Characterization of α-Glucosidase Inhibitors from Clinacanthus nutans Lindau Leaves by Gas Chromatography-Mass Spectrometry-Based Metabolomics and Molecular Docking Simulation
Abstract
1. Introduction
2. Results
2.1. α-Glucosidase Inhibitory Activity
2.2. GC-MS Chromatogram of C. nutans Leaves Extracts
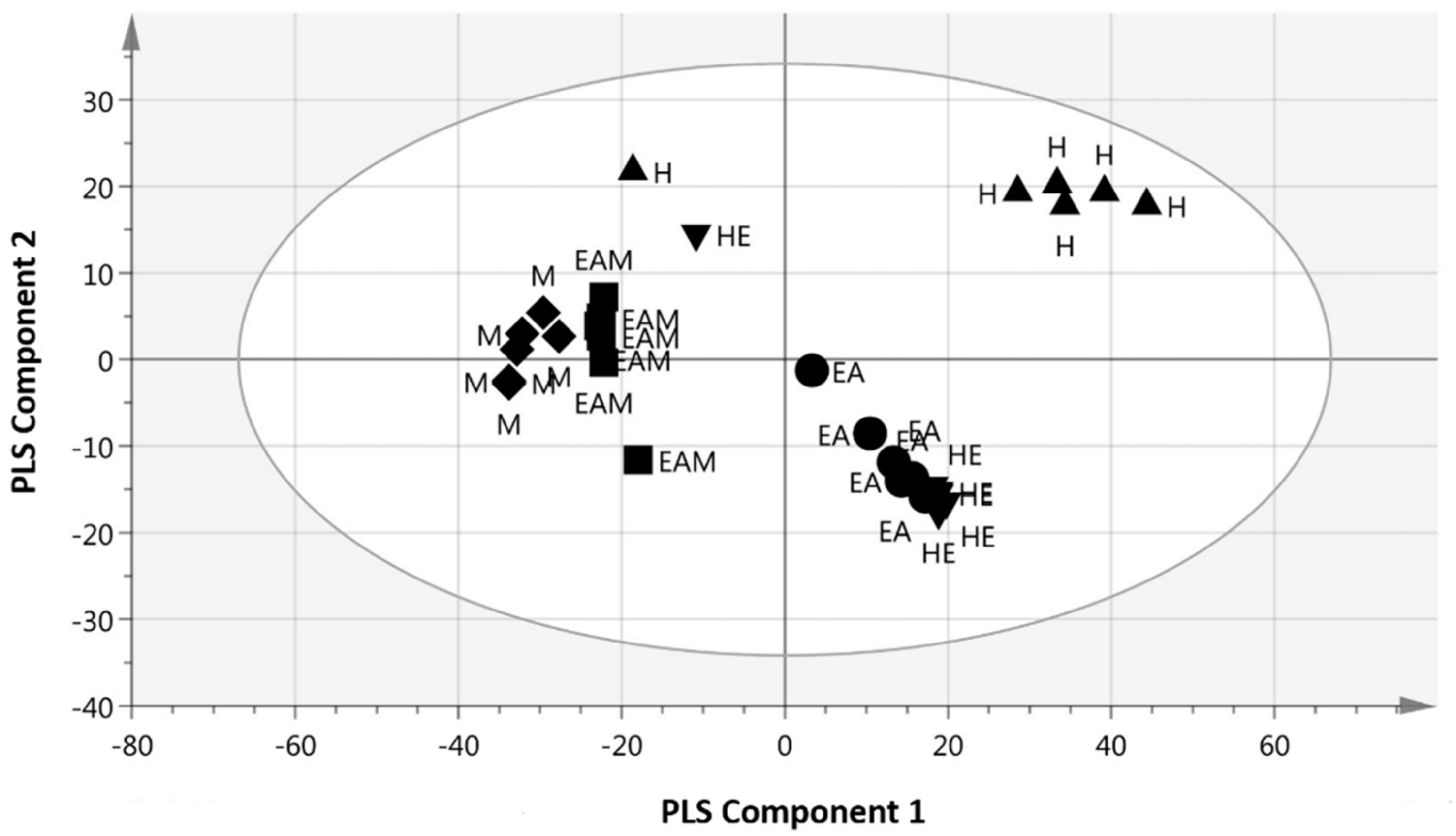
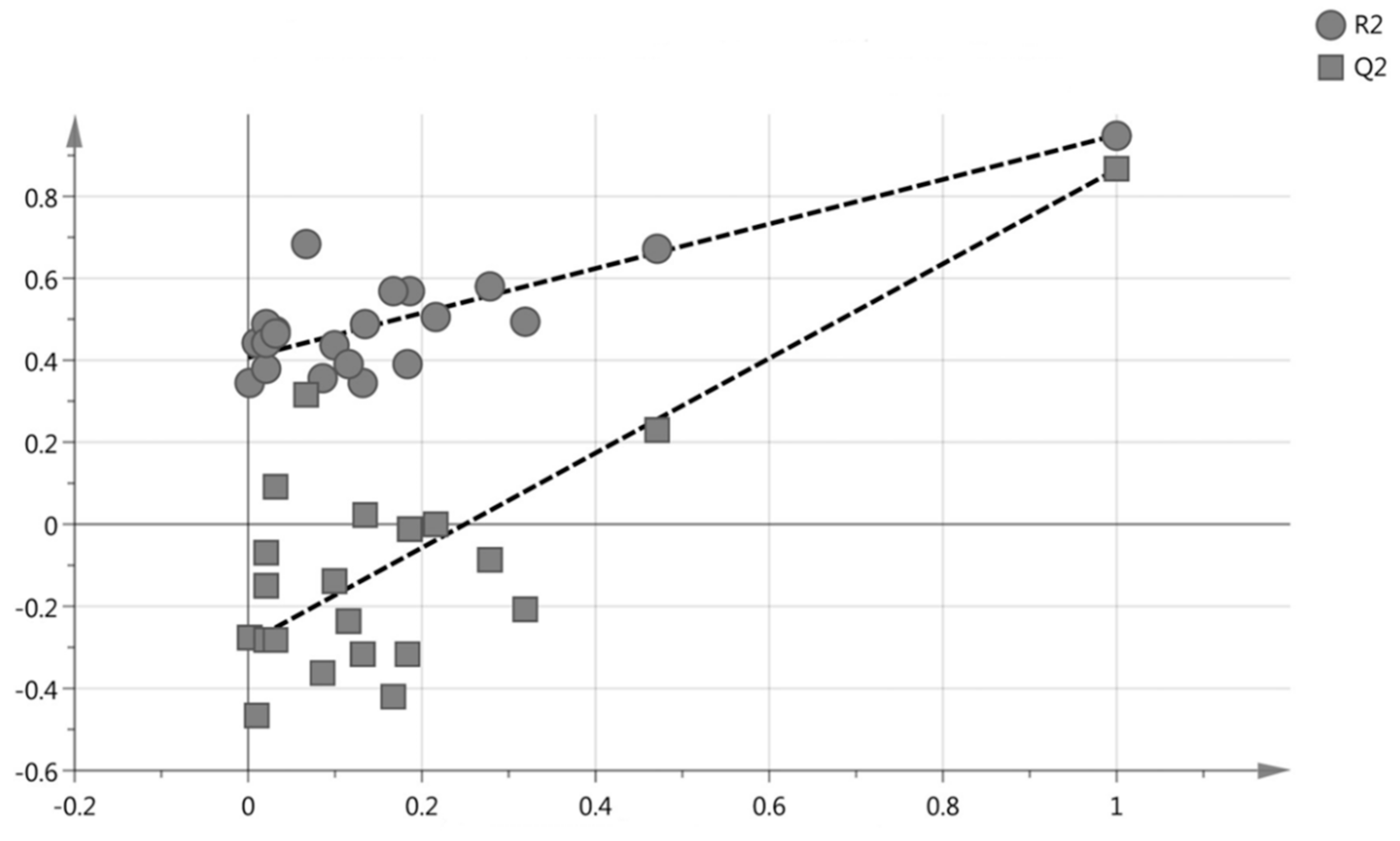
2.3. Multivariate Data Analysis
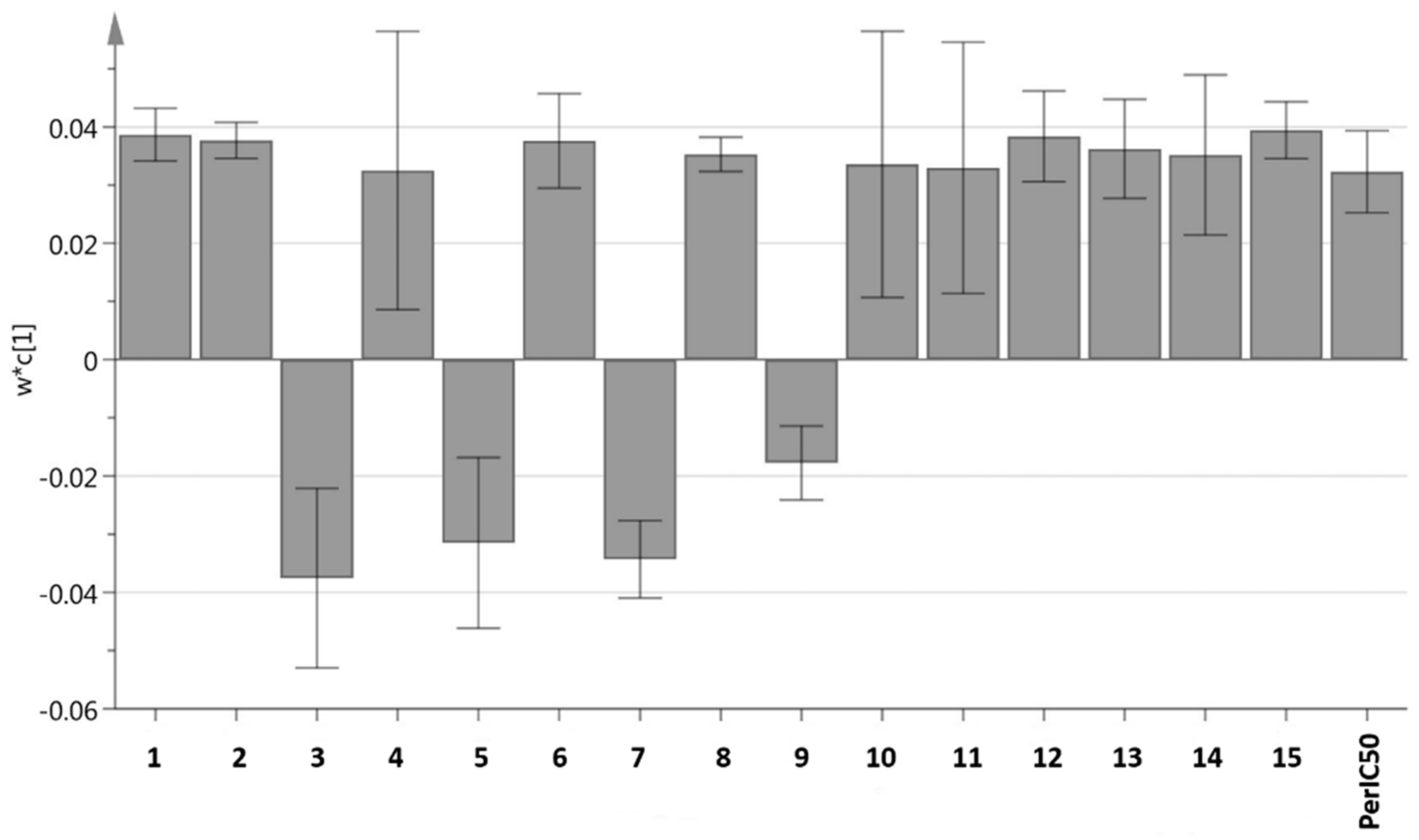
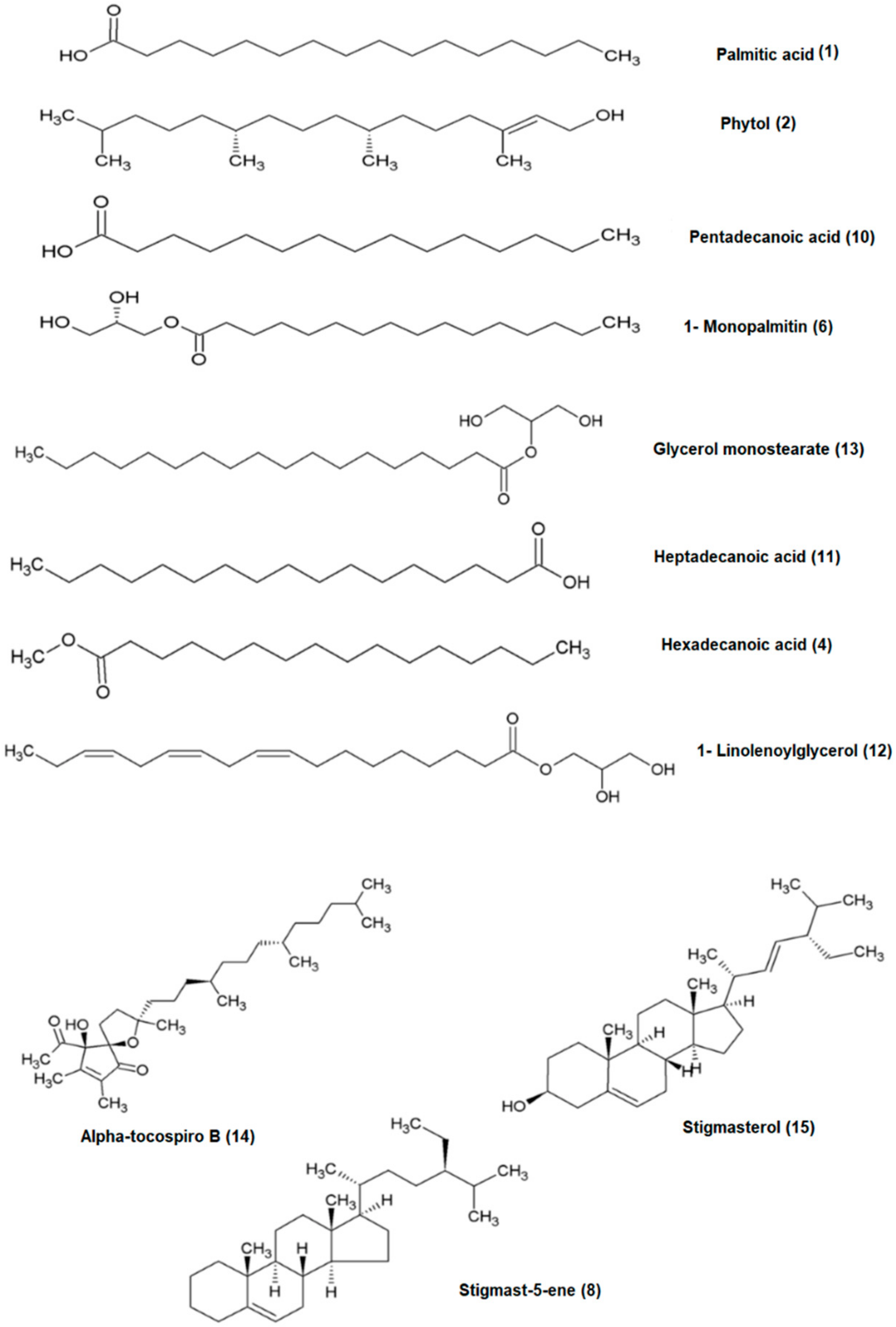
2.4. Bioactivity Confirmation and Quantification of Some Bioactive Compounds
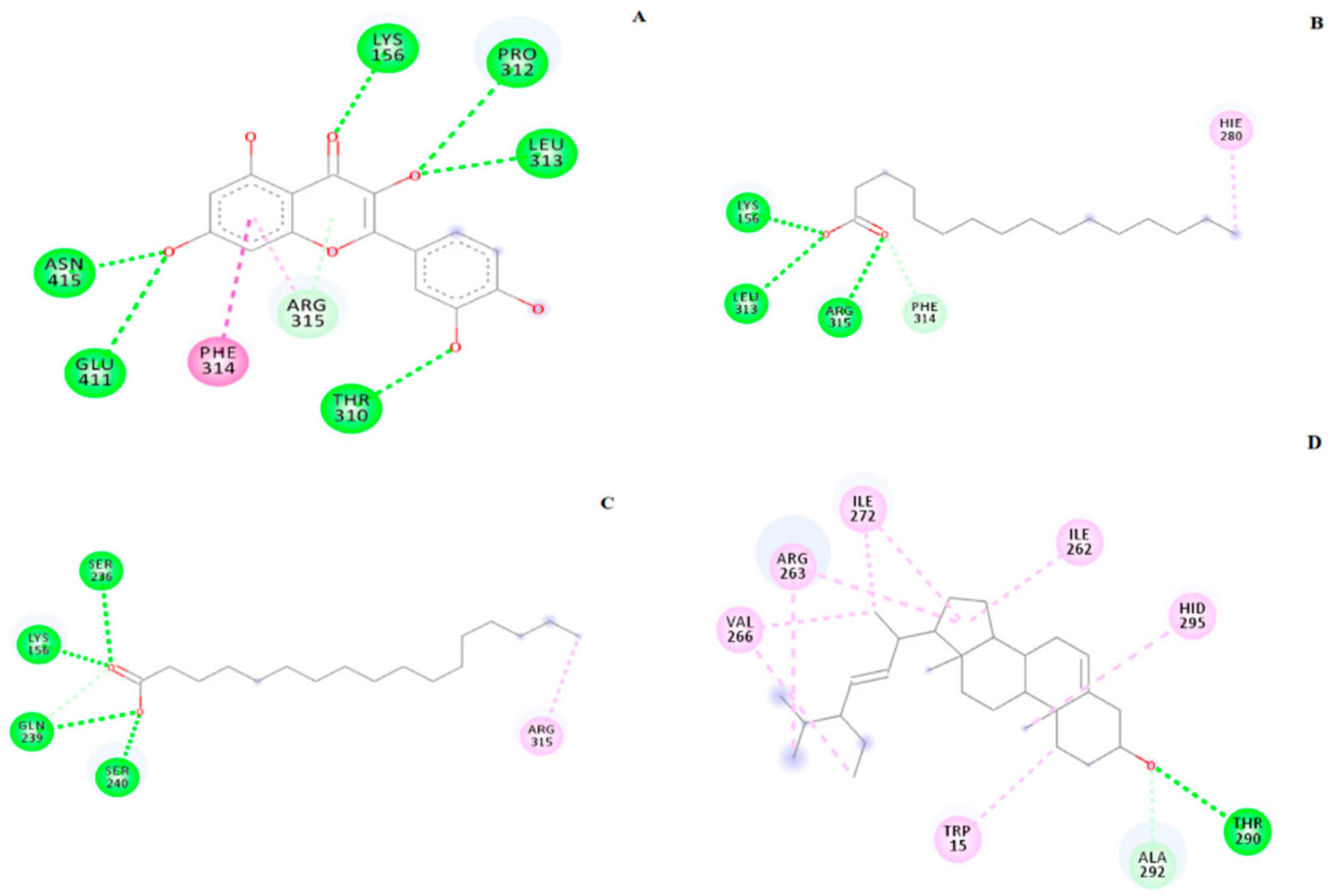
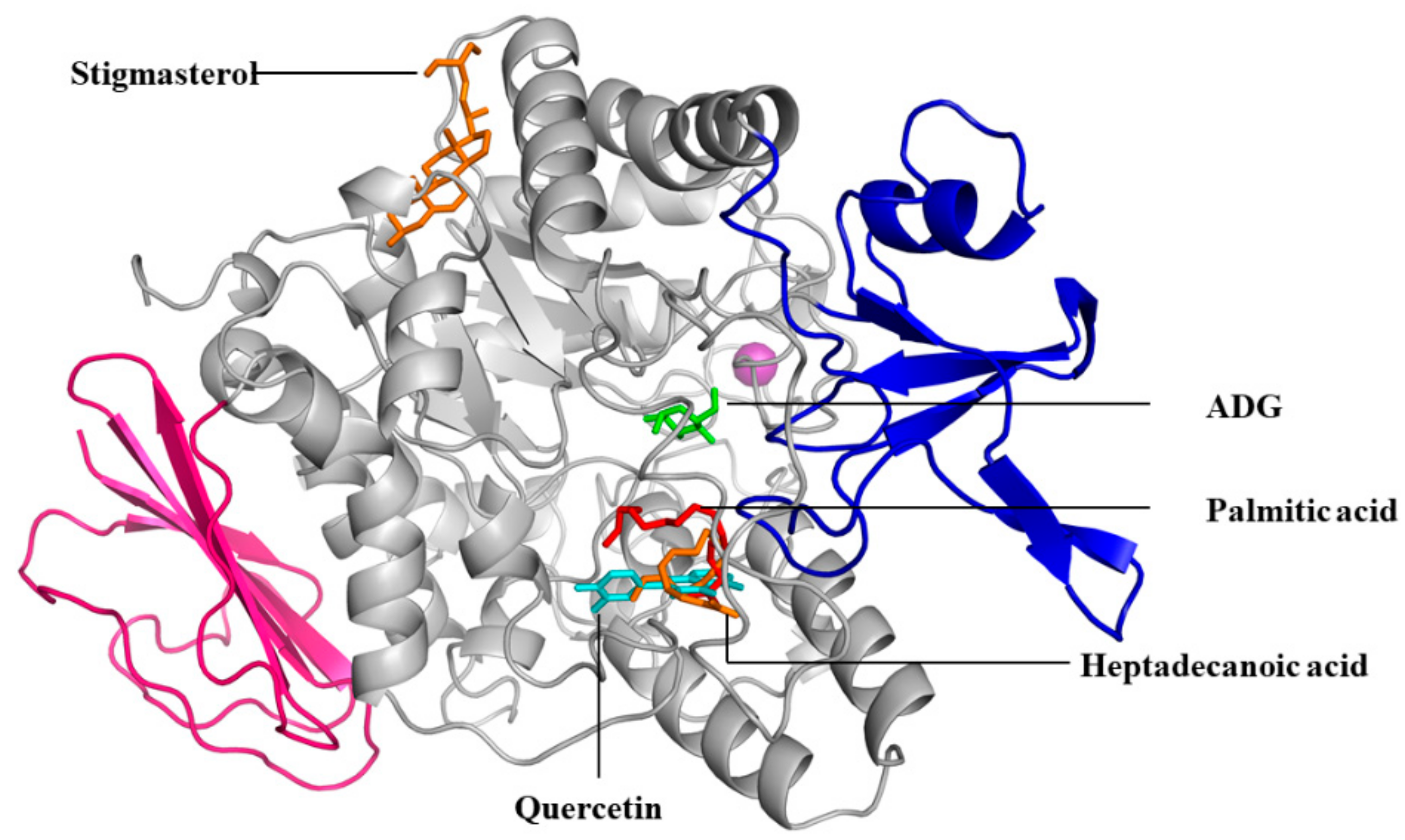
2.5. Molecular Docking
3. Discussion
4. Materials and Methodology
4.1. Materials
4.2. Sample Collection
4.3. Preparation of Extract
4.4. α-Glucosidase Inhibition Assay
4.5. Derivatization Procedure
4.6. GC-MS Analysis
4.7. Quantification of Active Phytoconstituents
4.7.1. Development of Standard Curve of the Reference
4.7.2. Determination of LOD and LOQ Concentration
4.7.3. Determination of Recovery of the Internal Standard (Methyl Nonadecanoate)
Development of Standard Curve of the Internal Standard
Calculation of the Recovery of the Internal Standards
4.7.4. Determination of Targeted Metabolites Concentration in the Sample
4.8. Molecular Docking
4.9. Data Processing and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Javadi, N.; Abas, Q.; Hamid, A.A.; Simoh, A.; Shaari, K.; Ismail, I.S.; Mediani, A.; Khatib, A. GC-MS-based metabolite profiling of Cosmos caudate Leaves possessing alpha-glucosidase inhibitory activity. J. Food Sci. 2014, 79, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, C.D.; Jain, A. Diabetes Mellitus: A Review. Int. J. Pure Appl. Biosci. 2015, 3, 224–230. [Google Scholar]
- Mohan, P.; Nandhakumar, L. Role of various flavonoids: Hypotheses on novel approach to treat diabetes. J. Med. Hypotheses Ideas 2014, 8, 1–6. [Google Scholar] [CrossRef]
