Involvement of Up-Regulation of DR5 Expression and Down-Regulation of c-FLIP in Niclosamide-Mediated TRAIL Sensitization in Human Renal Carcinoma Caki Cells
Abstract
1. Introduction
2. Results
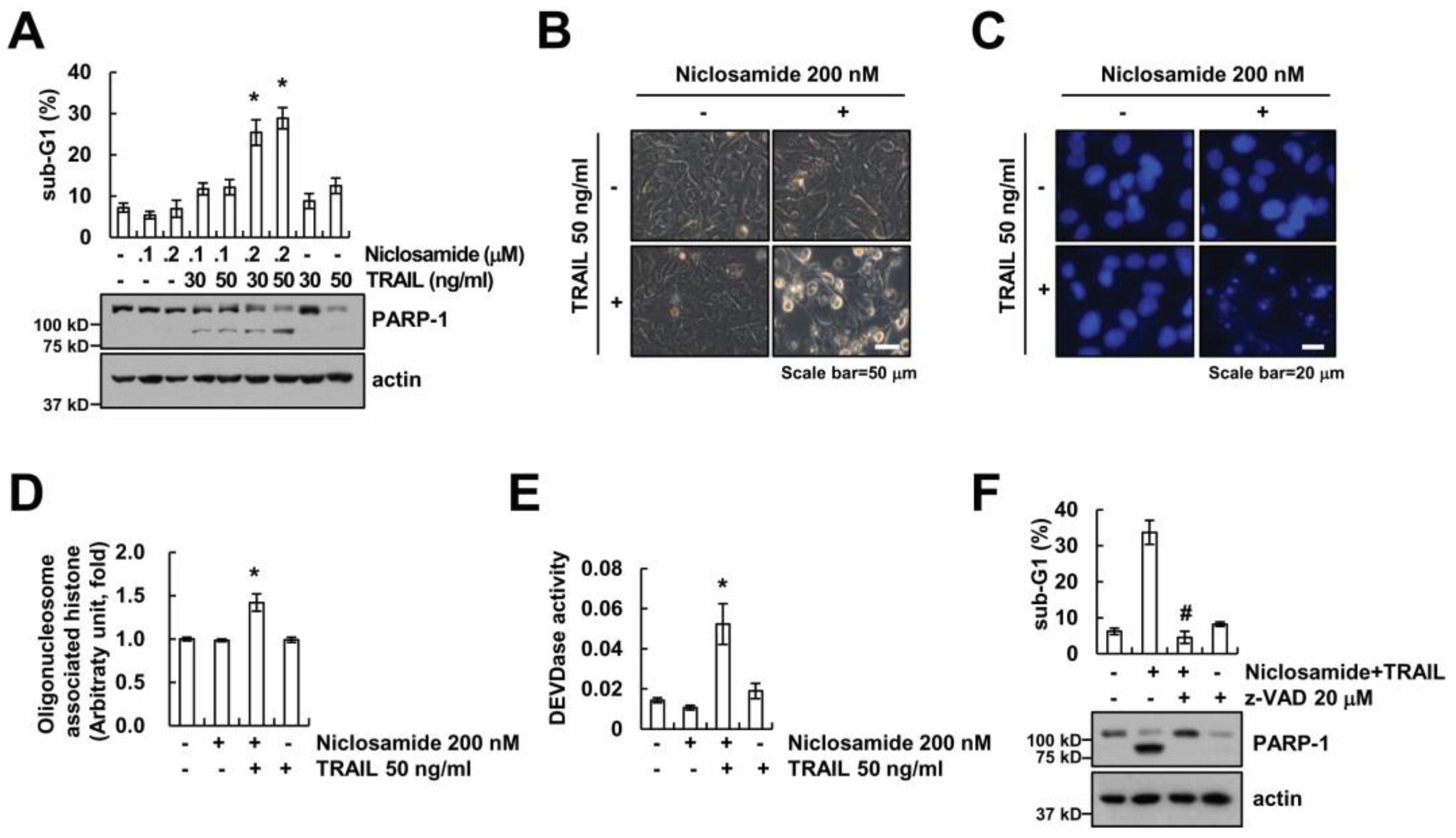
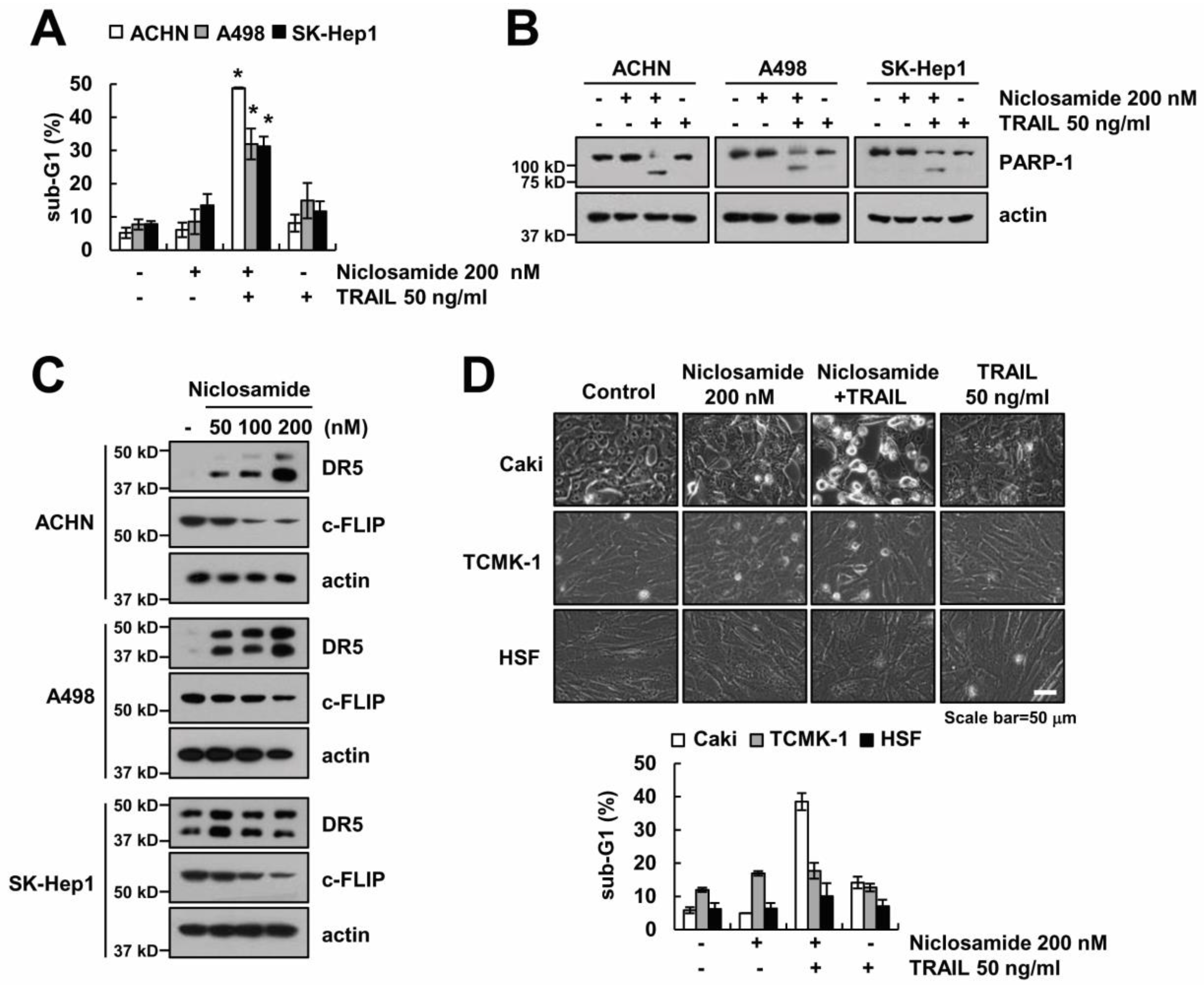
2.1. Niclosamide Sensitizes TRAIL-Mediated Apoptosis in Renal Carcinoma Caki Cells
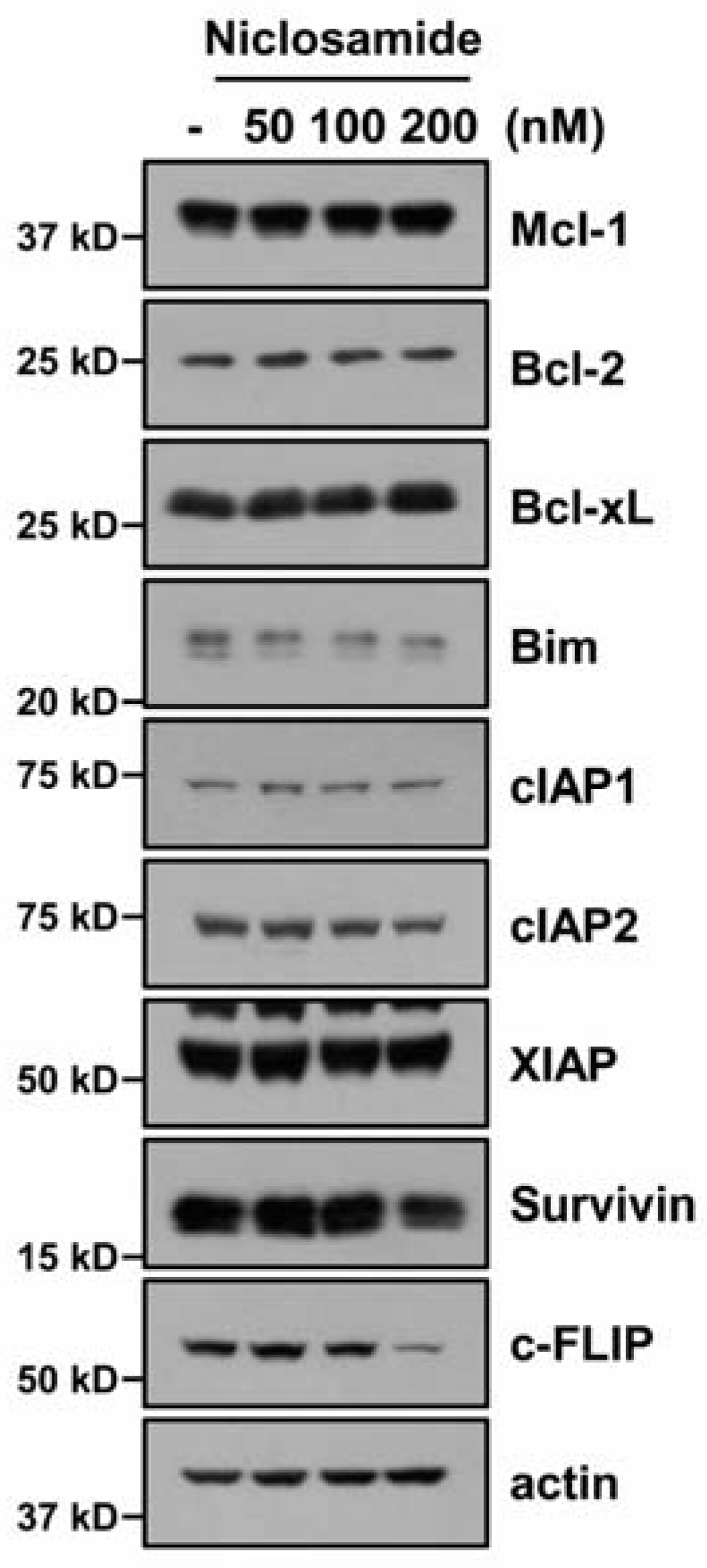
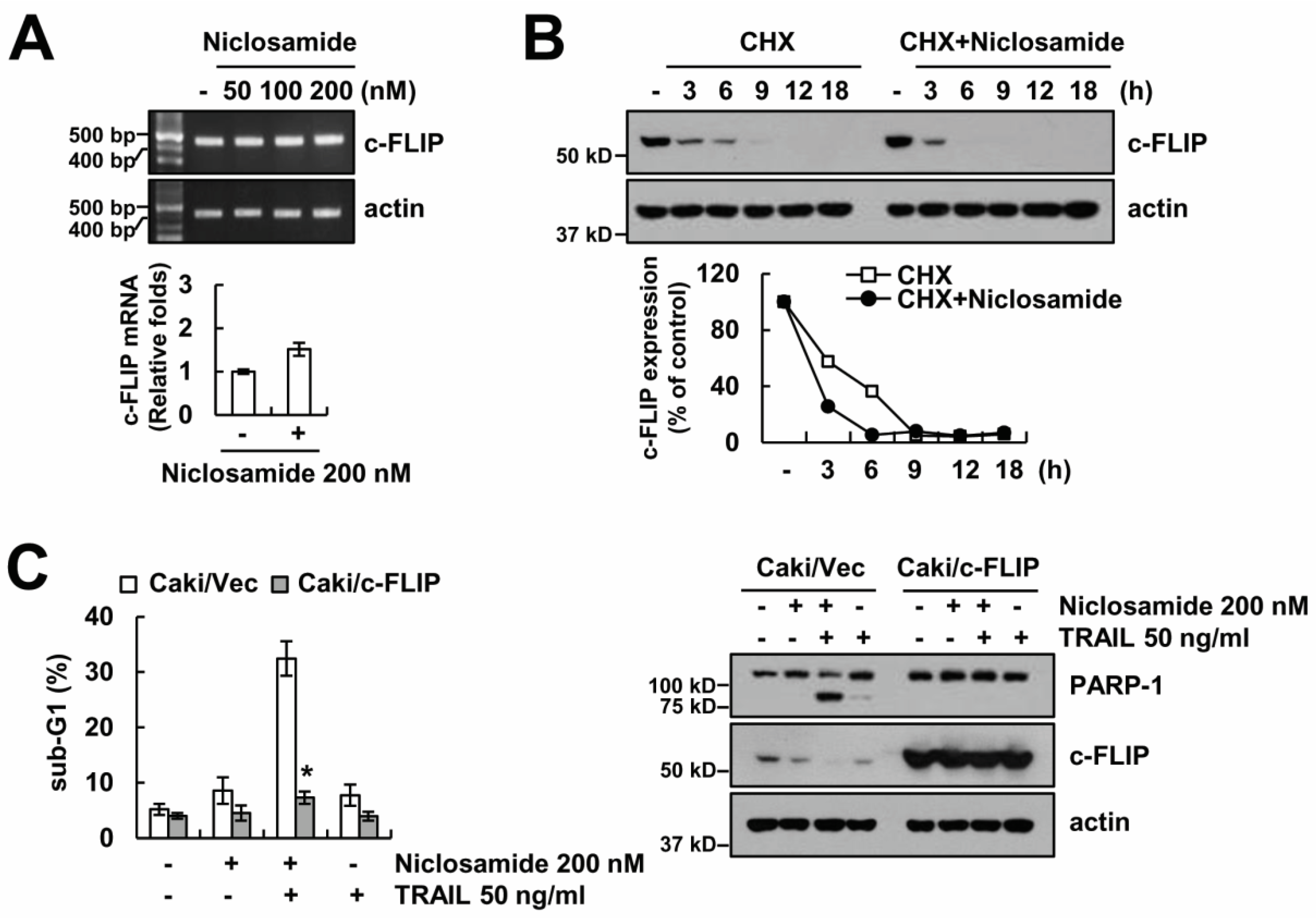
2.2. Down-Regulating c-FLIP Contributes to TRAIL Sensitization by Niclosamide
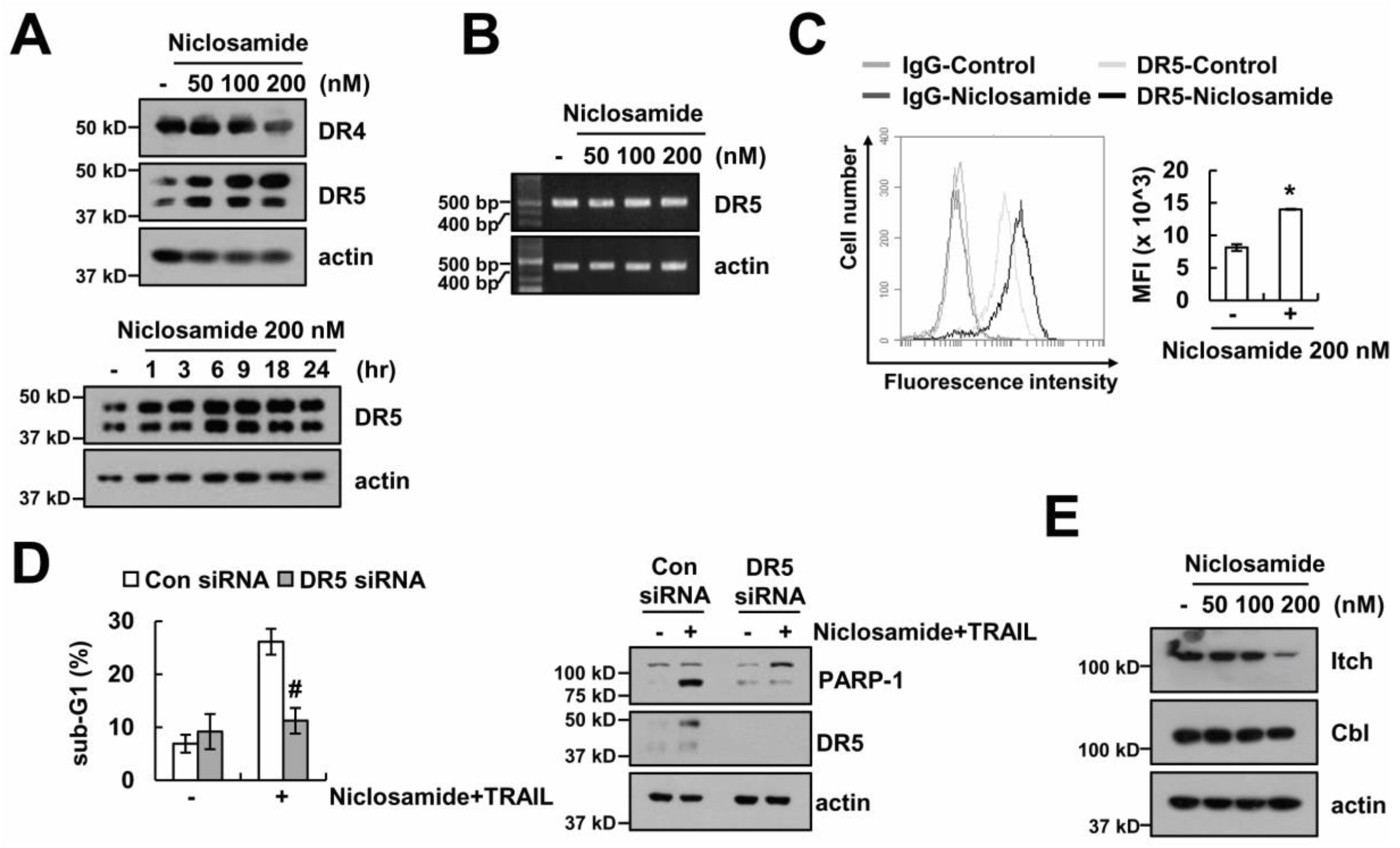
2.3. The Up-Regulating Death Receptor (DR)5 by Niclosamide is Associated with TRAIL Sensitization
2.4. Effect of Niclosamide on TRAIL Sensitization in Other Cancer and Normal Cells
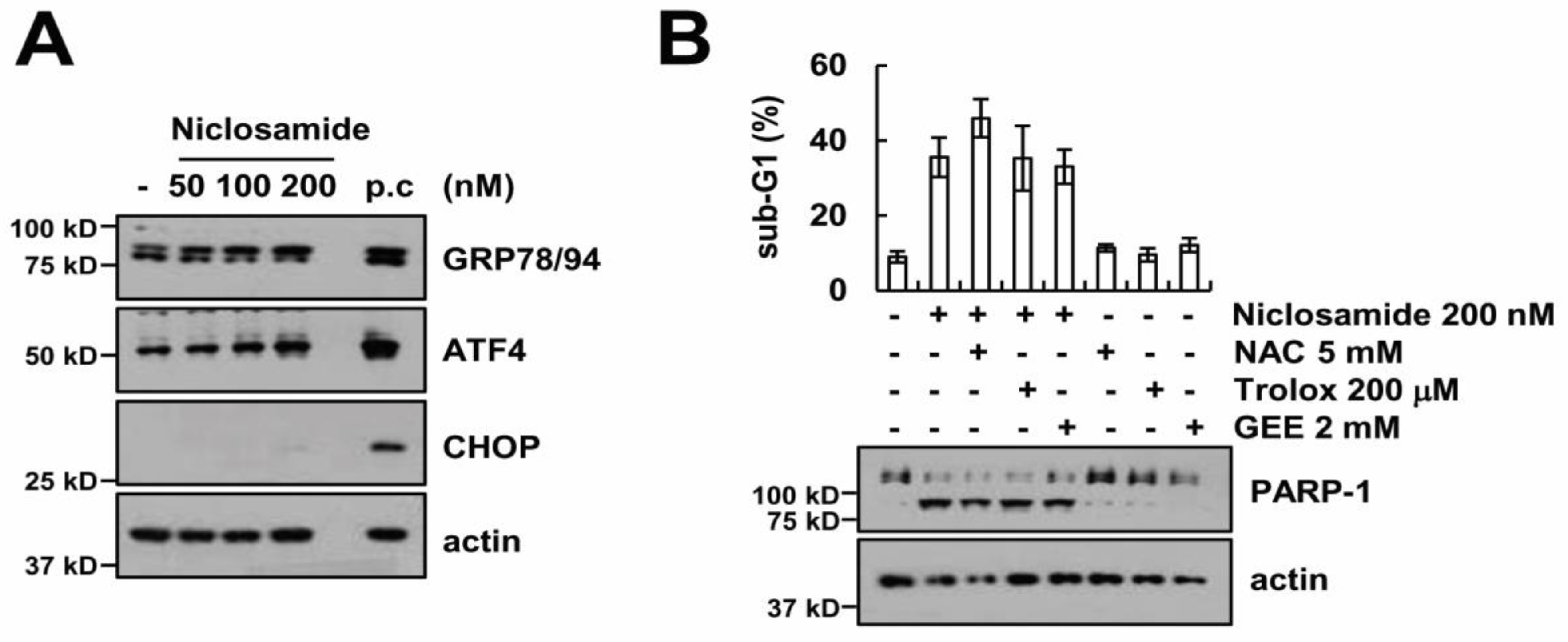
2.5. Endoplasmic Reticulum Stress and Reactive Oxygen Species Generation Are Not Involved in Niclosamide Plus TRAIL-Induced Apoptosis
3. Discussion
4. Materials and Methods
4.1. Cells and Materials
4.2. Western Blot Analysis and Flow Cytometry Analysis
4.3. DAPI Staining and Detection of DNA Fragmentation
4.4. DEVDase Activity Assay
4.5. RT-PCR and Quantitative PCR (qPCR)
4.6. Analysis of DR5 on the Cell Surface
4.7. RNA Interference
