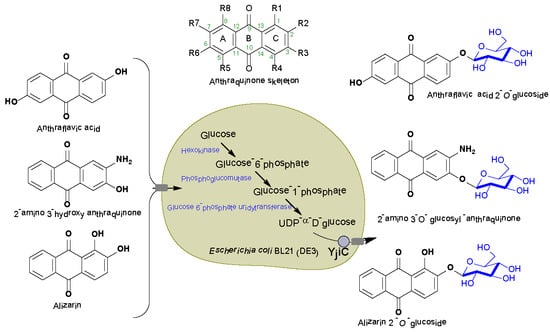
Microbial Synthesis of Non-Natural Anthraquinone Glucosides Displaying Superior Antiproliferative Properties
Abstract
1. Introduction
2. Results
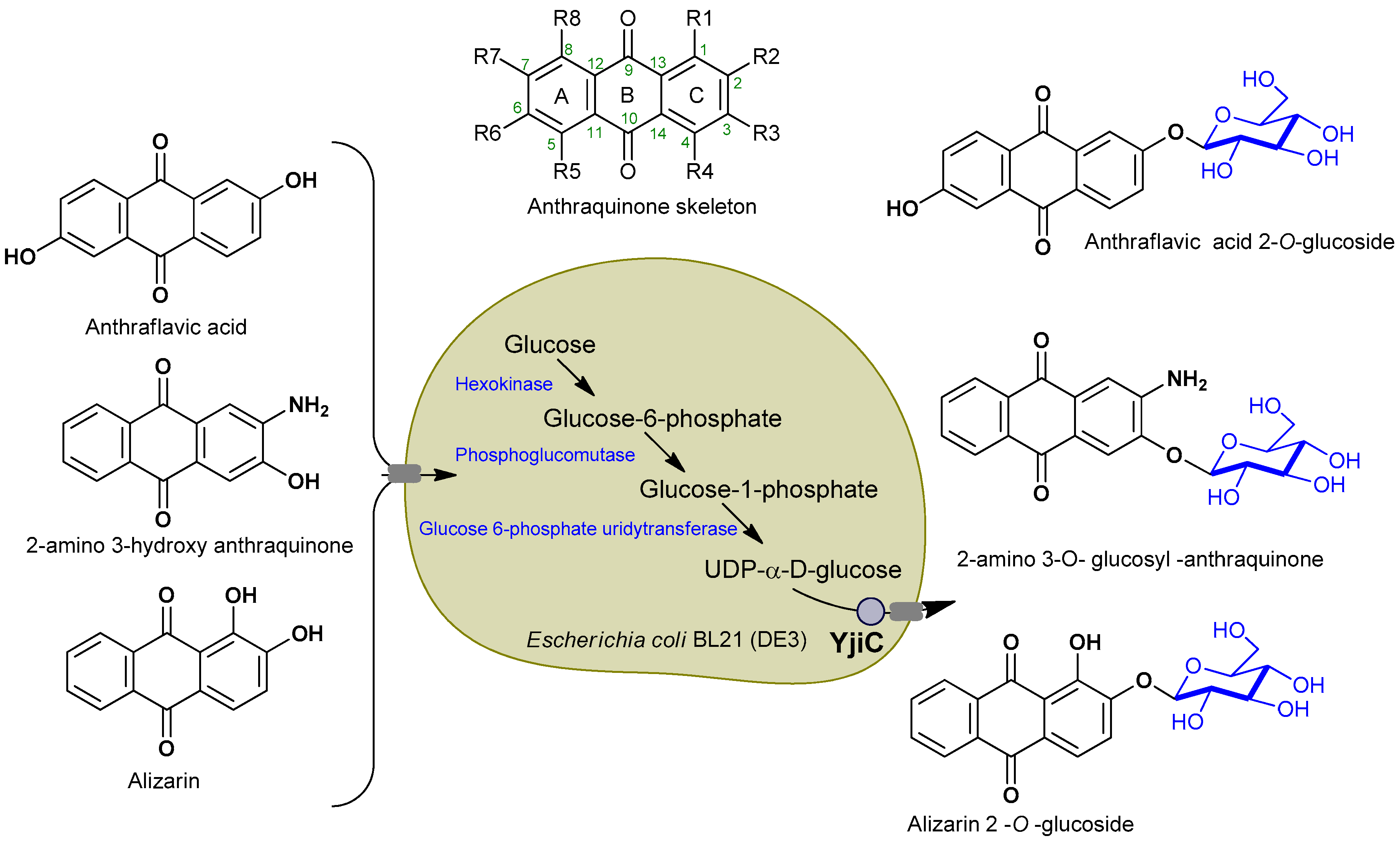
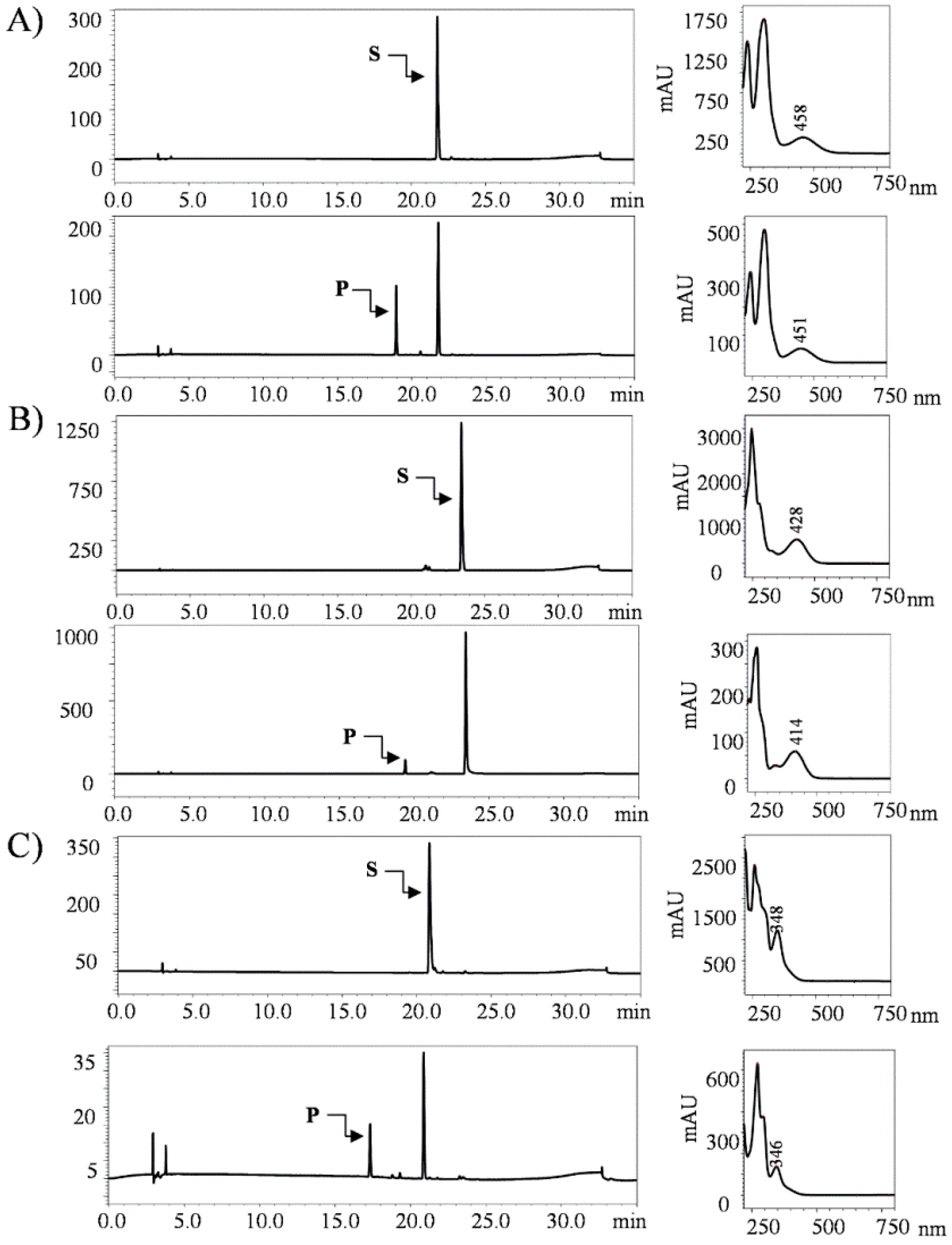
2.1. Biotransformation of Anthraquinones
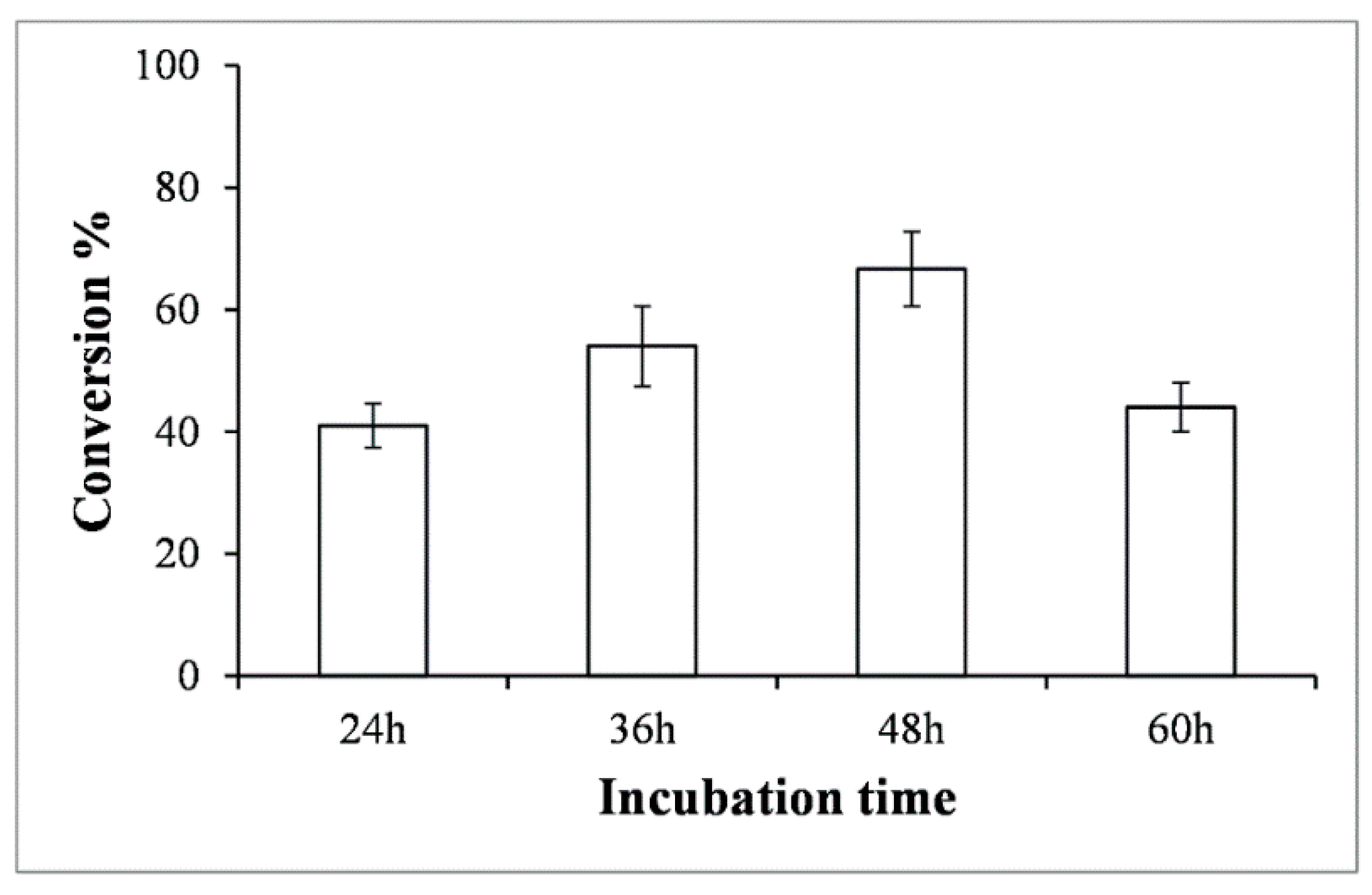
2.2. Glucose Supplementation for Production Optimization in Shake Flask
2.3. Comparative Anticancer Activity of Anthraquinone and Their Glucoside Derivatives
2.4. Scale up Production of Alizarin-O-Glucoside in Bioreactor
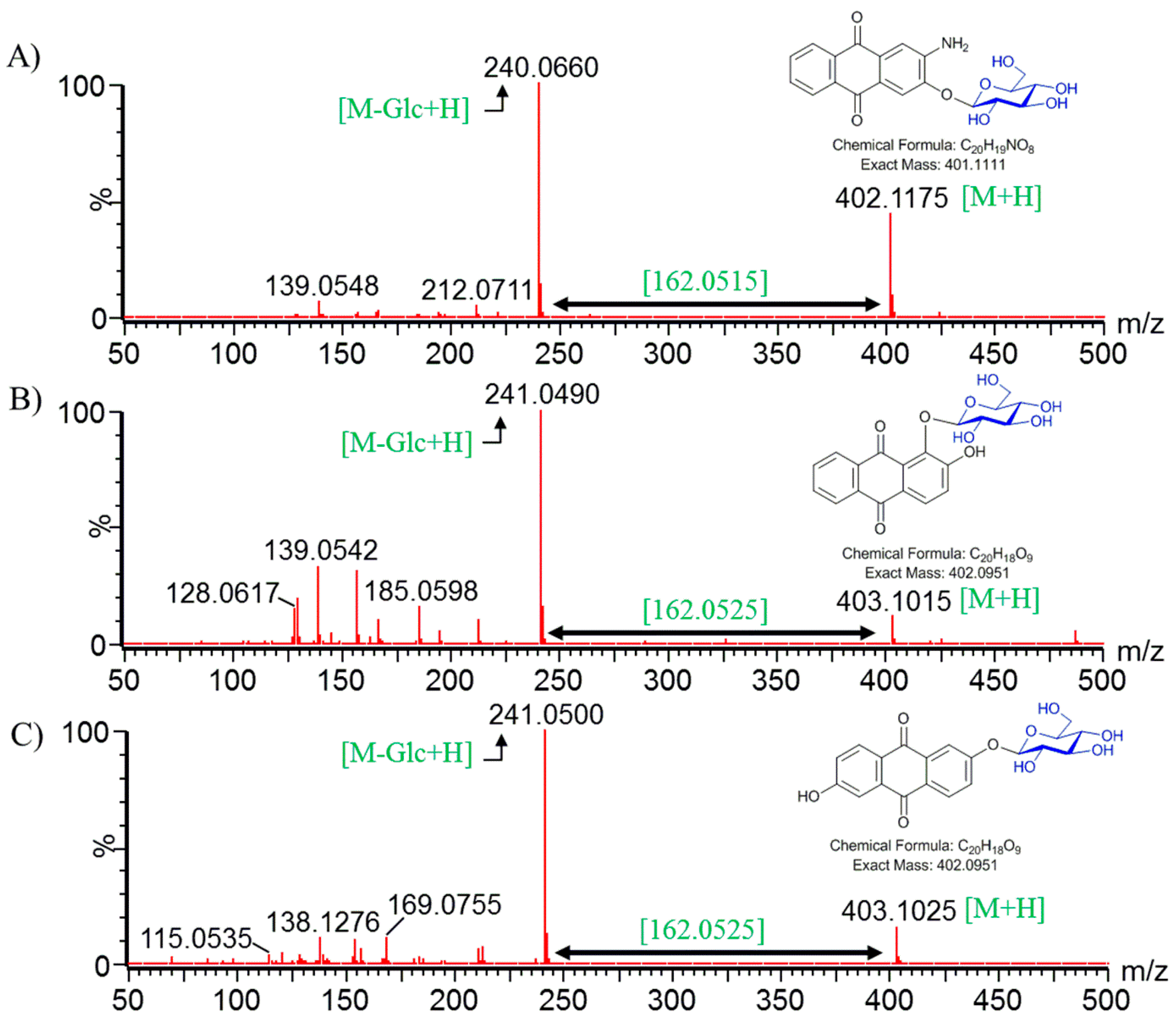
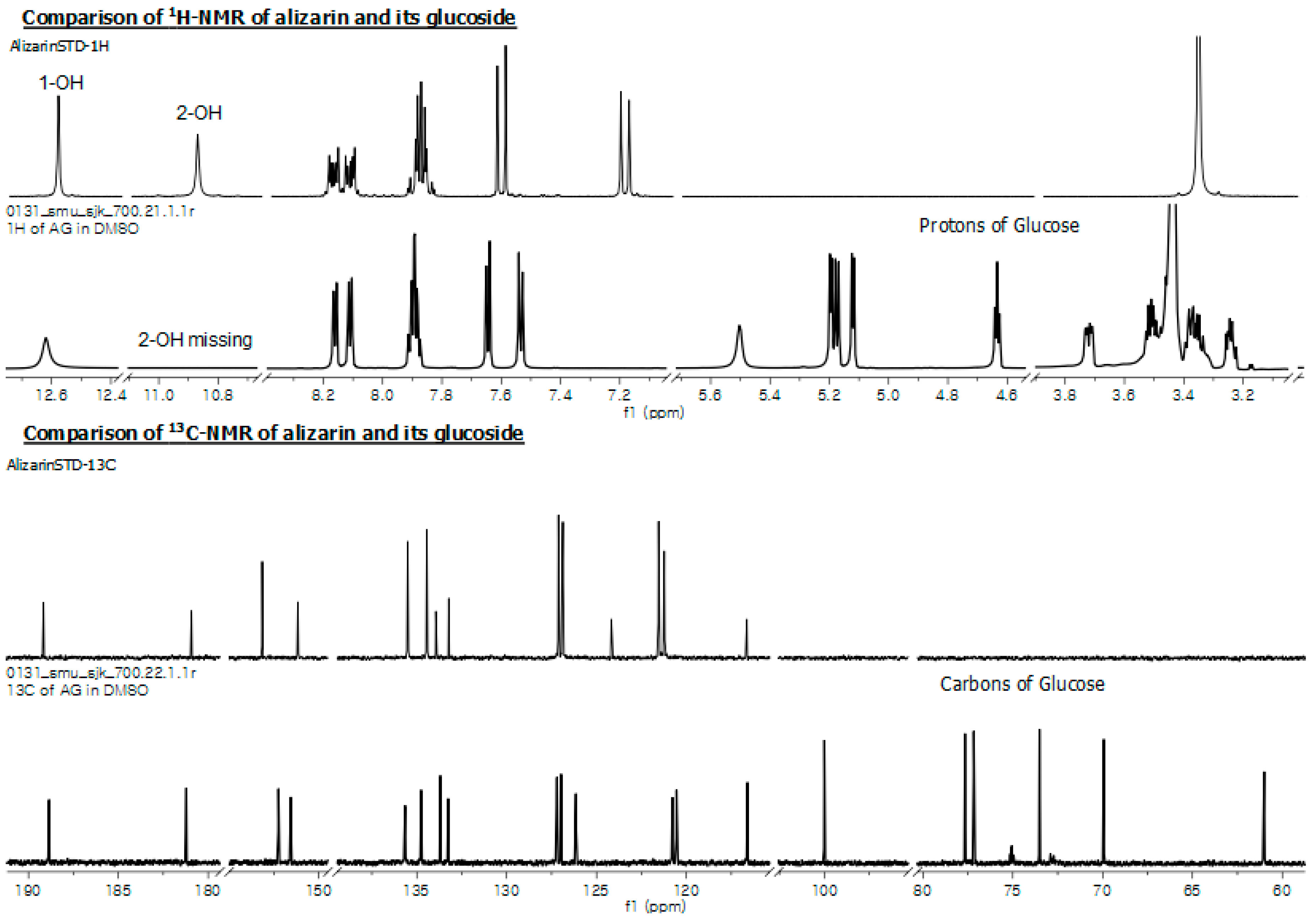
2.5. Structural Elucidation of Potential Anthraquinone Glucoside Derivative
3. Discussion
4. Materials and Methods
4.1. General Procedures
4.2. Vectors and Recombinant Strains
4.3. Culture Preparation and Whole Cell Biotransformation
4.4. Whole Cell Biocatalysis in Bioreactor
4.5. Analytical Procedures
4.6. Inhibitory Effects of Derivatives on Cancer Cell Growth
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dave, H.; Ledwani, L. A review on anthraquinones isolated from Cassia species and their applications. Indian J. Nat. Prod. Resour. 2012, 3, 291–319. [Google Scholar]