- Shihabudeen, M.S.H.; Priscilla, D.H.; Thirumurugan, K. Cinnamon extract inhibits α-glucosidase activity and damps postprandial glucose excursion in diabetic rats. Nutr. Metab. 2011, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Loh, S.P.; Hadira, O. In vitro inhibitory potential of selected Malaysian plants against key enzymes involved in hyperglycemia and hypertension. Malays. J. Nutr. 2011, 17, 77–86. [Google Scholar] [PubMed]
- Tee, E.S.; Yap, R.W.K. Type 2 diabetes mellitus in Malaysia: Current trends and risk factors. Eur. J. Clin. Nutr. 2017, 71, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Sudha, P.; Zinjarde, S.S.; Bhargava, S.Y.; Kumar, A.R. Potent Alpha-amylase inhibitory activity of Indian Ayurvedic medicinal plants. BMC Complement. Altern. Med. 2011, 11, 5. [Google Scholar] [CrossRef]
- Nair, A.S.; Kavrekar, V.; Mishra, A. In vitro studies on Alpha amylase and Alpha Glucosidase inhibitory activities of selected plant extracts. Eur. J. Exp. Biol. 2013, 3, 128–132. [Google Scholar]
- Arullappan, P.; Rajamanickam, P.; Thevar, N.; Kodimani, C.C. In vitro screening of cytotoxic, antimicrobial and antioxidant activities of Clinacanthus nutans (Acanthaceae) leaf extracts. Trop. J. Pharm. Res. 2014, 13, 1455–1461. [Google Scholar] [CrossRef]
- Putwatana, P.; Sanmanowong, P.; Oonprasertpong, L.; Junda, T.; Pitiporn, A.; Narkwong, L. Relief of radiation-induced oral mucositis in head and neck cancer. Cancer Nurs. 2009, 32, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Sakdarat, A.; Shuyprom, A. bioactive constituents from the leaves of Clinacanthus nutans Lindau. Bioorg. Med. Chem. 2009, 17, 1857–1860. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Ferdosh, A.; Ghafoor, K.; Hakim, A.; Juraimi, A.S.; Khatib, A. Clinacanthus nutans: A Review of the medicinal uses, pharmacology and phytochemistry. Asian Pac. J. Trop. Med. 2016, 9, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Uawonggul, N.; Chaveerach, A.; Thammasirirak, A.; Arkaravichien, T.; Chuachan, C.; Daduang, S. Screening of plants acting against Heterometrus Laoticus Scorpion venom activity on fibroblast cell lysis. J. Ethnopharmacol. 2006, 103, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Shim, A.Y.; Aziana, I.; Khoo, B.Y. Perspective and insight on Clinacanthus nutans Lindau in traditional medicine. Int J. Integr Biol 2013, 14, 7–9. [Google Scholar]
- Lusia M′ Barek, M.; Hasmadi, M.; Zaleha, A.Z.; Mohd Fadzelly, A.B. Effect of different drying methods on phytochemicals and antioxidant properties of unfermented and fermented teas from Sabah snake grass (Clinacanthus nutans Lind.) Leaves. Int. Food Res. J. 2015, 22, 661–670. [Google Scholar]
- Peng, T.W.; Wen, P.X.; He, C.J.; Akowuah, G.A. Effect of methanol extract of Clinacanthus nutans on serum biochemical parameters in rats. J. Appl. Pharm. Sci. 2014, 77, 77–86. [Google Scholar]
- Ghasemzadeh, A.; Nasiri, A.; Jaafar, H.Z.E.; Baghdadi, A.; Ahmad, I. Changes in phytochemical synthesis, chalcone synthase activity and pharmaceutical qualities of Sabah Snake Grass (Clinacanthus nutans L.) In relation to plant age. Molecules 2014, 19, 17632–17648. [Google Scholar] [CrossRef] [PubMed]
- Irondi, E.A.; Oboh, G.; Agboola, S.O.; Boligon, A.A.; Athayde, M.L. Phenolics Extract of Tetrapleura Tetraptera Fruit inhibits xanthine oxidase and Fe2+-induced lipid per oxidation in the kidney, liver, and lungs tissues of rats in vitro. Food Sci. Hum. Wellness 2016, 5, 17–23. [Google Scholar] [CrossRef]