4.8. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| TRAIL | Tumor necrosis factor-related apoptosis-inducing ligand |
| ROS | Reactive oxygen species |
| DR | Death receptor |
| ER | Endoplasmic reticulum |
| CHX | cycloheximide |
References
- French, L.E.; Tschopp, J. The TRAIL to selective tumor death. Nat. Med. 1999, 5, 146–147. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; El-Deiry, W.S. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 2003, 22, 8628–8633. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, H.N.; Ashkenazi, A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003, 10, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi, A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat. Rev. Cancer 2002, 2, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Chawla-Sarkar, M.; Bae, S.I.; Reu, F.J.; Jacobs, B.S.; Lindner, D.J.; Borden, E.C. Downregulation of Bcl-2, FLIP or IAPs (XIAP and survivin) by siRNAs sensitizes resistant melanoma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2004, 11, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Londono-Joshi, A.I.; Arend, R.C.; Aristizabal, L.; Lu, W.; Samant, R.S.; Metge, B.J.; Hidalgo, B.; Grizzle, W.E.; Conner, M.; Forero-Torres, A.; et al. Effect of niclosamide on basal-like breast cancers. Mol. Cancer Ther. 2014, 13, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Wieland, A.; Trageser, D.; Gogolok, S.; Reinartz, R.; Hofer, H.; Keller, M.; Leinhaas, A.; Schelle, R.; Normann, S.; Klaas, L.; et al. Anticancer effects of niclosamide in human glioblastoma. Clin. Cancer Res. 2013, 19, 4124–4136. [Google Scholar] [CrossRef] [PubMed]
- Arend, R.C.; Londono-Joshi, A.I.; Samant, R.S.; Li, Y.; Conner, M.; Hidalgo, B.; Alvarez, R.D.; Landen, C.N.; Straughn, J.M.; Buchsbaum, D.J. Inhibition of Wnt/beta-catenin pathway by niclosamide: A therapeutic target for ovarian cancer. Gynecol. Oncol. 2014, 134, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, Q.; Gong, Z.; Chen, S.; Cui, L. Niclosamide suppresses renal cell carcinoma by inhibiting Wnt/beta-catenin and inducing mitochondrial dysfunctions. Springerplus 2016, 5, 1436. [Google Scholar] [CrossRef] [PubMed]
- Suliman, M.A.; Zhang, Z.; Na, H.; Ribeiro, A.L.; Zhang, Y.; Niang, B.; Hamid, A.S.; Zhang, H.; Xu, L.; Zuo, Y. Niclosamide inhibits colon cancer progression through downregulation of the Notch pathway and upregulation of the tumor suppressor miR-200 family. Int. J. Mol. Med. 2016, 38, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; You, S.; Hu, Z.; Chen, Z.G.; Sica, G.L.; Khuri, F.R.; Curran, W.J.; Shin, D.M.; Deng, X. Inhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancer. PLoS ONE 2013, 8, e74670. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hu, Z.; Sun, S.Y.; Chen, Z.G.; Owonikoko, T.K.; Sica, G.L.; Ramalingam, S.S.; Curran, W.J.; Khuri, F.R.; Deng, X. Niclosamide overcomes acquired resistance to erlotinib through suppression of STAT3 in non-small cell lung cancer. Mol. Cancer Ther. 2013, 12, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Duan, L.; He, Q.; Zhang, Z.; Zhou, Y.; Wu, D.; Pan, J.; Pei, D.; Ding, K. Identification of Niclosamide as a New Small-Molecule Inhibitor of the STAT3 Signaling Pathway. ACS Med. Chem. Lett. 2010, 1, 454–459. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Li, R.; Park, D.; Xie, M.; Sica, G.L.; Cao, Y.; Xiao, Z.Q.; Deng, X. Disruption of STAT3 by niclosamide reverses radioresistance of human lung cancer. Mol. Cancer Ther. 2014, 13, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Gao, Y.; Zhang, X.; Wang, J.; Ding, D.; Zhang, Y.; Zhang, J.; Chen, H. Niclosamide sensitizes triple-negative breast cancer cells to ionizing radiation in association with the inhibition of Wnt/beta-catenin signaling. Oncotarget 2016, 7, 42126–42138. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, X.; Ward, T.; Pegram, M.; Shen, K. Combined niclosamide with cisplatin inhibits epithelial-mesenchymal transition and tumor growth in cisplatin-resistant triple-negative breast cancer. Tumour Biol. 2016, 37, 9825–9835. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Yang, D.; Yu, Y.; Xiang, M.; Li, H.; Yang, J.; Li, J.; Jiang, D.; Zhou, H.; Xu, Z.; et al. Niclosamide enhances the cytotoxic effect of cisplatin in cisplatin-resistant human lung cancer cells via suppression of lung resistance-related protein and c-myc. Mol. Med. Rep. 2018, 17, 3497–3502. [Google Scholar] [CrossRef] [PubMed]
- Kelley, S.K.; Ashkenazi, A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr. Opin. Pharmacol. 2004, 4, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, J.H.; Song, J.J. c-Cbl shRNA-expressing adenovirus sensitizes TRAIL-induced apoptosis in prostate cancer DU-145 through increases of DR4/5. Cancer Gene Ther. 2013, 20, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Tay, K.H.; Dong, L.; Thorne, R.F.; Jiang, C.C.; Yang, E.; Tseng, H.Y.; Liu, H.; Christopherson, R.; Hersey, P.; et al. Cystatin B inhibition of TRAIL-induced apoptosis is associated with the protection of FLIP(L) from degradation by the E3 ligase itch in human melanoma cells. Cell Death Differ. 