Discovery of High-Affinity Cannabinoid Receptors Ligands through a 3D-QSAR Ushered by Scaffold-Hopping Analysis †
Abstract
1. Introduction
2. Results and Discussion
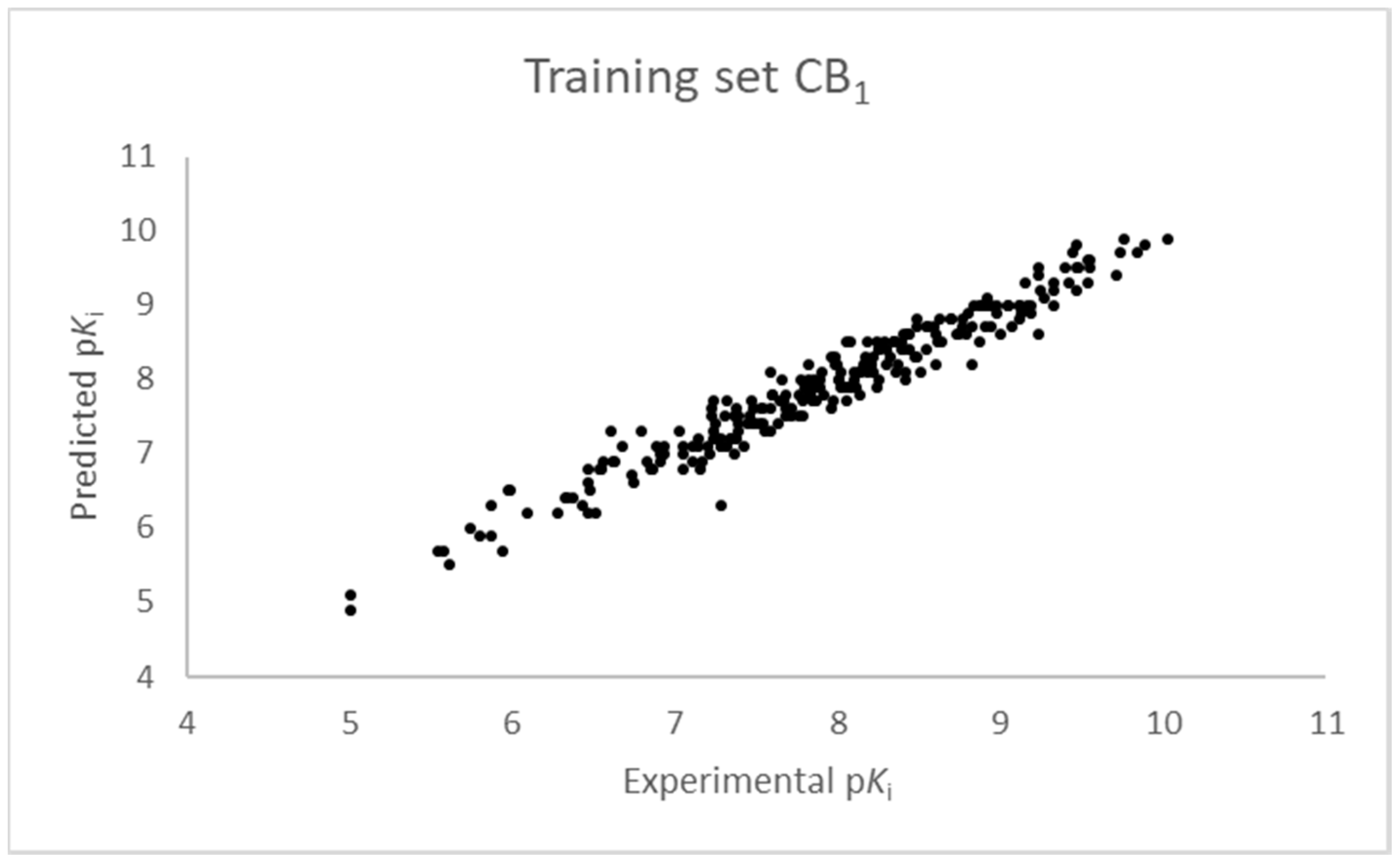
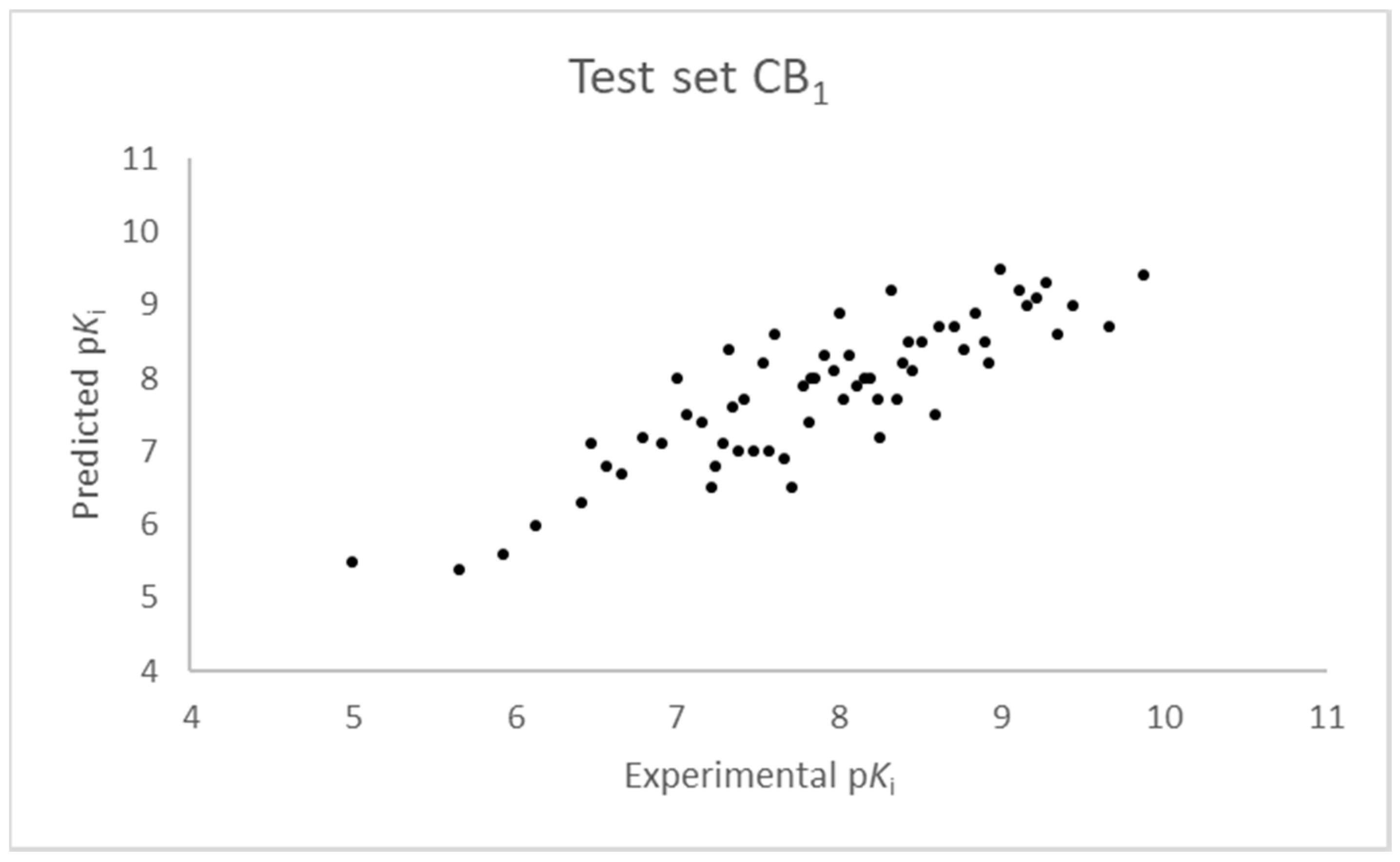
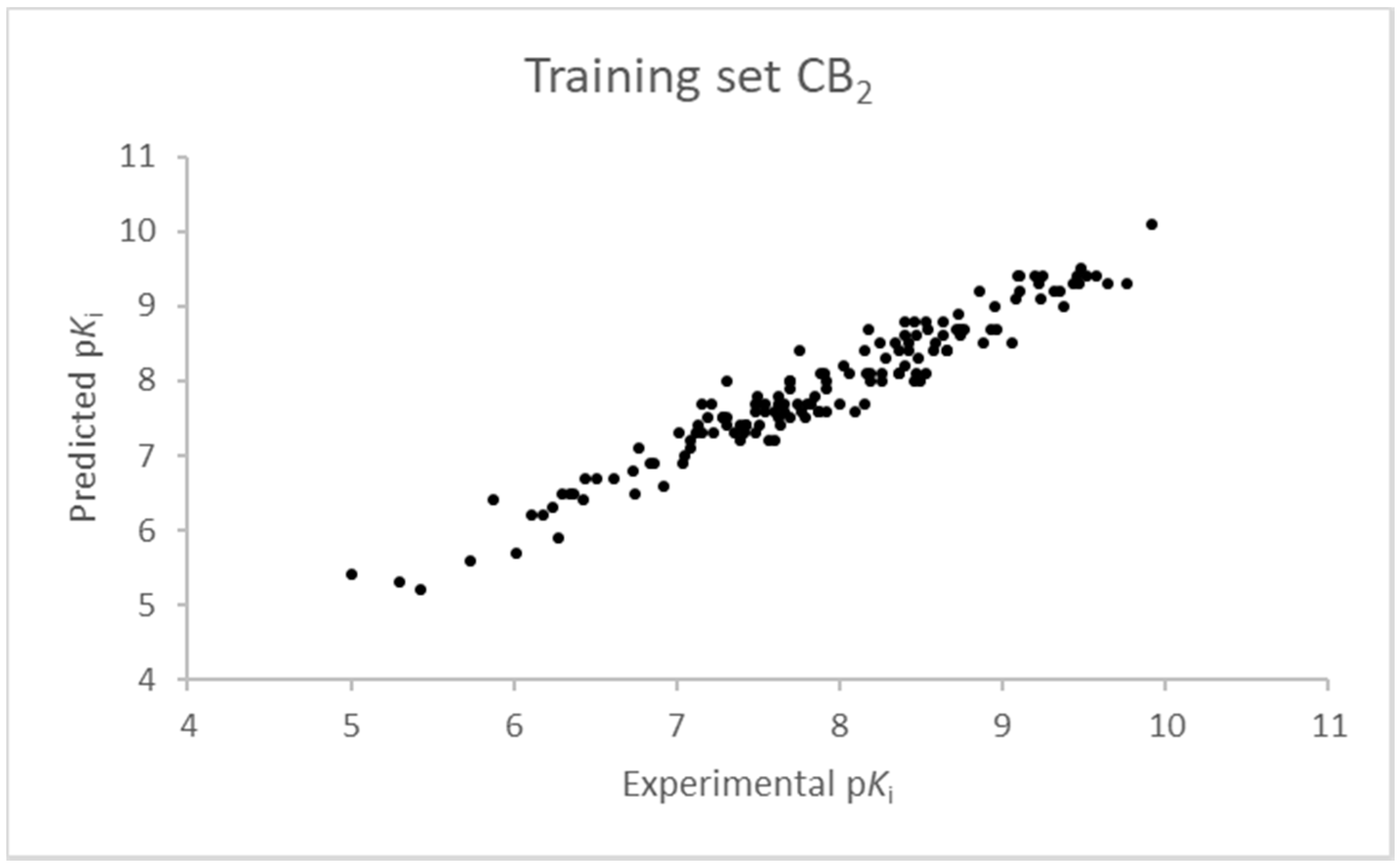
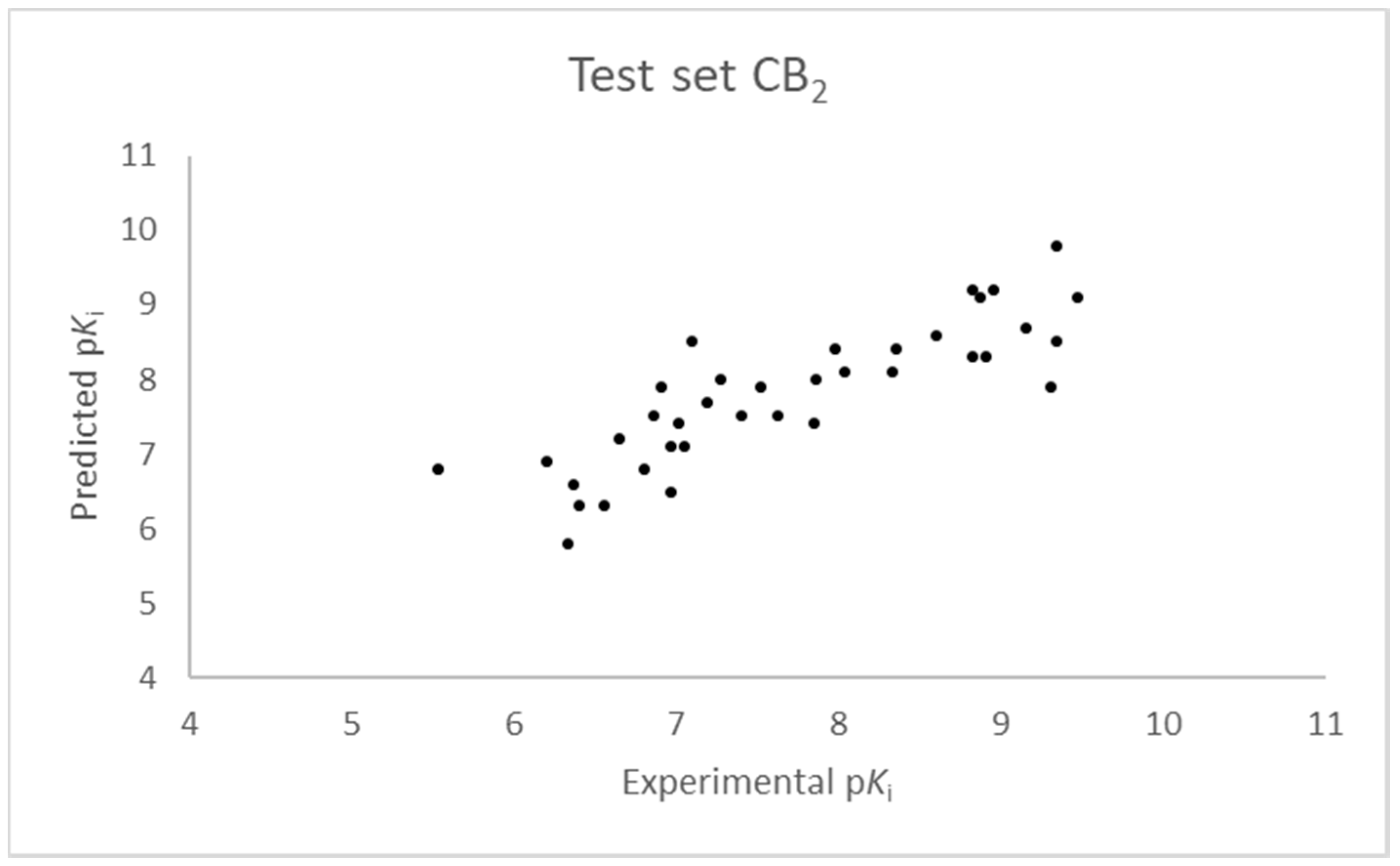
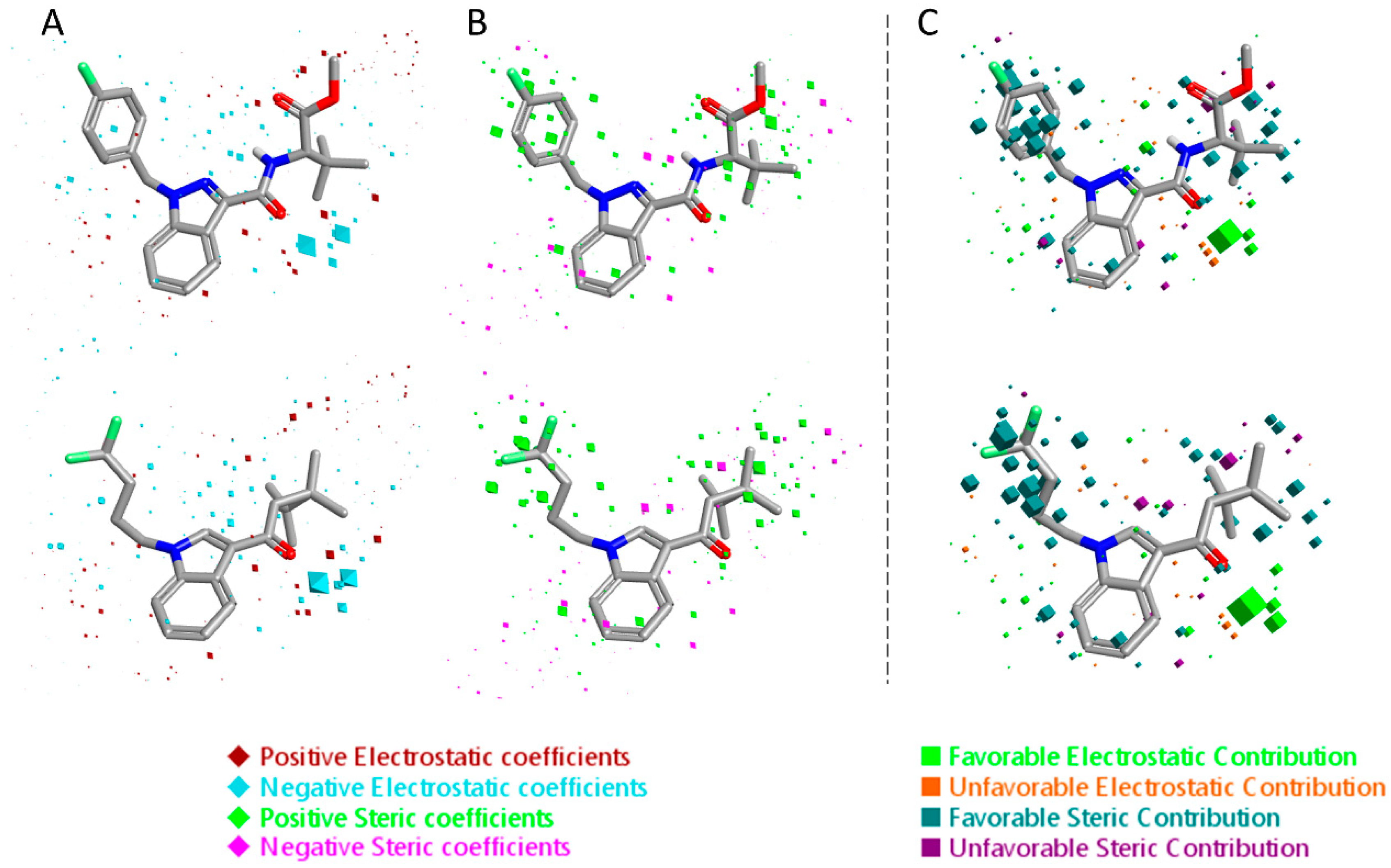
2.1. Statistical Analysis and Results
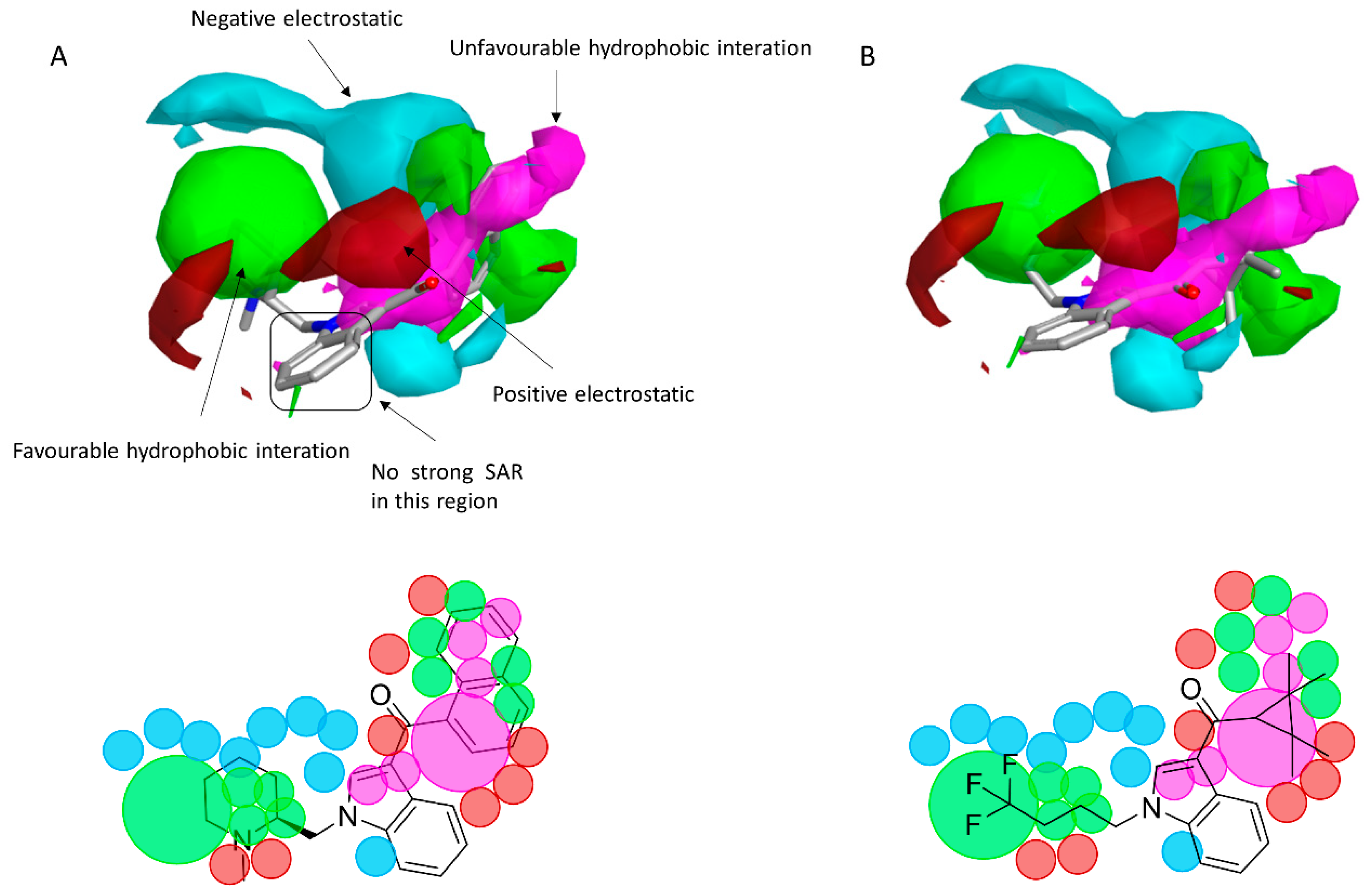
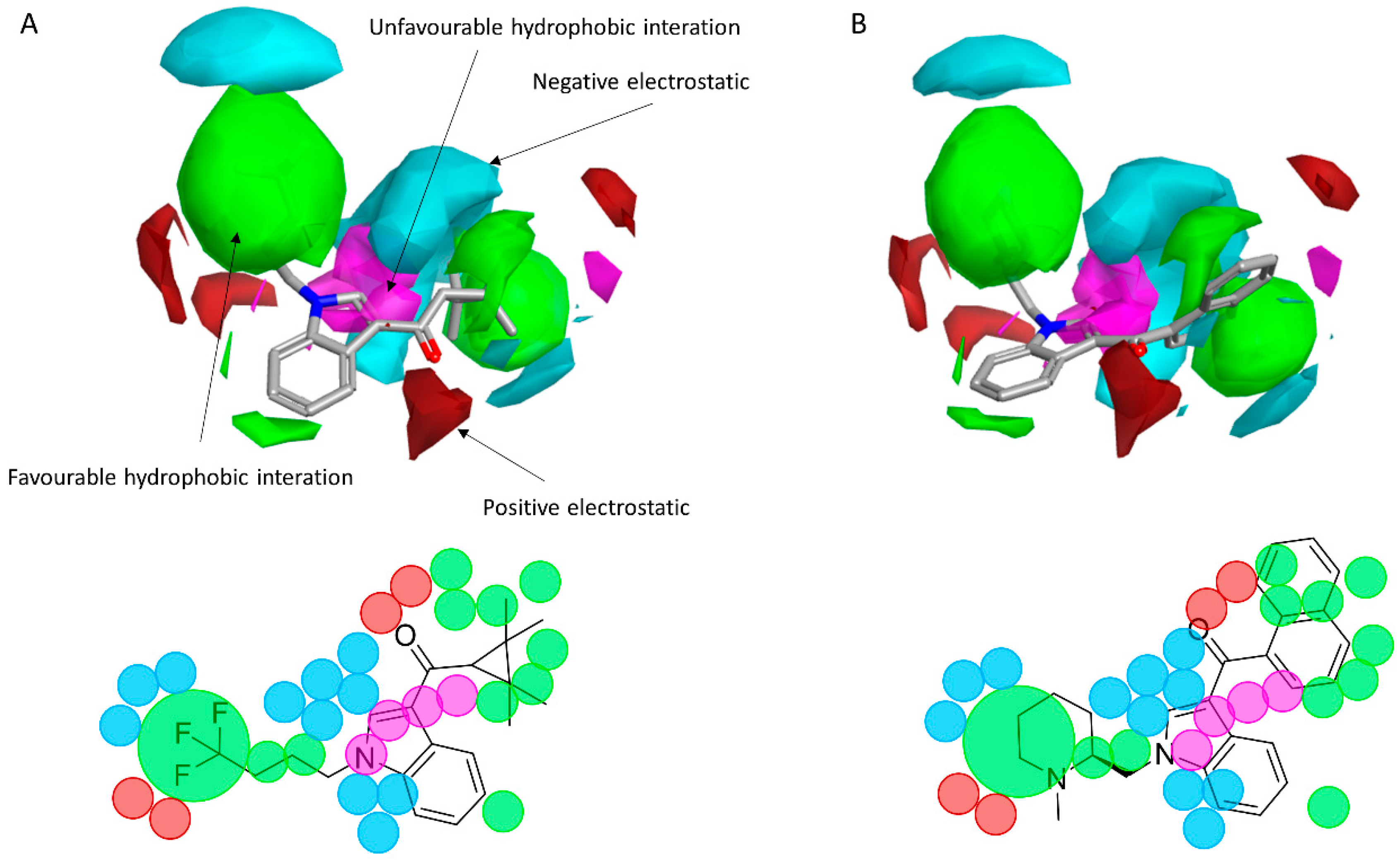
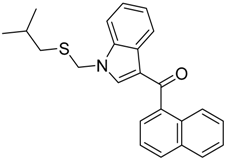
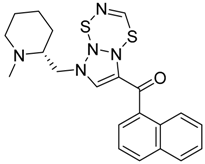
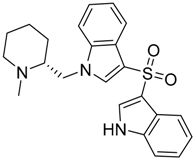
2.2. Finding Bioisosteres
3. Materials and Methods
3.1. Biological Data
3.2. Molecular Modeling
3.3. Compound Alignment
3.4. Bioisosteric Replacement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aizpurua-Olaizola, O.; Elezgarai, I.; Rico-Barrio, I.; Zarandona, I.; Etxebarria, N.; Usobiaga, A. Targeting the endocannabinoid system: Future therapeutic strategies. Drug Discov. Today 2017, 22, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R.G. Pharmacology of cannabinoid cb1 and cb2 receptors. Pharmacol. Ther. 1997, 74, 129–180. [Google Scholar] [CrossRef]
- Valdeolivas, S.; Satta, V.; Pertwee, R.G.; Fernandez-Ruiz, J.; Sagredo, O. Sativex-like combination of phytocannabinoids is neuroprotective in malonate-lesioned rats, an inflammatory model of huntington’s disease: Role of cb1 and cb2 receptors. ACS Chem. Neurosci. 2012, 3, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cabrerizo, R.; Garcia-Fuster, M.J. Opposite regulation of scannabinoid cb1 and cb2 receptors in the prefrontal cortex of rats treated with cocaine during adolescence. Neurosci. Lett. 2016, 615, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Clayton, N.; Marshall, F.H.; Bountra, C.; O’Shaughnessy, C.T. Cb1 and cb2 cannabinoid receptors are implicated in inflammatory pain. Pain 2002, 96, 253–260. [Google Scholar] [CrossRef]
- Elmes, S.J.; Winyard, L.A.; Medhurst, S.J.; Clayton, N.M.; Wilson, A.W.; Kendall, D.A.; Chapman, V. Activation of cb1 and cb2 receptors attenuates the induction and maintenance of inflammatory pain in the rat. Pain 2005, 118, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Mule, F.; Amato, A.; Baldassano, S.; Serio, R. Involvement of cb1 and cb2 receptors in the modulation of cholinergic neurotransmission in mouse gastric preparations. Pharmacol. Res. 2007, 56, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Herkenham, M.; Lynn, A.B.; Little, M.D.; Johnson, M.R.; Melvin, L.S.; de Costa, B.R.; Rice, K.C. Cannabinoid receptor localization in brain. Proc. Natl. Acad. Sci. USA 1990, 87, 1932–1936. [Google Scholar] [CrossRef] [PubMed]
