Selection and Identification of Novel Aptamers Specific for Clenbuterol Based on ssDNA Library Immobilized SELEX and Gold Nanoparticles Biosensor
Abstract
1. Introduction
2. Results
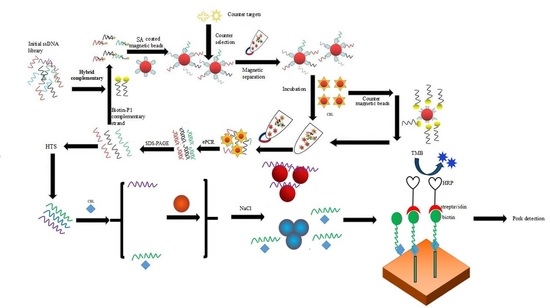
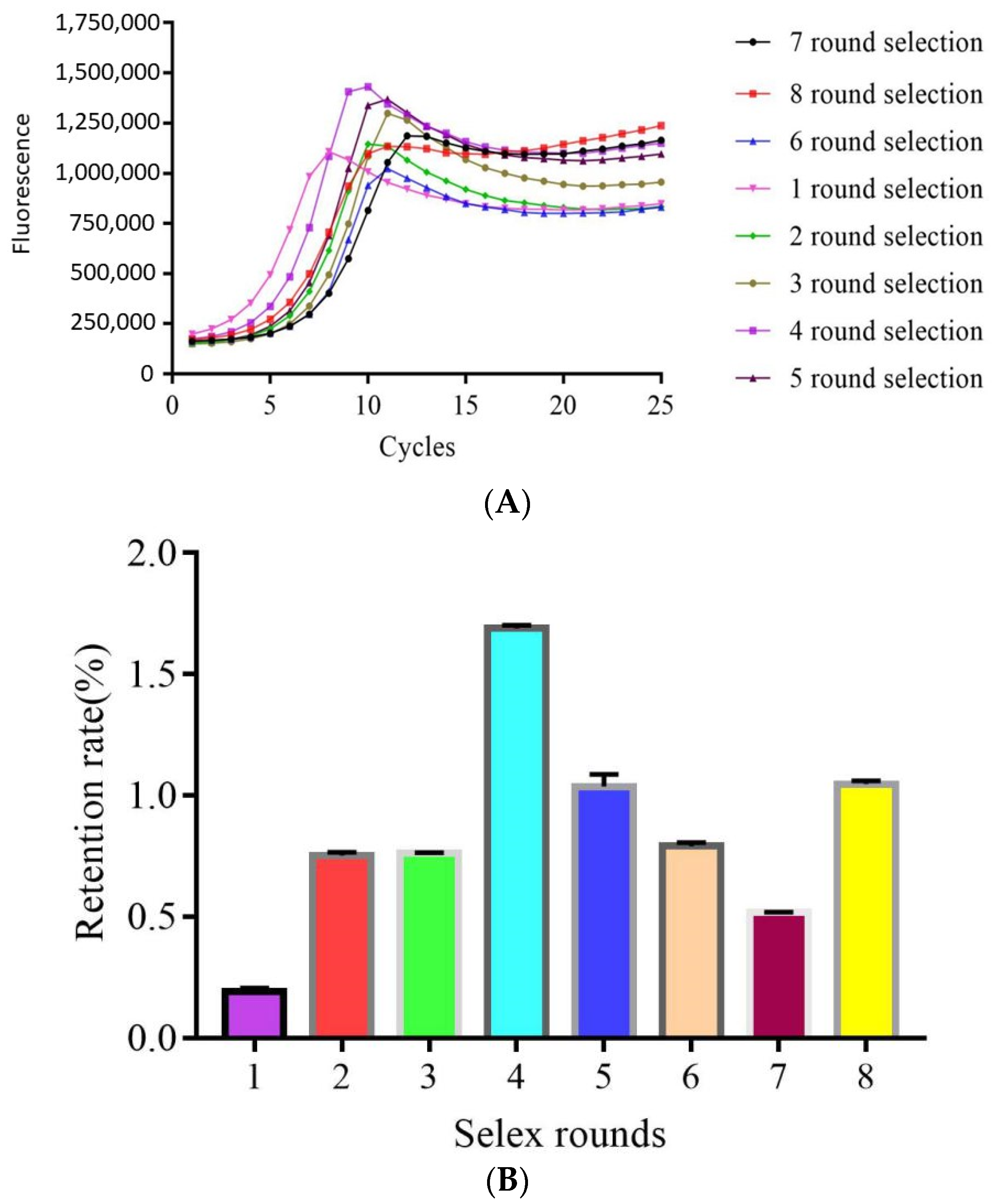
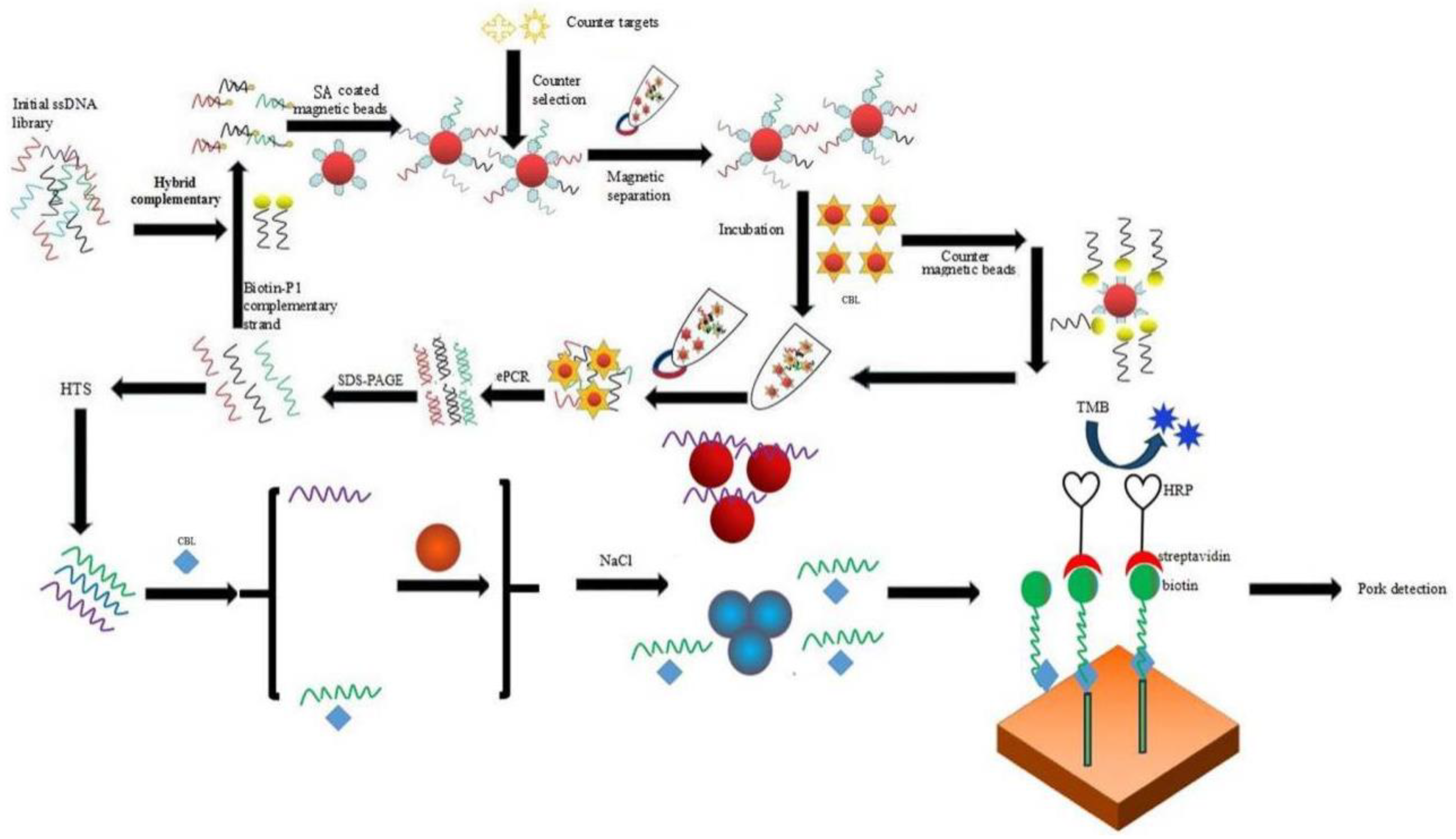
2.1. Magnetic Bead SELEX Selection of an Aptamer Specific for Clenbuterol
2.2. High-Throughput Sequencing and Sequence Analysis of the Enriched Library
2.3. Establishment of a Gold Nanoparticles Biosensor to Identify Active Aptamers
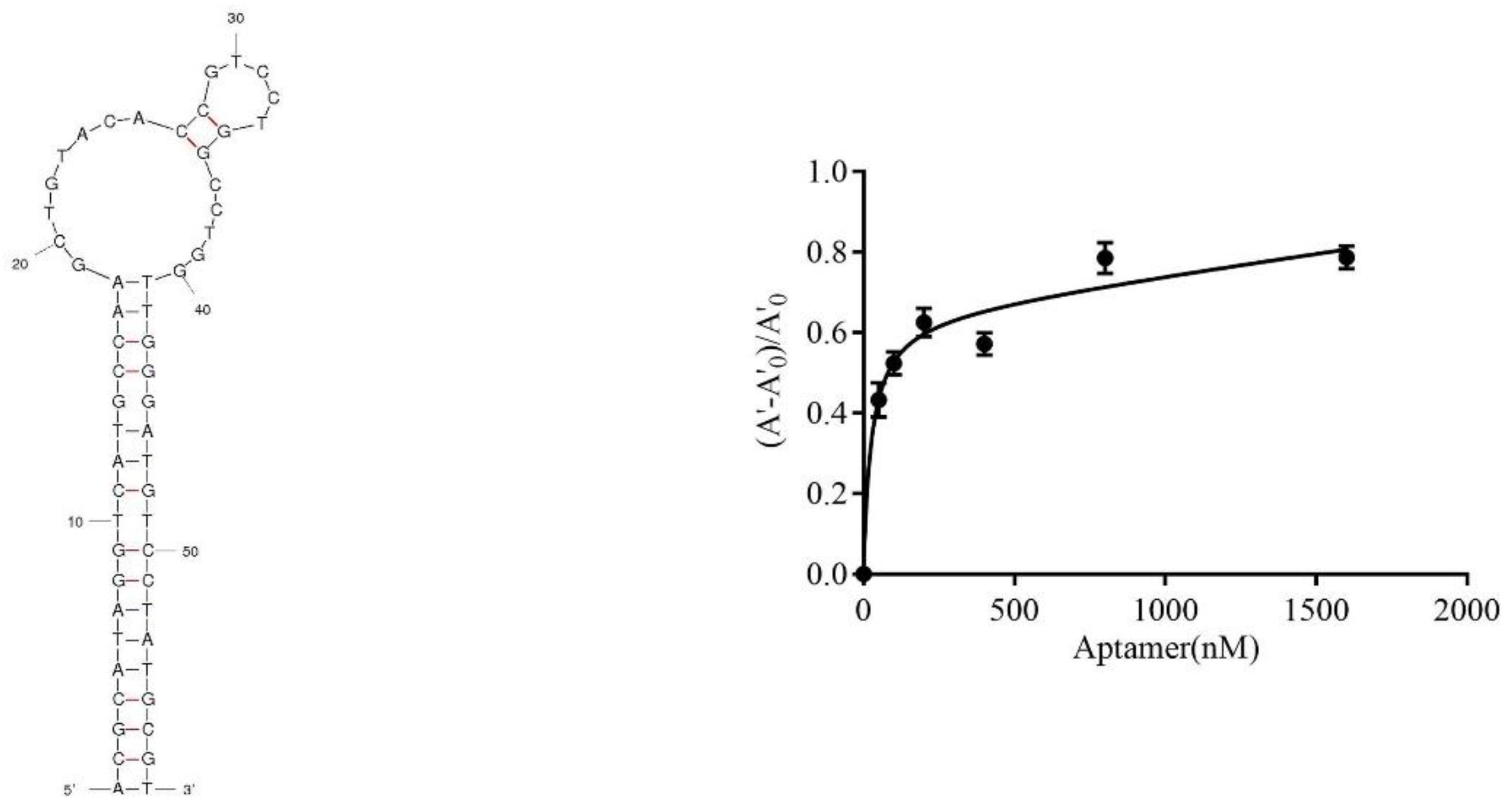
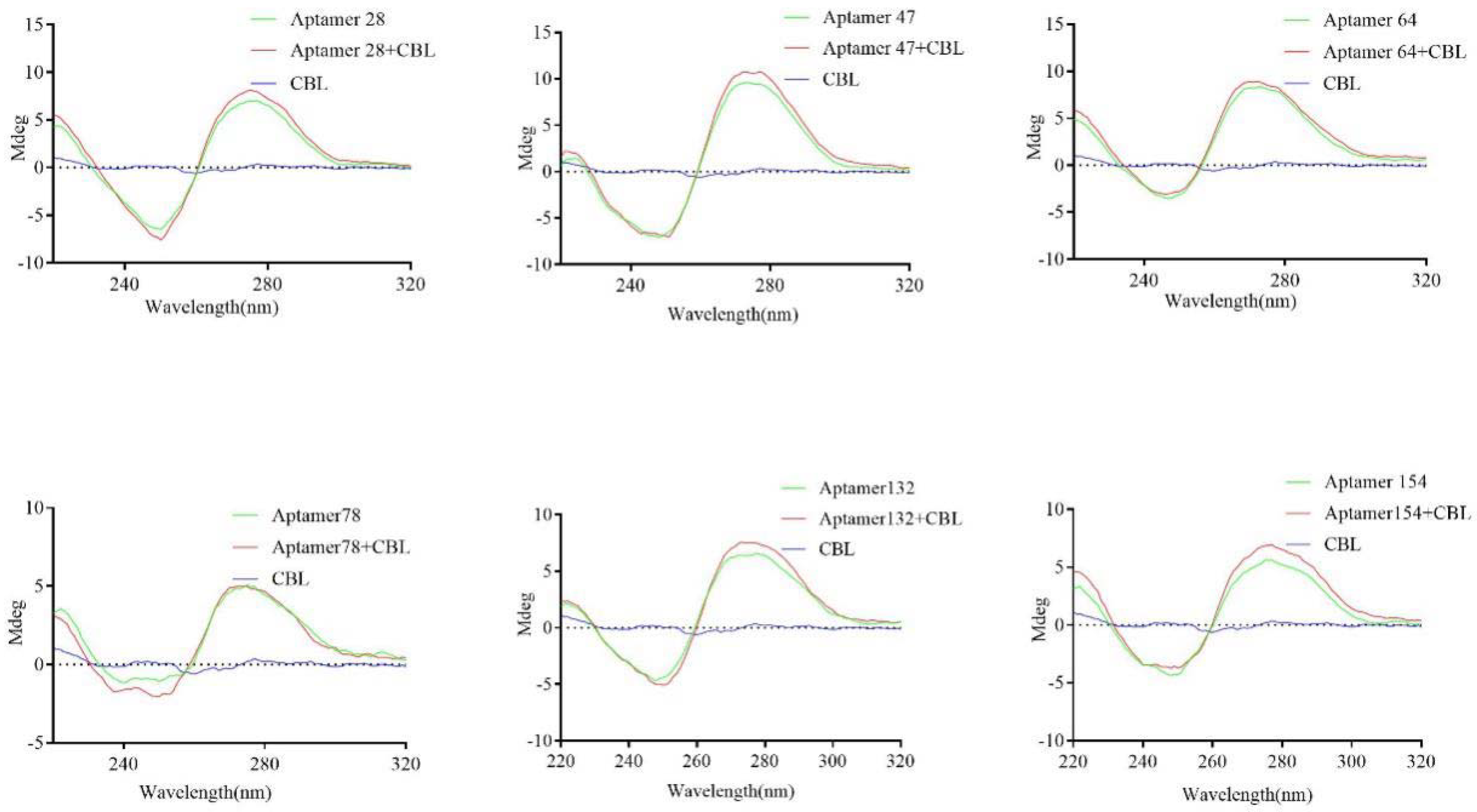
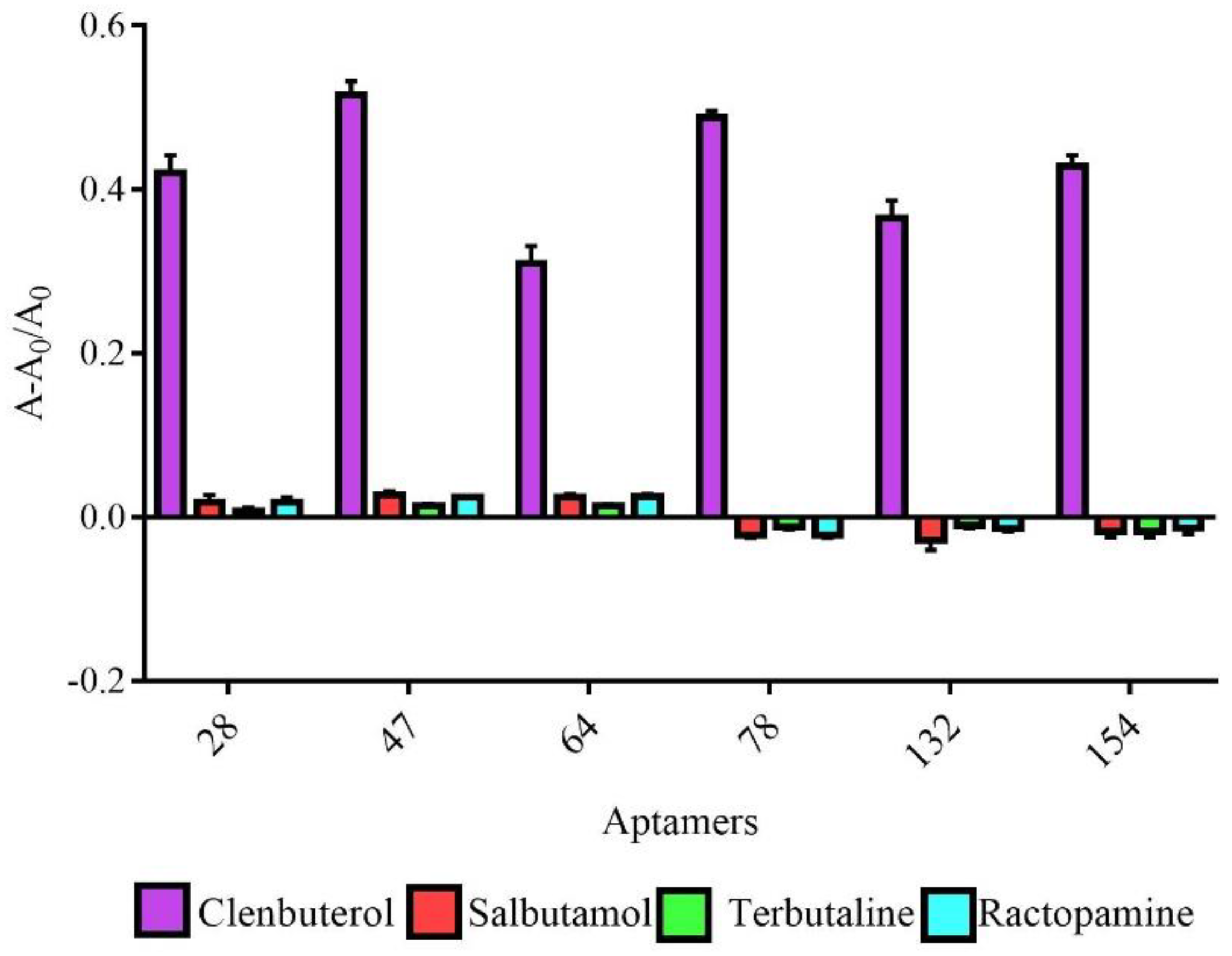
2.4. Characterization of Active Aptamers
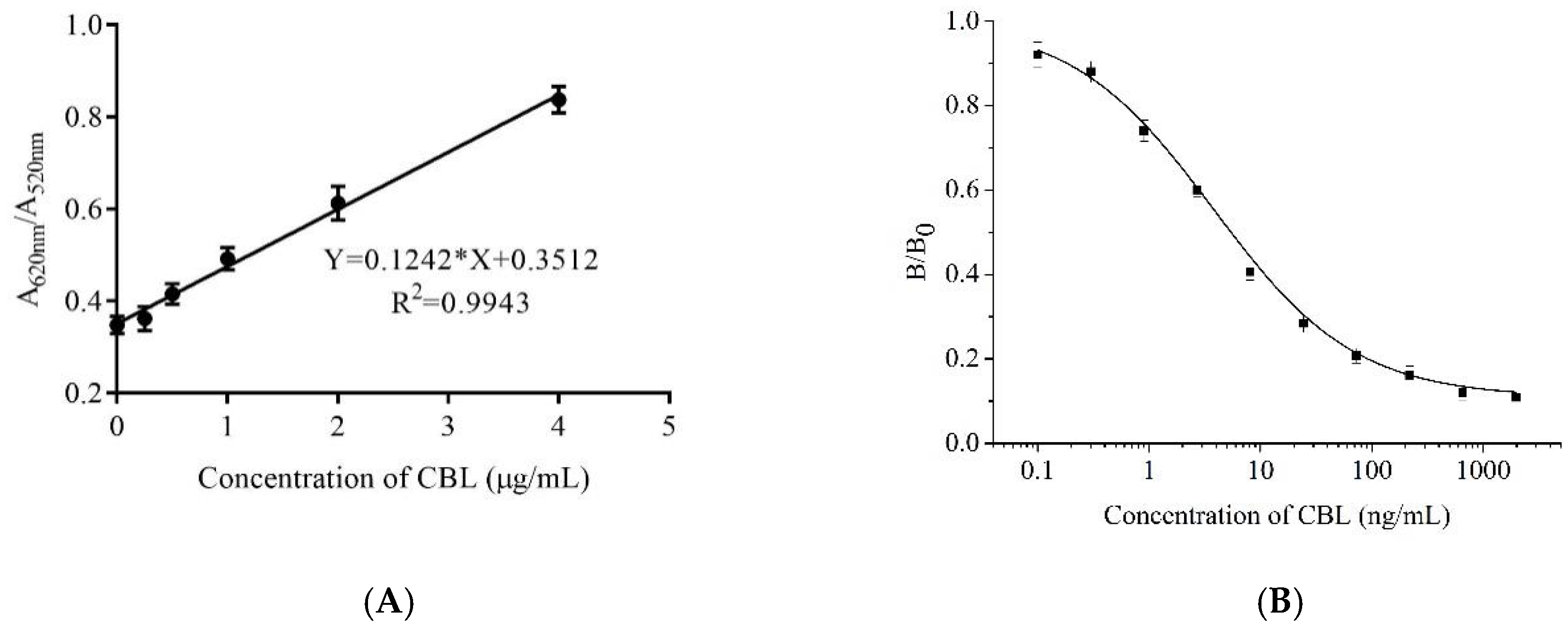
2.5. Detection of CBL Using a Gold Nanoparticles Biosensor and Indirect Competitive Enzyme Linked Aptamer Assay
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Selection of Aptamers Specific for CBL Based on ssDNA Library Immobilized SELEX
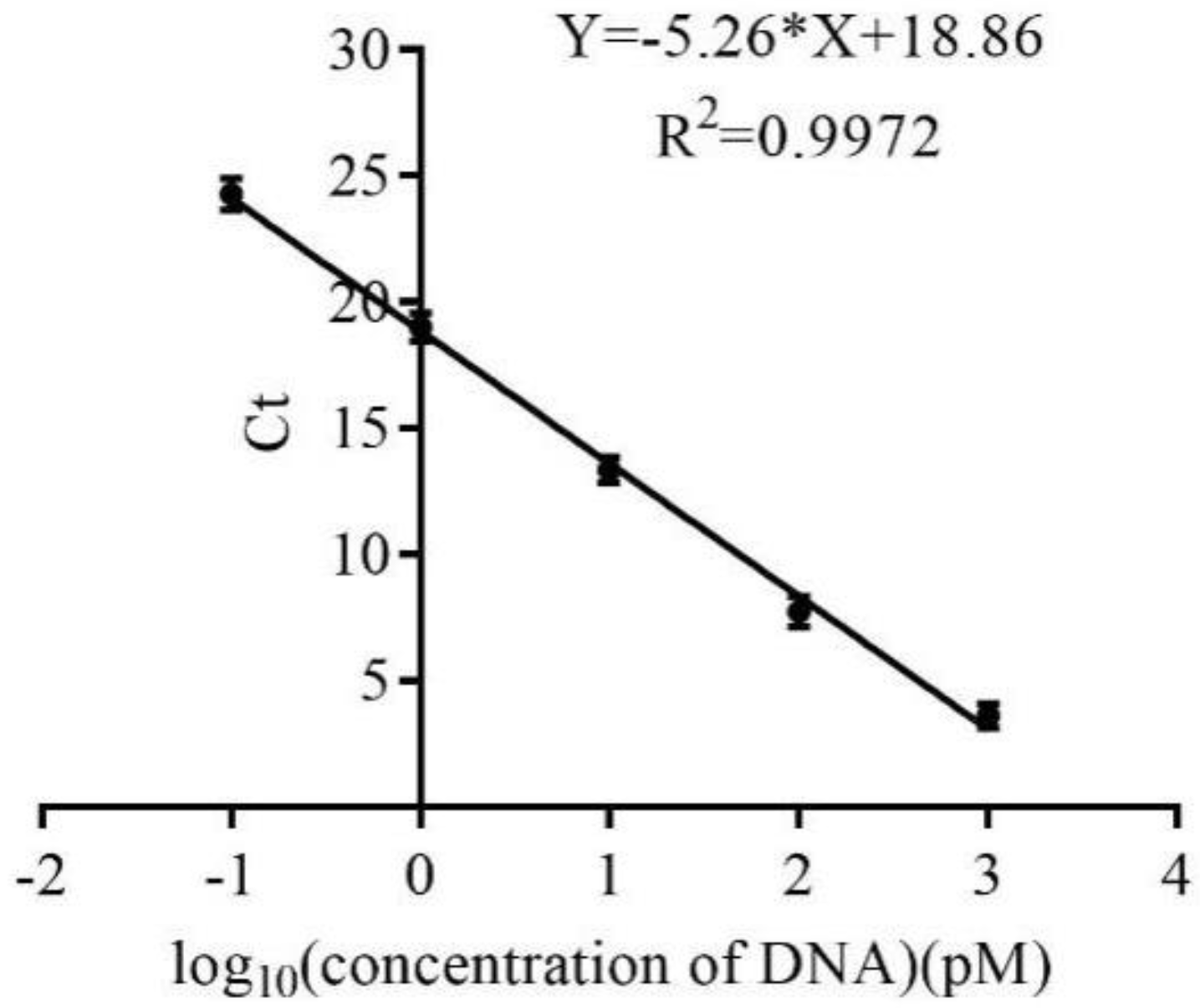
4.3. Establishment of a Real-Time Quantitative PCR Method for the Monitoring Selection Process
4.4. Preparation of Secondary Libraries
4.5. High-Throughput Sequencing and Sequence Analysis of the Enrichment Library
4.6. Determination of Binding Activity between Aptamers and CBL Using a Gold Nanoparticles Biosensor
4.7. Affinity Analysis of Active Aptamers
4.8. Circular Dichroism Spectrum Analysis of Active Aptamers
4.9. Specificity Analysis of Active Aptamers
4.10. Detection of CBL Using a Gold Nanoparticles Biosensor and ic-ELAA