- Kanokmedhakul, K.; Kanokmedhakul, S.; Phatchana, R. Biological activity of Anthraquinones and Triterpenoids from Prismatomeris fragrans. J. Ethnopharmacol. 2005, 100, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Kosalec, I.; Kremer, D.; Locatelli, M.; Epifano, F.; Genovese, S.; Carlucci, G.; Randic, M.; Zovko Koncic, M. Anthraquinone profile, antioxidant and antimicrobial activity of bark extracts of Rhamnus alaternus, R. fallax, R. intermedia and R. pumila. Food Chem. 2013, 136, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Sittie, A.A.; Lemmich, E.; Olsen, C.E.; Hviid, L.; Kharazmi, A.; Nkrumah, F.K.; Christensen, S.B. Structure-activity studies: In vitro antileishmanial and antimalarial activities of anthraquinones from Morinda lucida. Planta Med. 1999, 65, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Rath, G.; Ndonzao, M.; Hostettmann, K. Antifungal anthraquinones from Morinda lucia. Int. J. Pharmacogn. 1995, 33, 107–114. [Google Scholar] [CrossRef]
- Younos, C.; Rolland, A.; Fleurentin, J.; Lanhers, M.; Misslin, R.; Mortier, F. Analgesic and Behavioural Effects of Morinda citrifolia. Planta Med. 1990, 56, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Koumaglo, K.; Gbeassor, M.; Nikabu, O.; de Souza, C.; Werner, W. Effects of three compound extracted from Morinda lucida on Plasmodium falciparum. Planta Med. 1991, 58, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.N.; Zareen, S.; Yeap, S.K.; Ho, W.Y.; Lo, K.M.; Hasan, A.; Alitheen, N.B. Total synthesis, cytotoxic effects of damnacanthal, nordamnacanthal and related anthraquinone analogues. Molecules 2013, 18, 10042–10055. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.H.; Mohamad, H.; Mohidin, A.; Lajis, N.H. Antioxidant activity of anthraquinones from Morinda elliptica. Nat. Prod. Sci. 2002, 8, 48–51. [Google Scholar]