- Kim, C.S.; Subedi, L.; Oh, J.; Kim, A.Y.; Choi, A.U.; Lee, K.R. bioactive Triterpenoids from the twigs of Chaenomeles sinensis. J. Nat. Prod. 2017, 80, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Aaradhya, M.; Kodikonda, M.; Naik, P.R. Antihyperglycemic, antihyperlipidemic and antioxidant activity of phenolic rich extract of Brassica Oleraceae Var Gongylodes on streptozotocin induced Wistar rats. Springerplus 2015, 4, 212. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, D.; Li, N.; Hou, X.; Wang, D.; Li, D. Influence of environmental factors on the active substance production and antioxidant activity in Potentilla fruticosa L. and its quality assessment. Sci. Rep. 2016, 6, 118. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. J. Plant Chem. Biochem. Tech. 2010, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. MArse spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Easmin, A.; Zaidul, I.S.M.; Ghafoor, K.; Ferdosh, A.; Jaffri, J.; Ali, M.E. Rapid investigation of α-glucosidase inhibitory activity of Phaleria macrocarpa Extracts using FTIR-ATR based fingerprinting. J. Food Drug 2017, 25, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chang, W.; Wang, C.; Liang, Y. The use of calibration approaches for quantitative GC/MS analysis-secobarbital Example. Forensic Sci. J. 2006, 5, 13–19. [Google Scholar]
- Wang, K.; Bao, L.; Ma, K.; Zhang, J.; Chen, B.; He, J. A novel Class of α-glucosidase and HMG-CoA reductase inhibitors from Ganoderma Leucocontextum and the anti-diabetic properties of ganomycin in in KK-Aymice. Eur. J. Med. Chem. 2017, 127, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Yawadio Nsimba, R.; Kikuzaki, H.; Konishi, Y. Antioxidant activity of various extracts and fractions of Chenopodium Quinoa and Amaranthus spp. Seeds. Food Chem. 2008, 106, 760–766. [Google Scholar] [CrossRef]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikstrom, C. Multi-and Megavariate Data Analysis Basic Principles and Applications, 2nd ed.; Umetrics Academy: Umeå, Sweden, 2006. [Google Scholar]