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Torneke, K.; Larsson, C.I.; Appelgren, L.E. A comparison between clenbuterol, salbutamol and terbutaline in relation to receptor binding and in vitro relaxation of equine tracheal muscle. J. Vet. Pharmacol. Ther. 1998, 21, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qi, P.; Ma, X.; Zhong, J.G. Quick detection technique for clenbuterol hydrochloride by using surface plasmon resonance biosensor. Eur. Food Res. Technol. 2014, 239, 195–201. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, L.; Huang, Y.J.; Dandapat, A.; Dai, L.W.; Zhang, G.G.; Lu, X.F.; Zhang, J.W.; Lai, W.H.; Chen, T. Hollow Au-Ag Nanoparticles labeled immunochromatography strip for highly sensitive detection of clenbuterol. Sci. Rep.-UK 2017, 7, 41419. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.Y.; Nilghaz, A.; Choi, J.R.; Liu, X.Q.; Lu, X.N. Rapid detection of clenbuterol in milk using microfluidic paper-based ELISA. Food Chem. 2018, 246, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.Y.; Lin, C.; Yu, S.Y.; Gong, S.; Hu, P.; Li, Y.S.; Wu, Z.C.; Gao, Y.; Zhou, Y.; Liu, Z.S.; et al. Preparation of a specific ssDNA aptamer for brevetoxin-2 using SELEX. J. Anal. Methods Chem. 2016, 2016, 9241860. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Qiao, P.; Jing, L.; Song, Y.Z.; Zhang, J.Y.; Chen, Q.; Han, Q.Q. Screening and application of a new aptamer for the rapid detection of Sudan Dye III. Eur. J. Lipid Sci. Tech. 2018, 120, 1700112. [Google Scholar] [CrossRef]
- Wu, Z.Z.; Xu, E.; Jin, Z.Y.; Irudayaraj, J. An ultrasensitive aptasensor based on fluorescent resonant energy transfer and exonuclease-assisted target recycling for patulin detection. Food Chem. 2018, 249, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kou, F.X.; Xu, H.F.; Han, Y.J.; Yang, G.D.; Huang, X.J.; Chen, W.; Chi, Y.W.; Lin, Z.Y. Label-free ochratoxin A electrochemical aptasensor based on target-induced noncovalent assembly of peroxidase-like graphitic carbon nitride nanosheet. Sensor. Actuat. B-Chem. 2018, 270, 263–269. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, G.L.; Huang, Y.F.; Yan, J.; Chen, A.L. Aptamer based ultrasensitive determination of the beta-adrenergic agonist ractopamine using PicoGreen as a fluorescent DNA probe. Microchim. Acta 2017, 184, 439–444. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, L.H.; Gao, Z.Q.; Wang, H.S.; Zhang, H.X.; Li, H. An aptamer-based effective method for highly sensitive detection of chloramphenicol residues in animal-sourced food using real-time fluorescent quantitative PCR. Talanta 2017, 165, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yang, M.; Zheng, N.J.; Xie, N.; Liu, D.L.; Xie, C.F.; Yao, D.S. A novel aptasensor for electrochemical detection of ractopamine, clenbuterol, salbutamol, phenylethanolamine and procaterol. Biosens.Bioelectron. 2016, 80, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Li, X.Y.; Cui, F.; Qiu, Q.Y.; Chen, X.J.; Huang, H. Aflatoxin B1 electrochemical aptasensor based on tetrahedral DNA nanostructures functionalized three dimensionally ordered macroporous MoS2-AuNPs film. ACS Appl. Mater. Int. 2018, 10, 17551–17559. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.Y.; Tu, C.Y.; Dai, Y.Y.; Wang, W.P.; Wang, A.J.; Chen, J.R. A simple colorimetric analytical assay using gold nanoparticles for specific detection of tetracycline in environmental water samples. Anal. Methods-UK 2018, 10, 3402–3407. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Chen, M.L.; Chi, J.X.; Yu, X.; Chen, Y.X.; Lin, X.C.; Xie, Z.H. Aptamer-based polyhedral oligomeric silsesquioxane (POSS)-containing hybrid affinity monolith prepared via a “one-pot” process for selective extraction of ochratoxin A. J. Chromatogr. A 2018, 1563, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Li, W.; Ding, Z.; Deng, Y.; Liu, Y.; Zhu, X.R.; Cai, T.T.; Li, J.L.; Zheng, T.S. A competitive aptamer chemiluminescence assay for ochratoxin A using a single silica photonic crystal microsphere. Anal. Biochem. 