- Al-Otaibi, J.S.; EL Gogary, T.M. Synthesis of novel anthraquinones: Molecular structure, molecular chemical reactivity descriptors and interactions with DNA as antibiotic and anti-cancer drugs. J. Mol. Struct. 2017, 1130, 799–809. [Google Scholar] [CrossRef]
- Feng, Y.; Huang, S.; Dou, W.; Zhang, S.; Chen, J.H.; Shen, Y.; Shen, J.H.; Leng, Y. Emodin, a natural product, selectively inhibits 11β-hydroxysteroid dehydrogenase type 1 and ameliorates metabolic disorder in diet-induced obese mice. Br. J. Pharmacol. 2010, 161, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Alam, Z.; Al-Mahdi, Z.; Zhu, Y.; McKee, Z.; Parris, D.S.; Parikh, H.I.; Kellogg, G.E.; Kuchta, A.; McVoy, M.A. Anti-cytomegalovirus activity of the anthraquinone atanyl blue PRL. Antivir. Res. 2015, 114, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, G.; Babykutty, S.; Sathiadevan, P.P.; Srinivas, P. Molecular mechanism of emodin action: Transition from laxative ingredient to an antitumor agent. Med. Res. Rev. 2007, 27, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Twycross, R.; Sykes, N.; Mihalyo, M.; Wilcock, A. Stimulant Laxatives and Opioid-Induced Constipation. J. Pain Symptom Manag. 2012, 43, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Hunger, K. Industrial Dyes: Chemistry, Properties, Applications; Wiley-VCH: Weinheim, Germany, 2003. [Google Scholar]