- Cravatt, B.F.; Lichtman, A.H. The endogenous cannabinoid system and its role in nociceptive behavior. J. Neurobiol. 2004, 61, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Ruiz, J.; Romero, J.; Ramos, J.A. Endocannabinoids and neurodegenerative disorders: Parkinson’s disease, huntington’s chorea, alzheimer’s disease, and others. Handb. Exp. Pharmacol. 2015, 231, 233–259. [Google Scholar] [PubMed]
- Pryce, G.; Baker, D. Endocannabinoids in multiple sclerosis and amyotrophic lateral sclerosis. Handb. Exp. Pharmacol. 2015, 231, 213–231. [Google Scholar] [PubMed]
- Abbate, V.; Schwenk, M.; Presley Brandon, C.; Uchiyama, N. The ongoing challenge of novel psychoactive drugs of abuse. Part i. Synthetic cannabinoids (iupac technical report). Pure Appl. Chem. 2018, 90, 1255–1282. [Google Scholar] [CrossRef]
- Galiegue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carriere, D.; Carayon, P.; Bouaboula, M.; Shire, D.; Le Fur, G.; Casellas, P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Pavlopoulos, S.; Thakur, G.A.; Nikas, S.P.; Makriyannis, A. Cannabinoid receptors as therapeutic targets. Curr. Pharm. Des. 2006, 12, 1751–1769. [Google Scholar] [PubMed]
- Mackie, K. Cannabinoid receptors as therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Rescifina, A.; Floresta, G.; Marrazzo, A.; Parenti, C.; Prezzavento, O.; Nastasi, G.; Dichiara, M.; Amata, E. Sigma-2 receptor ligands qsar model dataset. Data Brief 2017, 13, 514–535. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Rescifina, A.; Marrazzo, A.; Dichiara, M.; Pistara, V.; Pittala, V.; Prezzavento, O.; Amata, E. Hyphenated 3d-qsar statistical model-scaffold hopping analysis for the identification of potentially potent and selective sigma-2 receptor ligands. Eur. J. Med. Chem. 2017, 139, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Amata, E.; Dichiara, M.; Marrazzo, A.; Salerno, L.; Romeo, G.; Prezzavento, O.; Pittala, V.; Rescifina, A. Identification of potentially potent heme oxygenase 1 inhibitors through 3d-qsar coupled to scaffold-hopping analysis. ChemMedChem 2018, 13, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Rescifina, A.; Floresta, G.; Marrazzo, A.; Parenti, C.; Prezzavento, O.; Nastasi, G.; Dichiara, M.; Amata, E. Development of a sigma-2 receptor affinity filter through a monte carlo based qsar analysis. Eur. J. Pharm. Sci. 2017, 106, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Mella-Raipan, J.; Hernandez-Pino, S.; Morales-Verdejo, C.; Pessoa-Mahana, D. 3d-qsar/comfa-based structure-affinity/selectivity relationships of aminoalkylindoles in the cannabinoid cb1 and cb2 receptors. Molecules 2014, 19, 2842–2861. [Google Scholar] [CrossRef] [PubMed]