2018, 554, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Al Rubaye, A.; Nabok, A.; Catanante, G.; Marty, J.L.; Takacs, E.; Szekacs, A. Detection of ochratoxin A in aptamer assay using total internal reflection ellipsometry. Sensor. Actuat. B-Chem. 2018, 263, 248–251. [Google Scholar] [CrossRef]
- Lin, C.; Liu, Z.S.; Wang, D.X.; Li, L.; Hu, P.; Gong, S.; Li, Y.S.; Cui, C.; Wu, Z.C.; Gao, Y.; et al. Generation of internal-image functional aptamers of okadaic acid via magnetic-bead SELEX. Mar. Drugs 2015, 13, 7433–7445. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple GO-SELEX for efficient screening of flexible aptamers. Chem. Commun. 2014, 50, 10513–10516. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.F.; Zhou, H.M.; Jiang, H.; Ou, H.C.; Li, X.; Zhang, L.Y. Development of a fraction collection approach in capillary electrophoresis SELEX for aptamer selection. Analyst 2015, 140, 2664–2670. [Google Scholar] [CrossRef] [PubMed]
- Setlem, K.; Monde, B.; Ramlal, S.; Kingston, J. Immuno affinity SELEX for simple, rapid, and cost-effective aptamer enrichment and identification against aflatoxin B1. Front. Microbiol. 2016, 7, 1909. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.G.; Zhan, S.S.; Wang, L.M.; Zhou, P. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst 2014, 139, 1550–1561. [Google Scholar] [CrossRef] [PubMed]