- Huang, Q.; Lu, G.; Shen, H.M.; Chung, M.C.M.; Choon, N.O. Anti-cancer properties of anthraquinones from rhubarb. Med. Res. Rev. 2007, 27, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Malik, E.M.; Müller, C.E. Anthraquionones as pharmacological tools and drugs. Med. Res. Rev. 2016, 36, 705–748. [Google Scholar] [CrossRef] [PubMed]
- Madje, B.R.; Shelke, K.F.; Sapkal, S.B.; Kakade, G.K.; Shingare, M.S. An efficient one-pot synthesis of anthraquinone derivatives catalyzed by alum in aqueous media. Green Chem. Lett. Rev. 2010, 3, 269–273. [Google Scholar] [CrossRef]
- Seidel, N.; Hahn, T.; Lirbing, S.; Seichter, W.; Kortus, J.; Weber, E. Synthesis and properties of new 9,10-anthraquinone derived compounds for molecular electronics. New J. Chem. 2013, 37, 601–610. [Google Scholar] [CrossRef]
- Pandey, R.P.; Li, T.F.; Kim, E.H.; Yamaguchi, T.; Park, Y.I.; Kim, J.S.; Sohng, J.K. Enzymatic synthesis of novel phloretin glucosides. Appl. Environ. Microbiol. 2013, 79, 3516–3521. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.P.; Malla, S.; Simkhada, D.; Kim, B.G.; Sohng, J.K. Production of 3-O-xylosyl quercetin in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Pandey, R.P.; Trang, N.T.H.; Chaudhary, A.K.; Sohng, J.K. Synthetic sugar cassettes for the efficient production of