- Mella-Raipan, J.A.; Lagos, C.F.; Recabarren-Gajardo, G.; Espinosa-Bustos, C.; Romero-Parra, J.; Pessoa-Mahana, H.; Iturriaga-Vasquez, P.; Pessoa-Mahana, C.D. Design, synthesis, binding and docking-based 3d-qsar studies of 2-pyridylbenzimidazoles--a new family of high affinity cb1 cannabinoid ligands. Molecules 2013, 18, 3972–4001. [Google Scholar] [CrossRef] [PubMed]
- Durdagi, S.; Kapou, A.; Kourouli, T.; Andreou, T.; Nikas, S.P.; Nahmias, V.R.; Papahatjis, D.P.; Papadopoulos, M.G.; Mavromoustakos, T. The application of 3d-qsar studies for novel cannabinoid ligands substituted at the c1’ position of the alkyl side chain on the structural requirements for binding to cannabinoid receptors cb1 and cb2. J. Med. Chem. 2007, 50, 2875–2885. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.; Han, X.W.; Liu, Q.; Makriyannis, A.; Wang, J.; Xie, X.Q. 3d-qsar studies of arylpyrazole antagonists of cannabinoid receptor subtypes cb1 and cb2. A combined nmr and comfa approach. J. Med. Chem. 2006, 49, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Salo, O.M.; Savinainen, J.R.; Parkkari, T.; Nevalainen, T.; Lahtela-Kakkonen, M.; Gynther, J.; Laitinen, J.T.; Jarvinen, T.; Poso, A. 3d-qsar studies on cannabinoid cb1 receptor agonists: G-protein activation as biological data. J. Med. Chem. 2006, 49, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Romero-Parra, J.; Chung, H.; Tapia, R.A.; Faundez, M.; Morales-Verdejo, C.; Lorca, M.; Lagos, C.F.; Di Marzo, V.; David Pessoa-Mahana, C.; Mella, J. Combined comfa and comsia 3d-qsar study of benzimidazole and benzothiophene derivatives with selective affinity for the cb2 cannabinoid receptor. Eur. J. Pharm. Sci. 2017, 101, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Aung, M.M.; Griffin, G.; Huffman, J.W.; Wu, M.; Keel, C.; Yang, B.; Showalter, V.M.; Abood, M.E.; Martin, B.R. Influence of the n-1 alkyl chain length of cannabimimetic indoles upon cb(1) and cb(2) receptor binding. Drug Alcohol Depend. 2000, 60, 133–140. [Google Scholar] [CrossRef]
- Wiley, J.L.; Compton, D.R.; Dai, D.; Lainton, J.A.; Phillips, M.; Huffman, J.W.; Martin, B.R. Structure-activity relationships of indole- and pyrrole-derived cannabinoids. J. Pharmacol. Exp. Ther. 1998, 285, 995–1004. [Google Scholar] [PubMed]
- Huffman, J.W.; Mabon, R.; Wu, M.J.; Lu, J.; Hart, R.; Hurst, D.P.; Reggio, P.H.; Wiley, J.L.; Martin, B.R. 3-indolyl-1-naphthylmethanes: New cannabimimetic indoles provide evidence for aromatic stacking interactions with the cb(1) cannabinoid receptor. Bioorg. Med. Chem. 2003, 11, 539–549. [Google Scholar] [CrossRef]