- He, J.A.; Liu, Y.A.; Fan, M.T.; Liu, X.J. Isolation and identification of the DNA aptamer target to acetamiprid. J. Agr. Food Chem. 2011, 59, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Gong, W.H.; Wu, S.J.; Wang, Z.P. Selection and application of ssDNA aptamers against clenbuterol hydrochloride based on ssDNA library immobilized SELEX. J. Agric. Food Chem. 2017, 65, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Gong, W.H.; Wu, S.J.; Wang, Z.P. An ssDNA library immobilized SELEX technique for selection of an aptamer against ractopamine. Anal. Chim. Acta. 2017, 961, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.W.; Yang, J.Y.; Ye, J.; Xu, L.R.; Xu, H.C.; Zhan, S.S. Colorimetric detection of bisphenol A based on unmodified aptamer and cationic polymer aggregated gold nanoparticles. Anal. Biochem. 2016, 499, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, Y.; Martyc, J.L.; Yang, X.R. Aptamer-based colorimetric biosensing of Ochratoxin A using unmodified gold nanoparticles indicator. Biosens. Bioelectron. 2011, 26, 2724–2727. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.F.; He, L.; Wang, J.J.; Fang, X.N.; Zhang, L.Y. Developing a combined strategy for monitoring the progress of aptamer selection. Analyst 2017, 142, 3136–3139. [Google Scholar] [CrossRef] [PubMed]
- Ramana, M.M.V.; Betkar, R.; Nimkar, A.; Ranade, P.; Mundhe, B.; Pardeshi, S. In vitro DNA binding studies of antiretroviral drug nelfinavir using ethidium bromide as fluorescence probe. J. Photoch. Photobio. B 2015, 151, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.M.; Jia, L.P.; Zhang, J.J. Study on the interaction between amikacin and DNA by molecular spectroscopy. J. Liaocheng Univer. 2008, 21, 67–70. [Google Scholar]
- Mencin, N.; Smuc, T.; Vranicar, M.; Mavri, J.; Hren, M.; Galesa, K.; Krkoc, P.; Ulrich, H.; Solar, B. Optimization of SELEX: Comparison of different methods for monitoring the progress of in vitro selection of aptamers. J. Pharmaceyt Biomed. 