flavonol glycosides in Escherichia coli. Microb. Cell Fact. 2015, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Parajuli, P.; Shin, J.Y.; Lee, J.; Lee, S.; Hong, Y.S.; Park, Y.I.; Kim, J.S.; Sohng, J.K. Enzymatic biosynthesis of novel resveratrol glucoside and glycoside derivatives. Appl. Environ. Microbiol. 2014, 80, 7235–7243. [Google Scholar] [CrossRef] [PubMed]
- Kalidhar, S.B. Structural elucidation in anthraquinones using 1H NMR glycosylation and alkylation shifts. Phytochemistry 1989, 28, 3459–3463. [Google Scholar] [CrossRef]
- Manojlovic, N.T.; Solujic, S.; Sukdolak, S.; Krstic, L.J. Isolation and antimicrobial activity of anthraquinone from some species of the lichen genus Xanthoria. J. Serb. Chem. Soc. 2000, 65, 555–560. [Google Scholar] [CrossRef]
- Chien, S.C.; Wu, Y.C.; Chen, Z.W.; Yang, W.C. Naturally occurring anthraquinones: Chemistry and therapeutic potential in autoimmune diabetes. Evid. Based Complement. Altern. Med. 2015, 2015, 357357. [Google Scholar] [CrossRef] [PubMed]
- Gürses, A.; Açıkyıldız, M.; Güneş, K.; Sadi Gürses, M. Dyes Pigments; Springer: Basel, Switzerland, 2016; p. 88. [Google Scholar]
- Chen, Q. Development of an anthraquinone process for the production of hydrogen peroxide in a trickle bed reactor-From bench scale to industrial scale. Chem. Eng. Process. Process Intensif. 2008, 47, 787–792. [Google Scholar] [CrossRef]