2010, 17, 1354–1367. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Lu, Z.; Ding, K.; Li, J.; Du, X.; Chen, C.; Sun, X.; Wu, Y.; Zhou, J.; Pan, J. Antineoplastic mechanisms of niclosamide in acute myelogenous leukemia stem cells: Inactivation of the NF-κB pathway and generation of reactive oxygen species. Cancer Res. 2010, 70, 2516–2527. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.; Son, A.R.; Ahn, J.; Song, J.Y. Niclosamide enhances ROS-mediated cell death through c-Jun activation. Biomed. Pharmacother. 2014, 68, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, L.; Shen, H.; Lin, H.; Li, D. Anthelminthic drug niclosamide sensitizes the responsiveness of cervical cancer cells to paclitaxel via oxidative stress-mediated mTOR inhibition. Biochem. Biophys. Res. Commun. 2017, 484, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.R.; Min, K.J.; Woo, S.M.; Choe, M.; Choi, K.S.; Lee, Y.K.; Yoon, G.; Kwon, T.K. Inhibition of Cathepsin S Induces Mitochondrial ROS That Sensitizes TRAIL-Mediated Apoptosis Through p53-Mediated Downregulation of Bcl-2 and c-FLIP. Antioxid. Redox Signal. 2017, 27, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Min, K.J.; Choi, K.S.; Kubatka, P.; Kruzliak, P.; Kim, D.E.; Kwon, T.K. Chloroquine enhances TRAIL-mediated apoptosis through up-regulation of DR5 by stabilization of mRNA and protein in cancer cells. Sci. Rep. 2016, 6, 22921. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.U.; Kim, T.H.; Kim, D.E.; Min, K.J.; Kwon, T.K. NOX4-mediated ROS production induces apoptotic cell death via down-regulation of c-FLIP and Mcl-1 expression in combined treatment with thioridazine and curcumin. Redox Biol. 2017, 13, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Kwon, Y.J.; Chun, Y.J. CYP1B1 Activates Wnt/beta-Catenin Signaling through Suppression of Herc5-Mediated ISGylation for Protein Degradation on beta-Catenin in HeLa Cells. Toxicol. Res. 2017, 33, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Shin, D.Y. Repression of the F-box protein Skp2 is essential for actin damage-induced tetraploid G1 arrest. BMB Rep. 2017, 50, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.; Shin, S.M. Induction of Lipin1 by ROS-Dependent SREBP-2 Activation. Toxicol. Res. 2017, 33, 219–224. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, J.M.; Woo, S.M.; Seo, S.U.; Min, K.