2014, 91, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Vanbrabant, J.; Leirs, K.; Vanschoenbeek, K.; Lammertyn, J.; Michiels, L. Remelting curve analysis as a tool for enrichment monitoring in the SELEX process. Analyst 2014, 139, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Heiat, M.; Ranjbar, R.; Latifi, A.M.; Rasaee, M.J.; Farnoosh, G. Essential strategies to optimize asymmetric PCR conditions as a reliable method to generate large amount of ssDNA aptamers. Biotechnol. Appl. Bioc. 2017, 64, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Citartan, M.; Tang, T.H.; Tan, S.C.; Gopinath, S.C.B. Conditions optimized for the preparation of single-stranded DNA (ssDNA) employing lambda exonuclease digestion in generating DNA aptamer. World J. Microb. Biot. 2011, 27, 1167–1173. [Google Scholar] [CrossRef]
- Williams, K.P.; Bartel, D.P. PCR product with strands of unequal length. Nucleic. Acids Res. 1995, 23, 4220–4221. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.K.; Shi, X.H.; Zhu, X.J.; Cui, L.L.; Shao, Q.X.; Ma, D. Construction and optimization of an efficient amplification method of a random ssDNA library by asymmetric emulsion PCR. Biotechnol. Appl. Bioc. 2017, 64, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Li, Q.; Duan, N.; Ma, H.L.; Wang, Z.P. DNA aptamer selection and aptamer-based fluorometric displacement assay for the hepatotoxinmicrocystin-RR. Microchim. Acta 2016, 183, 2555–2562. [Google Scholar] [CrossRef]
- Zhou, X.T.; Wang, L.M.; Shen, G.Q.; Zhang, D.W.; Xie, J.L.; Mamut, A.; Huang, W.W.; Zhou, S.S. Colorimetric determination of ofloxacin using unmodified aptamers and the aggregation of goldnanoparticles. Microchim. Acta 2018, 185, 355. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.A.; Shi, Y.P.; Pan, Y.; Yue, Z.F.; Zhang, H.; Yi, C.Q. Colorimetric and bare eye determination of urinary methylamphetamine based on the use of aptamers and the salt-induced aggregation of unmodified gold nanoparticles. Microchim. Acta 2015, 182, 505–511. [Google Scholar] [CrossRef]