- Wink, M. Biochemistry of Plant Secondary Metabolism. In Annual Plant Reviews, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2010; Volume 40, pp. 483–485. [Google Scholar]
- Duval, J.; Pecher, V.; Poujol, M.; Lesellier, E. Research advances for the extraction, analysis and uses of anthraquinones: A review. Ind. Crops Prod. 2016, 94, 812–833. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Joyce, S.A.; Kodama, H.J.; Schwar, G.; Clarke, D.J.; Bode, H.B. A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. ChemBioChem 2007, 8, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.P.; Parajuli, P.; Koffas, M.A.G.; Sohng, J.K. Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 2016, 34, 634–662. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.P.; Gurung, R.B.; Parajuli, P.; Koirala, N.; Tuoi, L.T.; Sohng, J.K. Assessing acceptor substrate promiscuity of YjiC-mediated glycosylation toward flavonoids. Carbohydr. Res. 2014, 393, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Pandey, R.P.; Pokhrel, A.R.; Ghimire, G.P.; Sohng, J.K. Enzymatic glycosylation of the topical antibiotic mupirocin. Glycoconj. J. 2014, 31, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, G.P.; Koirala, N.; Pandey, R.P.; Jung, H.J.; Sohng, J.K. Modification of emodin and aloe-emodin by glycosylation in engineered Escherihia coli. World J. Microbiol. Biotechnol. 