- Trivedi, D.K.; Iles, R.K. The Application of SIMCA P+ in Shotgun Metabolomics Analysis of ZICⓇ HILIC-MS Spectra of Human urine-Experience with the Shimadzu IT-T of and profiling Solutions Data extraction Software. J. Chromatogr. Sep. Tech. 2012, 3, 1–5. [Google Scholar] [CrossRef]
- Kuhlisch, C.; Pohnert, G. Metabolomics in chemical ecology. Nat. Prod. Rep. 2015, 32, 937–955. [Google Scholar] [CrossRef] [PubMed]
- Mukesh, B.; Rakesh, K. Review Article Molecular Docking: A Review. Int. J. Res. Ayurveda Pharm. 2011, 2, 1746–1751. [Google Scholar]
- Seong, A.H.; Roy, A.; Jung, H.A.; Jung, H.J.; Choi, J.S. Protein tyrosine phosphatase 1b and α-glucosidase inhibitory activities of Pueraria lobata Root and its constituents. J. Ethnopharmacol. 2016, 194, 706–816. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Kim, D.; Seong, A.; Kim, H.R.; Jung, H.; Choi, J. α-Glucosidase and protein tyrosine phosphatase 1b inhibitory activity of plastoquinones from Marine brown alga Sargassum serratifolium. Mar. Drugs 2017, 15, 368. [Google Scholar] [CrossRef] [PubMed]
- Lavle, N.; Shukla, P.; Panchal, A. ROLE of flavonoids and saponins in the treatment of diabetes mellitus. J. Pharm. Sci. Bioscientific Res. 2016, 6, 535–541. [Google Scholar]
- Sharma, N.; Verma, M.K.; Gupta, D.K.; Satti, N.K.; Khajuria, R.K. Isolation and quantification of Pinitol in Argyrolobium Rose Plant, by 1H-NMR. J. Saudi Chem. Soc. 2016, 20, 81–87. [Google Scholar] [CrossRef]
- Cencic, A.; Chingwaru, W. The role of functional foods, nutraceuticals, and food supplements in intestinal health. Nutrients 2010, 2, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Aykul, A.; Martinez-Hackert, E. Determination of half-maximal inhibitory concentration using biosensor-based protein interaction analysis. Anal. Biochem. 2016, 508, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Deseo, M.A.; Morris, C.; Winter, K.M.; Leach, D.N. Investigation of α-glucosidase inhibitory activity of wheat bran and germ. Food Chem. 2011, 126, 553–561. [Google Scholar] [CrossRef]