- Han, Q.Q.; Qiao, P.; Song, Y.Z.; Zhang, J.Y.; Chen, Q. A Aptamer Specific CBL and Its Application. Patent 201510485760.0, 23 December 2015. [Google Scholar]
- Zhao, W.A.; Chiuman, W.; Lam, J.C.F.; McManus, S.A.; Chen, W.; Cui, Y.G.; Pelton, R.; Brook, M.A.; Li, Y.F. DNA aptamer folding on gold nanoparticles: From colloid chemistry to biosensors. J. Am. Chem. Soc. 2008, 130, 3610–3618. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Ma, X.D.; Ding, C.; Jia, L. Colorimetric determination of lysozyme based on the aggregation of gold nanoparticlescontrolled by a cationic polymer and an aptamer. Microchim. Acta 2016, 183, 2353–2359. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of nanoparticles from UV-Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Kim, Y.S. Effects of active immunization against clenbuterol on the growth-promoting effect of clenbuterolin rats. J. Anim. Sci. 1997, 75, 446–453. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
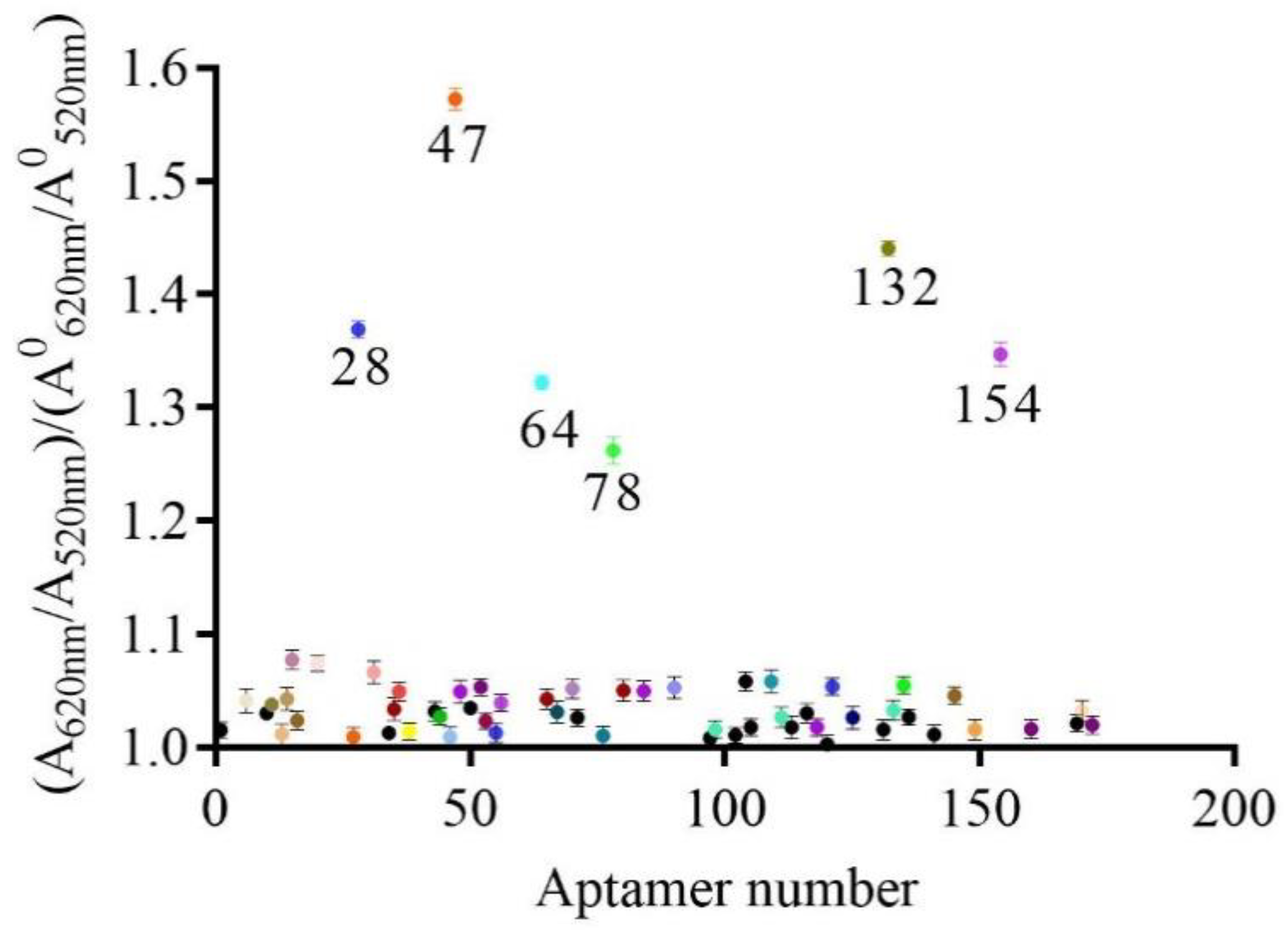
| No. | Sequences | Kd (nM) | dG |
|---|---|---|---|
| 28 | 5′ACGCATAGGATGCAACAACGCGAATGCCACCAATTCGTGCTTGTCGTGTCCTATGCGT3′ | 321 ± 104.5 | −15.09 |
| 47 | 5′ACGCATAGGTCATGCCAAGCTGTACACCGTCCTGGCCTGGTTGGGATGTCCTATGCGT3′ | 42.17 ± 8.98 | −13.76 |
| 64 | 5′ACGCATAGGTACACCACATCGGATCGTAATGCGTATGTGACTCAGTGTGCCTATGCGT3′ | 123.3 ± 14.28 | −16.26 |
| 78 | 5′ACGCATAGGGTACGCCACAGATCGACCTCTGGTTGGTCCTGTTGTGTGGCCTATGCGT3′ | 201.1 ± 42.42 | −16.00 |
| 132 | 5′ACGCATAGGGACCATGCGCAGTGAACTTGGTTCTTTGTGCTCATGTGTGCCTATGCGT3′ | 221.6 ± 41.87 | −14.77 |
| 154 | 5′ACGCATAGGGCATCACACATGTCCTTGCCATTGCTGACTTGTTTGGTGTCCTATGCGT3′ | 268.8 ± 40.43 | −15.40 |