2015, 31, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ye, M.; Zhan, J.; Chen, Y.; Guo, D. Microbial glycosylation of four free anthraquinones by Absidia coerulea. Biotechnol. Lett. 2004, 26, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Nam, W.; Kim, S.; Nam, S.; Friedman, M. Structure-Antioxidative and Anti-Inflammatory Activity Relationships of Purpurin and Related Anthraquinones in Chemical and Cell Assays. Molecules 2017, 22, 265. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |


| Position | 1H-NMR | 13C-NMR | ||
|---|---|---|---|---|
| Alizarin | Alizarin 2-O-β-d-Glucoside | Alizarin | Alizarin 2-O-β-d-Glucoside | |
| 1-OH | 12.59 (s, 1H) | 12.62 (s, 1H) | 153.15 | 155.25 |
| 2-OH | 10.88 (s, 1H) | - | 151.17 | 151.56 |
| 3 | 7.20 (d, J = 8.3Hz, 1H) | 7.53 (d, J = 8.55Hz, 1H) | 121.55 | 120.78 |
| 4 | 7.62 (d, J = 8.3Hz, 1H) | 7.64 (d, J = 8.45Hz, 1H) | 121.23 | 120.55 |
| 5 | 8.15 (m, 1H) | 8.14 (ddd, J = 35.05, 7.19, 1.82 Hz, 1H) | 126.88 | 126.97 |
| 6 | 7.89 (m, 1H) | 7.89 (td, J = 6.81, 6.78, 1.72 Hz, 1H) | 133.44 | 134.76 |
| 7 | 7.89 (m, 1H) | 7.89 (td, J = 6.81, 6.78, 1.72 Hz, 1H) | 135.51 | 135.65 |
| 8 | 8.15 (m, 1H) | 8.14 (ddd, J = 35.05, 7.19, 1.82 Hz, 1H) | 127.12 | 127.20 |
| 9 | - | - | 180.93 | 181.21 |
| 10 | - | - | 189.16 | 188.85 |
| 11 | - | - | 133.22 | 133.26 |
| 12 | - | - | 133.94 | 133.69 |
| 13 | - | - | 116.64 | 116.60 |
| 14 | - | - | 124.17 | 126.14 |
| 1′ | - | 5.17 (d, J = 7.4 Hz, 1H) | - | 100.00 |
| 2′ | - | 3.36 (ddd, J = 17.8, 15.0, 8.8 Hz, 1H) | - | 73.51 |