- Huffman, J.W.; Padgett, L.W. Recent developments in the medicinal chemistry of cannabimimetic indoles, pyrroles and indenes. Curr. Med. Chem. 2005, 12, 1395–1411. [Google Scholar] [CrossRef] [PubMed]
- Huffman, J.W.; Zengin, G.; Wu, M.J.; Lu, J.; Hynd, G.; Bushell, K.; Thompson, A.L.; Bushell, S.; Tartal, C.; Hurst, D.P.; et al. Structure-activity relationships for 1-alkyl-3-(1-naphthoyl)indoles at the cannabinoid cb(1) and cb(2) receptors: Steric and electronic effects of naphthoyl substituents. New highly selective cb(2) receptor agonists. Bioorg. Med. Chem. 2005, 13, 89–112. [Google Scholar] [CrossRef] [PubMed]
- Wiley, J.L.; Smith, V.J.; Chen, J.; Martin, B.R.; Huffman, J.W. Synthesis and pharmacology of 1-alkyl-3-(1-naphthoyl)indoles: Steric and electronic effects of 4- and 8-halogenated naphthoyl substituents. Bioorg. Med. Chem. 2012, 20, 2067–2081. [Google Scholar] [CrossRef] [PubMed]
- Hess, C.; Schoeder, C.T.; Pillaiyar, T.; Madea, B.; Muller, C.E. Pharmacological evaluation of synthetic cannabinoids identified as constituents of spice. Forensic Toxicol. 2016, 34, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.A.; Brockie, H.C.; Stevenson, L.A.; Murphy, V.L.; Templeton, F.; Makriyannis, A.; Pertwee, R.G. Agonist-inverse agonist characterization at cb1 and cb2 cannabinoid receptors of l759633, l759656, and am630. Br. J. Pharmacol. 1999, 126, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Huffman, J.W.; Szklennik, P.V.; Almond, A.; Bushell, K.; Selley, D.E.; He, H.; Cassidy, M.P.; Wiley, J.L.; Martin, B.R. 1-pentyl-3-phenylacetylindoles, a new class of cannabimimetic indoles. Bioorg. Med. Chem. Lett. 2005, 15, 4110–4113. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, N.; Kikura-Hanajiri, R.; Goda, Y. Identification of a novel cannabimimetic phenylacetylindole, cannabipiperidiethanone, as a designer drug in a herbal product and its affinity for cannabinoid cb(1) and cb(2) receptors. Chem. Pharm. Bull. 2011, 59, 1203–1205. [Google Scholar] [CrossRef] [PubMed]
- Wiley, J.L.; Marusich, J.A.; Lefever, T.W.; Antonazzo, K.R.; Wallgren, M.T.; Cortes, R.A.; Patel, P.R.; Grabenauer, M.; Moore, K.N.; Thomas, B.F. Ab-chminaca, ab-pinaca, and fubimina: Affinity and potency of novel synthetic cannabinoids in producing delta9-tetrahydrocannabinol-like effects in mice. J. Pharmacol. Exp. Ther. 2015, 354, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Showalter, V.M.; Compton, D.R.; Martin, B.R.; Abood, M.E. Evaluation of binding in a transfected cell line expressing a peripheral cannabinoid receptor (cb2): Identification of cannabinoid receptor subtype selective ligands. J. Pharmacol. Exp. Ther. 1996, 278, 989–999. [Google Scholar] [PubMed]