- Marella, A.; Tanwar, O.P.; Saha, R.; Ali, M.R.; Srivastava, A.; Akhter, M.; Shaquiquzzaman, M.; Alam, M.M. Quinoline: A versatile heterocyclic. Saudi Pharm. J. 2013, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- De Moraes, J.; De Oliveira, R.N.; Costa, J.P.; Junior, A.L.G.; De Sousa, D.P.; Freitas, R.M. Phytol, a diterpene alcohol from chlorophyll, as a drug against neglected tropical disease Schistosomiasis mansoni. PLoS Negl. Trop. Dis. 2014, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Tundis, R.; Loizzo, M.R.; Menichini, Q. NAtural products as α-amylase and α-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini-Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Verma, A.; Mishra, A.P. A pLant rEview: Butea Monosperma (Lam.) Kuntze. Res. J. Pharm. Biol. Chem. Sci. 2012, 3, 700–714. [Google Scholar]
- Nualkaew, A.; Padee, P.; Talubmook, C. Hypoglycemic activity in diabetic rats of Stigmasterol and Sitosterol-3-O-α-d-glucopyranoside isolated from Pseuderanthemum Palatiferum Nees Radlk. Leaf extract. J. Med. Plants Res. 2015, 9, 629–635. [Google Scholar] [CrossRef]
- Ghosh, T.; Ghosh, S.; Maity, T.K. Antihyperglycemic activity of Stigmasterol isolated from Bacopa Monnieri Linn. Aerial parts against Alloxan induced diabetic rats. Int. J. Nat. Prod. Res. 2014, 4, 40–46. [Google Scholar]
- Zareen, S.; Choudhary, M.I.; Akhtar, M.N.; Khan, S.N. α-Glucosidase inhibitory Activity of Triterpenoids from Cichorium Intybus. J. Nat. Prod. 2008, 392, 910–913. [Google Scholar] [CrossRef]
- Human Metabolome Database (HMDB). Available online: http://www.hmdb.ca/metabolites/HMDB0031074 (accessed on 10 December 2017).