| Samples | Spiked Concentration (ng/g) | Detection Concentration (ng/g) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Pork | 1 | 0.83 ± 0.02 | 83.33 ± 1.53 | 1.83 |
| 5 | 4.66 ± 0.09 | 93.27 ± 1.70 | 1.82 | |
| 10 | 9.70 ± 0.05 | 97.03 ± 0.45 | 0.46 |
| Name | Sequences (5′-3′) |
|---|---|
| Biotin-P | 5′-CCTATGCGTGGAGTGCCAAT-3′-biotin |
| FAM-Forward | 6-FAM-5′-ATTGGCACTCCACGCATAGG-3′ |
| polyA-Reverse | 5′-AAAAAAAAAAAAAAAAAAAAspacer18TTCACGGTAGCACGCATAGG-3′ |
| Q-Forward | 5′-ATTGGCACTCCACGCATAGG-3′ |
| Q-Reverse | 5′-TTCACGGTAGCACGCATAGG-3′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Lu, Q.; Chen, S.; Wang, F.; Hou, J.; Xu, Z.; Meng, C.; Hu, T.; Hou, Y. Selection and Identification of Novel Aptamers Specific for Clenbuterol Based on ssDNA Library Immobilized SELEX and Gold Nanoparticles Biosensor. Molecules 2018, 23, 2337. https://doi.org/10.3390/molecules23092337
Liu X, Lu Q, Chen S, Wang F, Hou J, Xu Z, Meng C, Hu T, Hou Y. Selection and Identification of Novel Aptamers Specific for Clenbuterol Based on ssDNA Library Immobilized SELEX and Gold Nanoparticles Biosensor. Molecules. 2018; 23(9):2337. https://doi.org/10.3390/molecules23092337
Chicago/Turabian StyleLiu, Xixia, Qi Lu, Sirui Chen, Fang Wang, Jianjun Hou, Zhenlin Xu, Chen Meng, Tianyuan Hu, and Yaoyao Hou. 2018. "Selection and Identification of Novel Aptamers Specific for Clenbuterol Based on ssDNA Library Immobilized SELEX and Gold Nanoparticles Biosensor" Molecules 23, no. 9: 2337. https://doi.org/10.3390/molecules23092337
APA StyleLiu, X., Lu, Q., Chen, S., Wang, F., Hou, J., Xu, Z., Meng, C., Hu, T., & Hou, Y. (2018). Selection and Identification of Novel Aptamers Specific for Clenbuterol Based on ssDNA Library Immobilized SELEX and Gold Nanoparticles Biosensor. Molecules, 23(9), 2337. https://doi.org/10.3390/molecules23092337