- Huffman, J.W.; Padgett, L.W.; Isherwood, M.L.; Wiley, J.L.; Martin, B.R. 1-alkyl-2-aryl-4-(1-naphthoyl)pyrroles: New high affinity ligands for the cannabinoid cb1 and cb2 receptors. Bioorg. Med. Chem. Lett. 2006, 16, 5432–5435. [Google Scholar] [CrossRef] [PubMed]
- Franz, F.; Angerer, V.; Moosmann, B.; Auwarter, V. Phase i metabolism of the highly potent synthetic cannabinoid mdmb-chmica and detection in human urine samples. Drug Test. Anal. 2017, 9, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Pryce, G.; Baker, D. Antidote to cannabinoid intoxication: The cb1 receptor inverse agonist, am251, reverses hypothermic effects of the cb1 receptor agonist, cb-13, in mice. Br. J. Pharmacol. 2017, 174, 3790–3794. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi-Carmona, M.; Barth, F.; Heaulme, M.; Shire, D.; Calandra, B.; Congy, C.; Martinez, S.; Maruani, J.; Neliat, G.; Caput, D.; et al. Sr141716a, a potent and selective antagonist of the brain cannabinoid receptor. FEBS Lett. 1994, 350, 240–244. [Google Scholar] [CrossRef]
- Gatch, M.B.; Forster, M.J. Delta(9)-tetrahydrocannabinol-like effects of novel synthetic cannabinoids in mice and rats. Psychopharmacology 2016, 233, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M.; Deng, H.; Zvonok, A.; Cockayne, D.A.; Kwan, J.; Mata, H.P.; Vanderah, T.W.; Lai, J.; Porreca, F.; Makriyannis, A.; et al. Activation of cb2 cannabinoid receptors by am1241 inhibits experimental neuropathic pain: Pain inhibition by receptors not present in the cns. Proc. Natl. Acad. Sci. USA 2003, 100, 10529–10533. [Google Scholar] [CrossRef] [PubMed]
- Buchler, I.P.; Hayes, M.J.; Hegde, S.G.; Hockerman, S.L.; Jones, D.E.; Kortum, S.W.; Rico, J.G.; Tenbrink, R.E.; Wu, K.K. Indazole Derivatives. Patent No. WO2009106980, 3 September 2009. [Google Scholar]
- Makriyannis, A.; Deng, H. Cannabimimetic Indole Derivatives. Patent No. WO200128557, 26 April 2001. [Google Scholar]
- Makriyannis, A.; Deng, H. Cannabimimetic Indole Derivatives. US Patent No. 2008/0090871, 17 April 2008. [Google Scholar]