- Kaufman, P.B.; Cseke, L.J.; Warber, A.; Duke, J.A.; Brielmann, H.L. Natural Products from Plants; CRC Press: Boca Raton, FL, USA, 2000. [Google Scholar]
- Nussinov, R. Introduction to Protein ensembles and Allostery. Chem. Rev. 2016, 116, 6263–6266. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P. Insights into protein-ligand interaction: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal structures of Isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Huang, W.; Zhang, J. Recent computational advances in the identification of allosteric sites in proteins. Drug Discov. Today 2014, 19, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Guarnera, E.; Berezovsky, I.N. Allosteric sites: Remote control in regulation of protein activity. Curr. Opin. Struct. Biol. 2016, 37, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zang, G.; Pan, J.; Wang, Y. α-Glucosidase inhibition by luteolin: Kinetics, interaction and molecular dolling. Int. J. Biol. Macromol. 2014, 64, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Brindis, Q.; Rodríguez, R.; Bye, R.; González-Andrade, M.; Feed, R. (Z)-3-Butylidenephthalide from Ligusticum porteri, An α-glucosidase inhibitor. J. Nat. Prod. 2011, 74, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhou, S.; Zhang, C.; Wu, D.; Luo, H.B.; Wu, Y. Docking-assisted 3D-QSAR studies on Xanthones as α-glucosidase inhibitors. J. Mol. Model. 2017, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Abd Aziz, A.M.; Low, C.N.; Chai, L.C.; Abd Razak, A.S.N.; Selamat, J.; Son, R. Screening of selected Malaysian plants against several food-borne pathogen bacteria. Int. Food Res. J. 2011, 18, 1195–1201. [Google Scholar]
- Javadi, N.; Abas, F.; Mediani, A.; Abd Hamid, A.; Khatib, A.; Simoh, S.; Shaari, K. Effect of storage time on metabolite profile and alpha-glucosidase inhibitory activity of Cosmos caudate Leaves-GCMS based metabolomics approach. J. Food Drug 2015, 23, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, A.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Salimon, J.; Omar, T.A.; Salih, N. An accurate and reliable method for identification and quantification of fatty acids and trans fatty acids in food fats samples using gas chromatography. Arab J. Chem. 2017, 10, S1875–S1882. [Google Scholar] [CrossRef]
- Shailajan, A.; Joshi, H. Optimized Separation and Quantification of Pharmacologically active markets Quercetin, Kaempferol, β-Sitosterol and Lupeol from Cuscuta Reflex Roxb. J. Pharm. Res. 2011, 4, 1851–1853. [Google Scholar]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated Pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Lestari, W.; Dewi, R.T.; Kardono, L.B.S.; Yanuar, A. Dolling Sulochrin and Its derivative as α-Glucosidase inhibitors of Saccharomyces cerevisiae. Indones. J. Chem. 2017, 17, 144–150. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds namely; palmitic acid, heptadecanoic acid and stigmasterol are available from the authors. |

| Samples | IC50 (μg/mL) |
|---|---|
| Hexane (H) | 3.05 ± 0.05 d |
| Hexane: Ethyl acetate (HE) | 5.54 ± 0.09 d |
| Ethyl acetate (EA) | 8.42 ± 0.17 c |
| Ethyl acetate: Methanol (EAM) | 37.45 ± 0.90 b |
| Methanol (M) | 133.57 ± 0.30 a |
| Quercetin | 5.77 ± 1.01 (19.09 μM) |
| No. | RT (min) | % of Area | Probability | M+ | Molecular Formula | Tentative Metabolites |
|---|---|---|---|---|---|---|
| 1 | 17.24 | 10.50 | 99 | 256.24 | C16H32O2 | Palmitic acid |
| 2 | 26.64 | 1.42 | 95 | 296.31 | C20H40O | Phytol |
| 3 | 45.47 | 36.18 | 93 | 342.12 | C12H22O11 | Sucrose |
| 4 | 11.63 | 0.91 | 99 | 270.26 | C17H34O2 | Hexadecanoic acid |
| 5 | 49.48 | 0.28 | 93 | 342.12 | C12H22O11 | Maltose |
| 6 | 44.43 | 1.03 | 95 | 330.51 | C19H38O4 | 1-Monopalmitin |
| 7 | 10.31 | 19.04 | 91 | 180.06 | C6H12O6 | d-Glucose |
| 8 | 56.83 | 4.50 | 99 | 398.39 | C29H50 | Stigmast-5-ene |
| 9 | 14.30 | 3.42 | 93 | 196.16 | C6H12O7 | d-Gluconic acid |
