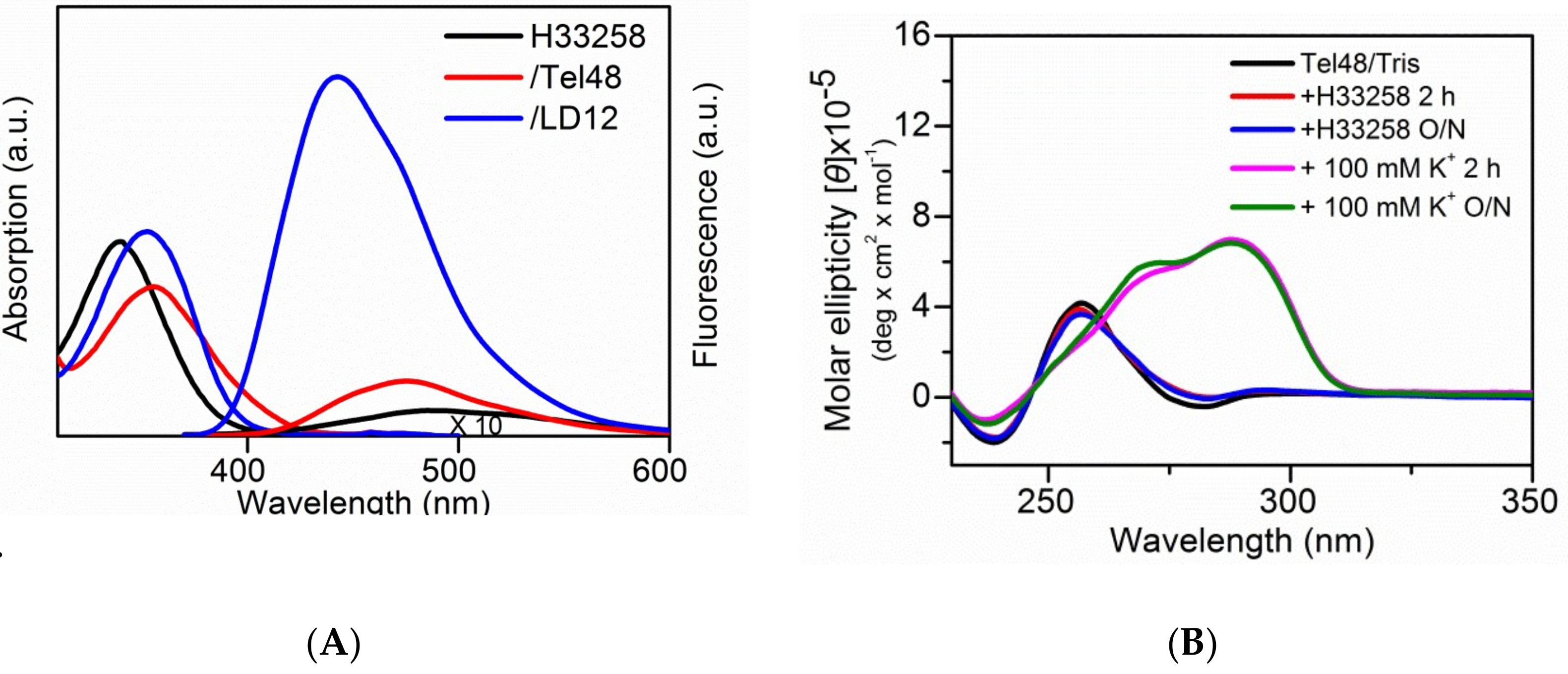
2.3. Study of the Effect of Hoechst 33258 on G4 Foci in Cells
3. Discussion
4. Materials and Methods
4.1. Chemical and Sample Preparation
4.2. Circular Dichroism (CD)
4.3. Kinetic Binding Experiment
4.4. Nuclear Magnetic Resonance (NMR) Spectroscopy
4.5. Polyacrylamide Gel Electrophoresis (PAGE)
4.6. Cell Cultures
4.7. Fluorescence Lifetime Imaging Microscopy (FLIM)
4.8. Quantitative Analysis of o-BMVC Foci
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 2008, 48, 2013–2018. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Lipps, H.J.; Rhodes, D. G-quadruplex structure: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Chien, C.-H.; Chu, J.-F.; Huang, W.-C.; Lin, M.-Y.; Chang, C.-C.; Chang, T.-C. Fluorescent probe for visualizing guanine-quadruplex DNA by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 2013, 18, 101309. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Chen, W.-W.; Chu, I.-T.; Wang, C.-L.; Chang, C.-C.; Lin, M.-C.; Lou, P.-J.; Chang, T.-C. The G-quadruplex fluorescent probe 3,6-bis(1-methyl-2-vinylpyridinium) carbazole diiodide as a biosensor for human cancers. Sci. Rep. 2018, 8, 16082. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Rankin, S.; Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Besnard, E. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012, 19, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-M.; Huang, F.-C.; Kuo, M.-H.; Wang, Z.-F.; Tseng, T.-Y.; Chang, L.-C.; Yen, S.-J.; Chang, T.-C.; Lin, J.-J. Inhibition of cancer cell migration and invasion through suppressing the Wnt1-mediating signal pathway by G-quadruplex structure stabilizers. J. Biol. Chem. 2014, 289, 14612–14623. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-T.; Wang, Z.-F.; Chu, I.-T.; Kuan, Y.-M.; Li, M.-H.; Huang, M.-C.; Chiang, P.-C.; Chang, T.-C.; Chen, C.-T. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J. Biol. Chem. 2017, 292, 20859–20870. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wang, W.; Yang, C.; Kang, T.-S.; Leung, C.-H.; Ma, D.-L. Iridium(III) complexes with 1,10-phenanthroline-based N∧N ligands as highly selective luminescent G-quadruplex probes and application for switch-on ribonuclease H. detection. J. Mater. Chem. B 2016, 4, 6791–6796. [Google Scholar] [CrossRef]
- Wheelhouse, R.T.; Sun, D.; Han, H.; Han, F.X.; Hurley, L.H. Cationic porphyrins as telomerase inhibitors: The interaction of tetra (N-methyl-4-pyridyl) porphyrin with quadruplex DNA. J. Am. Chem. Soc. 1998, 120, 3261–3262. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef]
- Huang, F.-C.; Chang, C.-C.; Wang, J.-M.; Chang, T.-C.; Lin, J.-J. Induction of senescence in cancer cells by a G-quadruplex stabilizer BMVC4 is independent of its telomerase inhibitory activity. Brit. J. Pharm. 2012, 167, 393–406. [Google Scholar] [CrossRef]
- Kim, M.Y.; Vankayalapati, H.; Shin-ya, K.; Wierzba, K.; Hurley, L.H. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G.-quadruplex. J. Am. Chem. Soc. 2002, 124, 2098–2099. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Promontorio, R.; Hampel, S.M.; Micco, M.; Neidle, S.; Parkinson, G.N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. [Google Scholar] [CrossRef]