- Cheeseright, T.; Mackey, M.; Rose, S.; Vinter, A. Molecular field extrema as descriptors of biological activity: Definition and validation. J. Chem. Inf. Model. 2006, 46, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, M.; Huang, B.; Ji, H.; Yuan, M. Comparative molecular field analysis (comfa) and comparative molecular similarity indices analysis (comsia) studies on alpha(1a)-adrenergic receptor antagonists based on pharmacophore molecular alignment. Int. J. Mol. Sci. 2011, 12, 7022–7037. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.Q.; Jin, H.X.; Yan, X.J.; Zhu, P.; Hu, G.X. 3d-qsar and 3d-qssr studies of thieno[2,3-d]pyrimidin-4-yl hydrazone analogues as cdk4 inhibitors by comfa analysis. Acta Pharmacol. Sin. 2014, 35, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Khan, F. 3d-qsar studies on maslinic acid analogs for anticancer activity against breast cancer cell line mcf-7. Sci. Rep. 2017, 7, 6019. [Google Scholar] [CrossRef] [PubMed]
- Fallarini, S.; Massarotti, A.; Gesu, A.; Giovarruscio, S.; Zabetta, G.C.; Bergo, R.; Giannelli, B.; Brunco, A.; Lombardi, G.; Sorba, G.; et al. In silico-driven multicomponent synthesis of 4,5-and 1,5-disubstituted imidazoles as indoleamine 2,3-dioxygenase inhibitors. MedChemComm 2016, 7, 409–419. [Google Scholar] [CrossRef]
- Lee, J.W.; Hirota, T.; Kumar, A.; Kim, N.J.; Irle, S.; Kay, S.A. Development of small-molecule cryptochrome stabilizer derivatives as modulators of the circadian clock. ChemMedChem 2015, 10, 1489–1497. [Google Scholar] [CrossRef] [PubMed]
- De Jong, S. Simpls: An alternative approach to partial least squares regression. Chemom. Intell. Lab. Syst. 1993, 18, 251–263. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. Pls-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Schoeder, C.T.; Hess, C.; Madea, B.; Meiler, J.; Muller, C.E. Pharmacological evaluation of new constituents of “spice”: Synthetic cannabinoids based on indole, indazole, benzimidazole and carbazole scaffolds. Forensic Toxicol. 2018, 36, 385–403. [Google Scholar] [CrossRef] [PubMed]
- Gamage, T.F.; Farquhar, C.E.; Lefever, T.W.; Marusich, J.A.; Kevin, R.C.; McGregor, I.S.; Wiley, J.L.; Thomas, B.F. Molecular and behavioral pharmacological characterization of abused synthetic cannabinoids mmb- and mdmb-fubinaca, mn-18, nnei, cumyl-pica, and 5-fluoro-cumyl-pica. J. Pharmacol. Exp. Ther. 2018, 365, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Barf, T.; Lehmann, F.; Hammer, K.; Haile, S.; Axen, E.; Medina, C.; Uppenberg, J.; Svensson, S.; Rondahl, L.; Lundback, T. N-benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (a-fabp) inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 1745–1748. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J. Optimization of parameters for semiempirical methods iv: Extension of mndo, am1, and pm3 to more main group elements. J. Mol. Model. 2004, 10, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Alemán, C.; Luque, F.J.; Orozco, M. Suitability of the pm3-derived molecular electrostatic potentials. J. Comput. Chem. 1993, 14, 799–808. [Google Scholar] [CrossRef]