| 3′ | - | 3.36 (ddd, J = 17.8, 15.0, 8.8 Hz, 1H) | - | 77.17 |
| 4′ | - | 3.49 (m, 1H) | - | 77.66 |
| 5′ | - | 3.25 (dd, J = 9.21, 5.23 Hz, 1H) | - | 69.94 |
| 6′a | - | 3.72 (ddd, J = 11.92, 5.33, 2.13 Hz, 1H) | - | 61.01 |
| 6′b | 3.49 (m, 1H) | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.T.H.; Pandey, R.P.; Parajuli, P.; Han, J.M.; Jung, H.J.; Park, Y.I.; Sohng, J.K. Microbial Synthesis of Non-Natural Anthraquinone Glucosides Displaying Superior Antiproliferative Properties. Molecules 2018, 23, 2171. https://doi.org/10.3390/molecules23092171
Nguyen TTH, Pandey RP, Parajuli P, Han JM, Jung HJ, Park YI, Sohng JK. Microbial Synthesis of Non-Natural Anthraquinone Glucosides Displaying Superior Antiproliferative Properties. Molecules. 2018; 23(9):2171. https://doi.org/10.3390/molecules23092171
Chicago/Turabian StyleNguyen, Trang Thi Huyen, Ramesh Prasad Pandey, Prakash Parajuli, Jang Mi Han, Hye Jin Jung, Yong Il Park, and Jae Kyung Sohng. 2018. "Microbial Synthesis of Non-Natural Anthraquinone Glucosides Displaying Superior Antiproliferative Properties" Molecules 23, no. 9: 2171. https://doi.org/10.3390/molecules23092171
APA StyleNguyen, T. T. H., Pandey, R. P., Parajuli, P., Han, J. M., Jung, H. J., Park, Y. I., & Sohng, J. K. (2018). Microbial Synthesis of Non-Natural Anthraquinone Glucosides Displaying Superior Antiproliferative Properties. Molecules, 23(9), 2171. https://doi.org/10.3390/molecules23092171