-J.; Kim, D.E.; Kwon, T.K. Involvement of Up-Regulation of DR5 Expression and Down-Regulation of c-FLIP in Niclosamide-Mediated TRAIL Sensitization in Human Renal Carcinoma Caki Cells. Molecules 2018, 23, 2264. https://doi.org/10.3390/molecules23092264
Yun JM, Woo SM, Seo SU, Min K-J, Kim DE, Kwon TK. Involvement of Up-Regulation of DR5 Expression and Down-Regulation of c-FLIP in Niclosamide-Mediated TRAIL Sensitization in Human Renal Carcinoma Caki Cells. Molecules. 2018; 23(9):2264. https://doi.org/10.3390/molecules23092264
Chicago/Turabian StyleYun, Jeong Mi, Seon Min Woo, Seung Un Seo, Kyoung-Jin Min, Dong Eun Kim, and Taeg Kyu Kwon. 2018. "Involvement of Up-Regulation of DR5 Expression and Down-Regulation of c-FLIP in Niclosamide-Mediated TRAIL Sensitization in Human Renal Carcinoma Caki Cells" Molecules 23, no. 9: 2264. https://doi.org/10.3390/molecules23092264
APA StyleYun, J. M., Woo, S. M., Seo, S. U., Min, K.-J., Kim, D. E., & Kwon, T. K. (2018). Involvement of Up-Regulation of DR5 Expression and Down-Regulation of c-FLIP in Niclosamide-Mediated TRAIL Sensitization in Human Renal Carcinoma Caki Cells. Molecules, 23(9), 2264. https://doi.org/10.3390/molecules23092264