- Qiao, F.; Luo, L.; Peng, H.; Luo, S.; Huang, W.; Cui, J.; Li, X.; Kong, L.; Jiang, D.; Chitwood, D.J.; et al. Characterization of three novel fatty acid- and retinoid-binding protein genes (ha-far-1, ha-far-2 and hf-far-1) from the cereal cyst nematodes heterodera avenae and h. Filipjevi. PLoS ONE 2016, 11, e0160003. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.P.; Leonard, J.T.; Roy, K. Exploring the impact of size of training sets for the development of predictive qsar models. Chemom. Intell. Lab. Syst. 2008, 90, 31–42. [Google Scholar] [CrossRef]
- Hua, T.; Vemuri, K.; Nikas, S.P.; Laprairie, R.B.; Wu, Y.; Qu, L.; Pu, M.; Korde, A.; Jiang, S.; Ho, J.H.; et al. Crystal structures of agonist-bound human cannabinoid receptor cb1. Nature 2017, 547, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Chaudhaery, S.S.; Roy, K.K.; Saxena, A.K. Consensus superiority of the pharmacophore-based alignment, over maximum common substructure (mcs): 3d-qsar studies on carbamates as acetylcholinesterase inhibitors. J. Chem. Inf. Model. 2009, 49, 1590–1601. [Google Scholar] [CrossRef] [PubMed]
- Olesen, P.H. The use of bioisosteric groups in lead optimization. Curr. Opin. Drug Discov. Dev. 2001, 4, 471–478. [Google Scholar]
- Burger, A. Isosterism and bioisosterism in drug design. Prog. Drug. Res. 1991, 37, 287–371. [Google Scholar] [PubMed]
- Patani, G.A.; LaVoie, E.J. Bioisosterism: A rational approach in drug design. Chem. Rev. 1996, 96, 3147–3176. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. Zinc—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Kruger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The chembl bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.R.; Parry, D.M.; Perry, B.G.; Groom, C.R. Heteroaromatic rings of the future. J. Med. Chem. 2009, 52, 2952–2963. [Google Scholar] [CrossRef] [PubMed]
- Emcdda. European Drug Report 2018. Available online: Http://www.Emcdda.Europa.Eu/edr2018_en (accessed on 17 July 2018).
- Nayak, B.P.; Khajuria, H. Synthetic marijuana is no more marijuana. Asian J. Psychiatr. 2017. [Google Scholar] [CrossRef] [PubMed]
- Palamar, J.J.; Barratt, M.J. Synthetic cannabinoids: Undesirable alternatives to natural marijuana. Am. J. Drug Alcohol Abuse 2016, 42, 371–373. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds not available from the authors. |
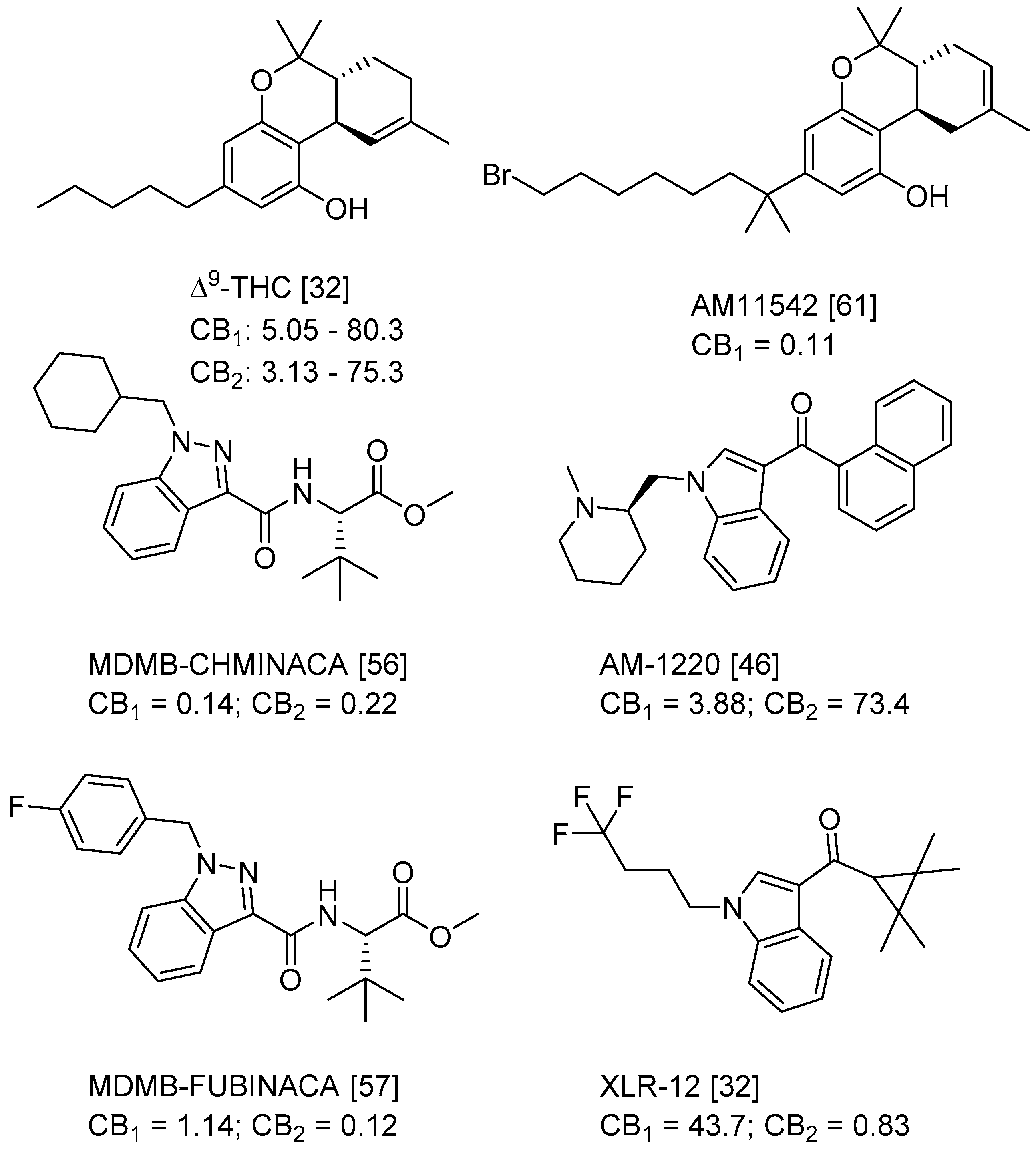