- Huang, W.-C.; Tseng, T.-Y.; Chen, Y.-T.; Chang, C.-C.; Wang, Z.-F.; Wang, C.-L.; Hsu, T.-N.; Li, P.-T.; Chen, C.-T.; Lin, J.-J.; et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015, 43, 10102–10113. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Maiti, S. Effect of loop orientation on quadruplex-TMPyP4 interaction. J. Phys. Chem. B 2008, 112, 8151–8159. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Chaudhury, N.K.; Chowdhury, S. Hoechst 33258 binds to G-quadruplex in the promoter region of human c-MYC. Biochem. Biophys. Res. Commun. 2003, 310, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-F.; Li, M.-H.; Chen, W.-W.; Hsu, S.-T.; Chang, T.-C. A novel transition pathway of ligand-induced topological conversion from hybrid forms to parallel forms of human telomeric G.-quadruplexes. Nucleic Acids Res. 2016, 44, 3958–3968. [Google Scholar] [CrossRef] [PubMed]
- Pagano, B.; Mattia, C.A.; Giancola, C. Applications of isothermal titration calorimetry in biophysical studies of G.-quadruplexes. Int. J. Mol. Sci. 2009, 10, 2935–2957. [Google Scholar] [CrossRef]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef]
- Kotar, A.; Wang, B.; Shivalingam, A.; Gonzalez-Garcia, J.; Vilar, R.; Plavec, J. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. Int. Ed. 2016, 55, 12508–12511. [Google Scholar] [CrossRef]
- Otsu, N.A. Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cys. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.-C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Hamon, F.; Teulade-Fichou, M.P. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal. Bioanal. Chem. 2011, 400, 3419–3427. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome Atlas. Cell Rep. 2018, 23, 239–254. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of o-BMVC and BMVC4 are available from the authors upon request. |



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, T.-Y.; Chu, I.-T.; Lin, S.-J.; Li, J.; Chang, T.-C. Binding of Small Molecules to G-quadruplex DNA in Cells Revealed by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci. Molecules 2019, 24, 35. https://doi.org/10.3390/molecules24010035
Tseng T-Y, Chu I-T, Lin S-J, Li J, Chang T-C. Binding of Small Molecules to G-quadruplex DNA in Cells Revealed by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci. Molecules. 2019; 24(1):35. https://doi.org/10.3390/molecules24010035
Chicago/Turabian StyleTseng, Ting-Yuan, I-Te Chu, Shang-Jyun Lin, Jie Li, and Ta-Chau Chang. 2019. "Binding of Small Molecules to G-quadruplex DNA in Cells Revealed by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci" Molecules 24, no. 1: 35. https://doi.org/10.3390/molecules24010035
APA StyleTseng, T.-Y., Chu, I.-T., Lin, S.-J., Li, J., & Chang, T.-C. (2019). Binding of Small Molecules to G-quadruplex DNA in Cells Revealed by Fluorescence Lifetime Imaging Microscopy of o-BMVC Foci. Molecules, 24(1), 35. https://doi.org/10.3390/molecules24010035