| 10 | 12.34 | 0.24 | 98 | 242.22 | C15H30O2 | Pentadecanoic acid |
| 11 | 23.95 | 0.38 | 93 | 270.26 | C17H34O2 | Heptadecanoic acid |
| 12 | 47.59 | 0.46 | 99 | 352.52 | C17H36O | 1-Linolenoylglycerol |
| 13 | 47.99 | 0.40 | 91 | 358.31 | C21H42O4 | Glycerol monostearate |
| 14 | 49.24 | 0.07 | 96 | 462.37 | C29H50O4 | Alpha-Tocospiro B |
| 15 | 55.59 | 2.70 | 99 | 412.37 | C29H48O | Stigmasterol |
| Samples | IC50 Value | |
|---|---|---|
| µg/mL | µM | |
| Palmitic acid | 24.09 ± 0.44 c | 93.95 |
| Heptadecanoic acid | 26.10 ± 0.77 b | 96.51 |
| Stigmasterol | 65.31 ± 0.37 a | 158.25 |
| Quercetin | 5.77 ± 1.01 | 19.09 |
| Analytes | R2 1 | Concentration (µg/mg Sample) 2 | LOD (µg/mg Sample) | LOQ (µg/mg Sample) |
|---|---|---|---|---|
| Palmitic acid | 0.99 | 433.25 ± 22.96 a | 2.50 | 5.00 |
| Heptadecanoic acid | 0.99 | 20.27 ± 1.20 c | 5.00 | 10.00 |
| Stigmasterol | 0.99 | 386.48 ± 21.03 b | 0.31 | 0.63 |
| Compound | Binding Energy (kcal/mol) | H-bond Interacting Residues | Other Interacting Residues |
|---|---|---|---|
| Control ligand | −6.00 | ASH69, HIE112, GLN182, ARG213, ASH215, GLH277, HIE351, ASP352, ARG442 | TYR72 |
| Quercetin | −8.15 | LYS156, THR310, PRO312, LEU313, GLU411, ASN415 | PHE314, ARG315 |
| Palmitic acid | −3.75 | LYS156, LEU313, ARG315 | HIE280, PHE314 |
| Heptadecanoic acid | −3.80 | LYS156, SER236, GLN239, SER240 | ARG315 |
| Stigmasterol | −8.66 | THR290 | TRP15, ILE262, ARG263, ILE272, VAL266, HID295 |
| Compound | Binding Energy (kcal/mol) | H-bond Interacting Residues | Other Interacting Residues |
|---|---|---|---|
| 1-linolenoylglycerol | −4.76 | LYS156, SER241 | TYR158, SER240, PHE314 |
| 1-monoplamitin | −4.01 | TYR158, SER240, ASP242 | PHE314, ILE419 |
| Alpha-tocospiro B | −7.99 | GLN279, GLU411 | LYS156, TYR158, PHE159, PHE178, HIE280, PHE314 |
| Glycerol monostearate | −3.95 | SER241 | LYS156, TYR158, SER240, ASP242, PHE314, ARG315, TYR316 |
| Hexadecenoic acid (methyl ester) | −4.00 | ALA292, ASN259, HID295 | VAL266 |
| Pentadecanoic acid | −4.42 | HID295 | VAL266 |
| Phytol | −5.51 | GLN239, SER240 | LYS156, TYR158, VAL232, LEU313, PHE314 |
| Stigmast-5-ene | −9.09 | - | TYR158, LYS156, HIE280, PRO312, PHE314, ARG315, TYR316 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murugesu, S.; Ibrahim, Z.; Ahmed, Q.-U.; Nik Yusoff, N.-I.; Uzir, B.-F.; Perumal, V.; Abas, F.; Saari, K.; El-Seedi, H.; Khatib, A. Characterization of α-Glucosidase Inhibitors from Clinacanthus nutans Lindau Leaves by Gas Chromatography-Mass Spectrometry-Based Metabolomics and Molecular Docking Simulation. Molecules 2018, 23, 2402. https://doi.org/10.3390/molecules23092402
Murugesu S, Ibrahim Z, Ahmed Q-U, Nik Yusoff N-I, Uzir B-F, Perumal V, Abas F, Saari K, El-Seedi H, Khatib A. Characterization of α-Glucosidase Inhibitors from Clinacanthus nutans Lindau Leaves by Gas Chromatography-Mass Spectrometry-Based Metabolomics and Molecular Docking Simulation. Molecules. 2018; 23(9):2402. https://doi.org/10.3390/molecules23092402
Chicago/Turabian StyleMurugesu, Suganya, Zalikha Ibrahim, Qamar-Uddin Ahmed, Nik-Idris Nik Yusoff, Bisha-Fathamah Uzir, Vikneswari Perumal, Faridah Abas, Khozirah Saari, Hesham El-Seedi, and Alfi Khatib. 2018. "Characterization of α-Glucosidase Inhibitors from Clinacanthus nutans Lindau Leaves by Gas Chromatography-Mass Spectrometry-Based Metabolomics and Molecular Docking Simulation" Molecules 23, no. 9: 2402. https://doi.org/10.3390/molecules23092402
APA StyleMurugesu, S., Ibrahim, Z., Ahmed, Q.-U., Nik Yusoff, N.-I., Uzir, B.-F., Perumal, V., Abas, F., Saari, K., El-Seedi, H., & Khatib, A. (2018). Characterization of α-Glucosidase Inhibitors from Clinacanthus nutans Lindau Leaves by Gas Chromatography-Mass Spectrometry-Based Metabolomics and Molecular Docking Simulation. Molecules, 23(9), 2402. https://doi.org/10.3390/molecules23092402