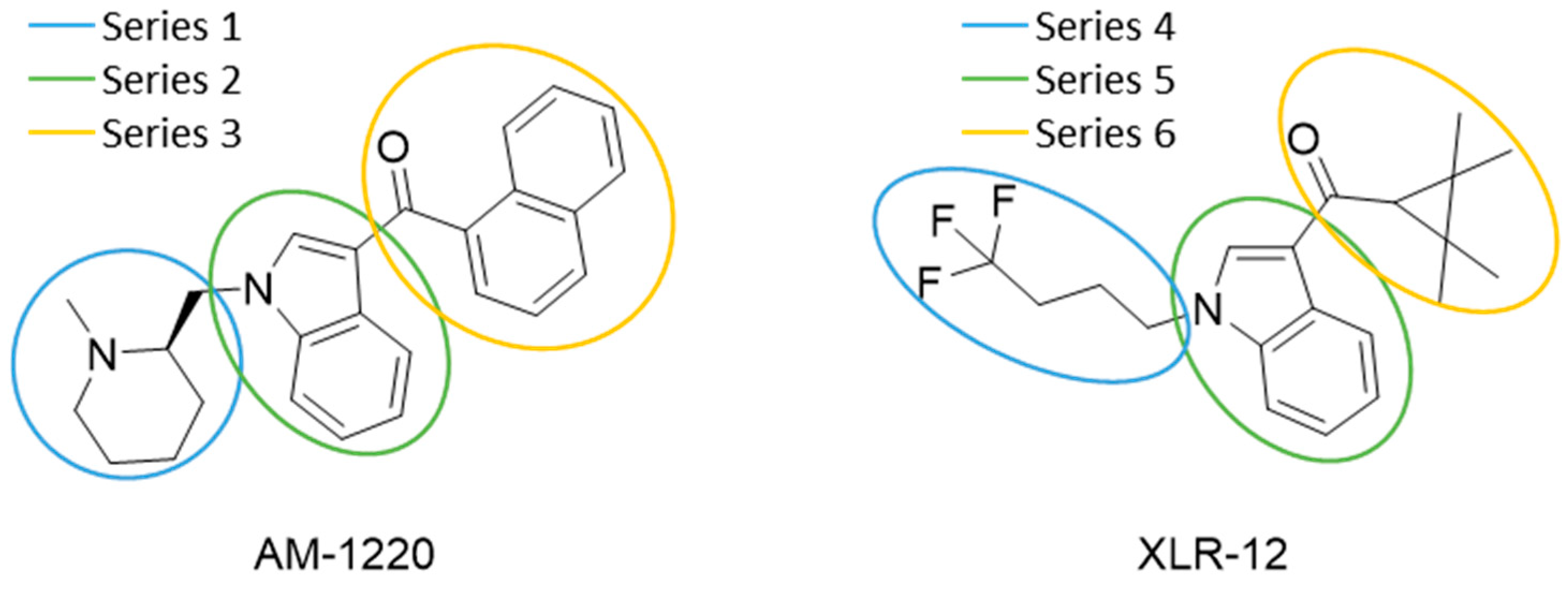
| Series 1 | Series 2 | Series 3 |
 |  |  |
| CB1 = 9.3 CB2 = 7.9 | CB1 = 8.8 CB2 = 7.9 | CB1 = 8.9 CB2 = 6.7 |
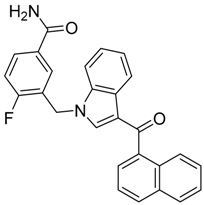 | 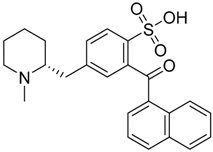 | 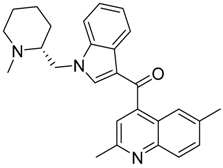 |
| CB1 = 9.2 CB2 = 8.3 | CB1 = 8.7 CB2 = 8.0 | CB1 = 8.8 CB2 = 8.0 |
| Series 4 | Series 5 | Series 6 |
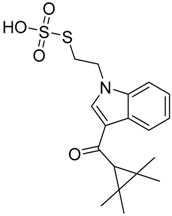 | 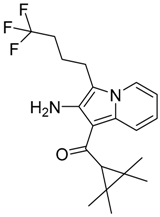 | 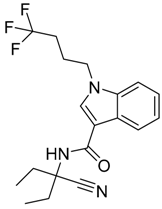 |
| CB1 = 8.5 CB2 = 9.9 | CB1 = 8.0 CB2 = 9.3 | CB1 = 8.3 CB2 = 8.9 |
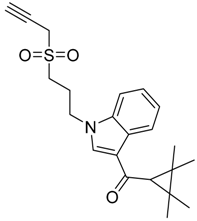 | 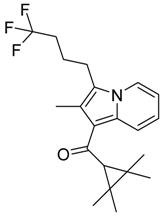 | 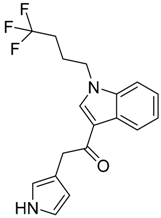 |
| CB1 = 8.5 CB2 = 9.2 | CB1 = 8.0 CB2 = 9.6 | CB1 = 8.2 CB2 = 8.9 |
| Series 1 | Series 2 | Series 3 |
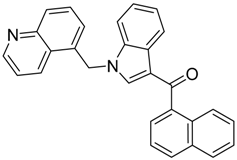 | 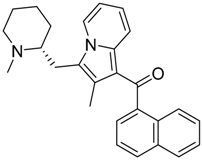 | 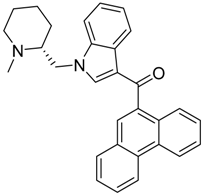 |
| CB2 = 9.4 CB1 = 8.6 | CB2 = 8.7 CB1 = 8.1 | CB2 = 8.3 CB1 = 8.1 |
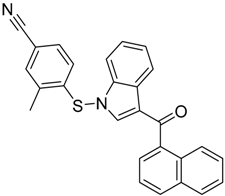 |  | 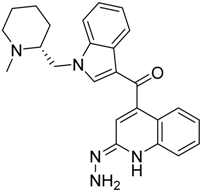 |
| CB2 = 9.4 CB1 = 7.7 | CB2 = 8.6 CB1 = 8.6 | CB2 = 8.2 CB1 = 7.8 |
| Series 4 | Series 5 | Series 6 |
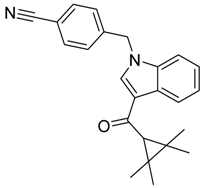 | 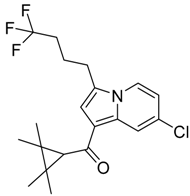 | 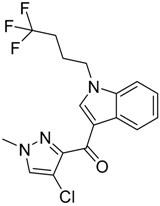 |
| CB2 = 10.2 CB1 = 7.9 | CB2 = 9.9 CB1 = 7.8 | CB2 = 10.2 CB1 = 7.8 |
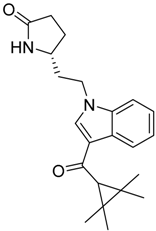 | 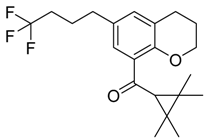 |  |
| CB2 = 10.1 CB1 = 7.8 | CB2 = 9.8 CB1 = 7.3 | CB2 = 10.1 CB1 = 7.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Floresta, G.; Apirakkan, O.; Rescifina, A.; Abbate, V. Discovery of High-Affinity Cannabinoid Receptors Ligands through a 3D-QSAR Ushered by Scaffold-Hopping Analysis. Molecules 2018, 23, 2183. https://doi.org/10.3390/molecules23092183
Floresta G, Apirakkan O, Rescifina A, Abbate V. Discovery of High-Affinity Cannabinoid Receptors Ligands through a 3D-QSAR Ushered by Scaffold-Hopping Analysis. Molecules. 2018; 23(9):2183. https://doi.org/10.3390/molecules23092183
Chicago/Turabian StyleFloresta, Giuseppe, Orapan Apirakkan, Antonio Rescifina, and Vincenzo Abbate. 2018. "Discovery of High-Affinity Cannabinoid Receptors Ligands through a 3D-QSAR Ushered by Scaffold-Hopping Analysis" Molecules 23, no. 9: 2183. https://doi.org/10.3390/molecules23092183
APA StyleFloresta, G., Apirakkan, O., Rescifina, A., & Abbate, V. (2018). Discovery of High-Affinity Cannabinoid Receptors Ligands through a 3D-QSAR Ushered by Scaffold-Hopping Analysis. Molecules, 23(9), 2183. https://doi.org/10.3390/molecules23092183







