Abstract
A rapid emergence of resistant bacteria is occurring worldwide, endangering the efficacy of antibiotics and reducing the therapeutic arsenal available for treatment of infectious diseases. In the present study, we developed a new class of compounds with antibacterial activity obtained by a simple, two step synthesis and screened the products for in vitro antibacterial activity against ATCC® strains using the broth microdilution method. The compounds exhibited minimum inhibitory concentrations (MIC) of 1–32 μg/mL against Gram-positive ATCC® strains. The structure–activity relationship indicated that the thiophenol ring is essential for antibacterial activity and the substituents on the thiophenol ring module, for antibacterial activity. The most promising compounds detected by screening were tested against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) clinical isolates. We found remarkable activity against VREF for compounds 7 and 16, were the MIC50/90 were 2/4 µg/mL and 4/4 µg/mL, respectively, while for vancomycin the MIC50/90 was 256/512 µg/mL. Neither compound affected cell viability in any of the mammalian cell lines at any of the concentrations tested. These in vitro data show that compounds 7 and 16 have an interesting potential to be developed as new antibacterial drugs against infections caused by VREF.
1. Introduction
Infectious diseases are a leading cause of death worldwide, and the increasing emergence of antibacterial resistance has contributed to rising rates of potentially fatal infections. It is estimated that by 2050, diseases caused by antibiotic-resistant microorganisms will be responsible for 10 million deaths per year [1]. The bacteria implicated in these infections include the so-called ESKAPE pathogens, such as Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and various Enterobacter sp. [2,3]. These microorganisms have become a global public health problem due to their ability to adapt to antibacterial agents. S. aureus-related skin infections, wound infections, and sepsis cases are the leading causes of healthcare-associated infections [4], and about 90% of S. aureus isolates were found to be methicillin-resistant in a study using data from 21 of the 35 countries in the Americas [4]. In the United States (US), these resistant strains are responsible for over 11,000 deaths per year [5]. In addition, Enterococcus species cause significant numbers of urinary tract, surgical site, and blood infections [4]. In the United Kingdom [6] and the US [7], up to 25% and 60% of E. faecium strains are resistant to vancomycin (VAN), respectively.
Traditionally, development of new antibacterial molecules has been based mainly on two strategies: modifying or adding a small chemical group to an antibiotic already in clinical use, to improve some aspect of its pharmacodynamic and/or pharmacokinetic profile; or seeking new molecules from natural products such as plants, bacteria, or fungi that have demonstrated activity against resistant bacteria. Both strategies involve structural modifications or additions that preserve the pharmacophore and therefore maintain both the mechanism and site of action. Optimizing these compounds may be initially effective, however, due to the structural similarity between novel and existing molecules, bacteria rapidly adapt their resistance mechanisms to thwart new antibiotics [8]. The traditional chemical approach is effective for identifying and optimizing compounds to treat pathologies such as hypertension, diabetes, dyslipidemia, inflammation, and allergies, in which the pharmacological targets do not adapt or generate resistance mechanisms. Infections, however, pose a different challenge, as bacteria are free-living organisms that seek to survive in the presence a harmful agent.
If we continue to rely exclusively on these traditional strategies, it is only a matter of time before our entire investment in the generation of antibiotics is overwhelmed by antibacterial resistance. The way forward should focus on rational, design-oriented development of new synthetic molecules capable of reducing the probability that exposed bacteria will generate a resistance phenotype. Antibacterial agents that have novel chemical structures and that act on unexplored bacterial targets are less likely to be subject to existing compound- or target-based resistance mechanisms. Of course, even new classes of antibiotics may be subject to general mechanisms of resistance, such as increased efflux, reduced influx, or target-site resistance mutations [8].
New approaches should consider a target that is different from existing targets, essential for microbial cell survival, highly conserved in clinically relevant species, absent or radically different in human cells, and easy to assay and approach biochemically [9,10]. However, structural modifications based on the traditional medicinal chemistry approach will also be needed to optimize effectiveness, and rational design will require synthesis of multiple compounds in order to determine the relationship between structure and activity. Therefore, it is critical to use a simple, versatile, and low-cost process to synthesize these molecules.
In this regard, various activities have been attributed to quinonic compounds. These compounds present two important characteristics for drug design. First is the versatility of synthetic processes, which allows active compounds to be obtained in a few stages; the second is the broad spectrum of biological activities described, which shows that the choice of substituents is critical to guiding the objective of biological activity, as shown by the work of Gordaliza et al. [11]. Quinonic compounds exert interesting antibacterial effects [12,13,14] and have already been integrated into antibacterial compounds, such as alkannin [15] and renierone [16]. These quinone-based antibiotics have been found to have activity against S. aureus, E. faecium, and Bacillus subtilis [17,18]. Promisingly, Tandon et al. have shown that thioaryl substitution in naphtoquinone results in good antibacterial activity [19,20]. However, it has not been further studied how the substituents of quinone compounds are related to their antibacterial activity. This information can be used to guide the rational design of new antibacterial quinone compounds.
In this study, our group develop a new kind of antibacterial agent based structurally on the quinonic core. We synthesized and assessed a set of 17 compounds against American Type Culture Collection (ATCC®) bacterial strains and addressed the study of the structure–activity relationship based on lipophilicity (logP), half wave potential (E1/2), and volume (MR) parameters. We also analyzed the crystallographic structure of the two compounds with the best antibacterial performance and tested their activity against multidrug-resistant clinical isolates, to calculate minimum inhibitory concentration (MIC)50/90 and minimum bactericidal concentration (MBC)50/90 values against S. aureus and E. faecium. Finally, the toxicity of these two compounds against the HeLa, HTC-116, SHSY-5Y, and Vero cell lines was evaluated to assess their safety and suitability for use as a treatment.
2. Results and Discussion
2.1. Synthesis and Determination of Physicochemical Parameters
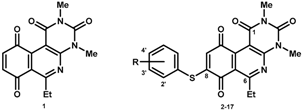
The final compounds were formed in two steps. First, we used activated benzoquinone, generated in situ from the corresponding hydroquinone with silver (I) oxide, reacting with enaminone through an ionic [3+3] process to produce the corresponding core 1 with a yield of 84% by a further in situ oxidation. During the second step, compounds 2–17 were obtained by nucleophilic addition of the corresponding thiophenol derivatives to core 1 with subsequent aerobic oxidation of the hydroquinone formed by Michael addition (Figure 1). Yields ranged from 47% to 88% for the 2′-, 3′-, and 4′-series. The lipophilic (LogP), half-wave potential (E1/2) and molar refractivity (MR) parameters (Table 1) are discussed in Section 2.2. It should be noted that formation of the final products was highly favorable, with reactions produced under mild reaction conditions at room temperature. The total production time for each compound was 48 h. In addition, each product had a characteristic color that was distinct from that of the starting agent (yellow), facilitating purification by enabling rapid visual detection during the column chromatographic separation.

Figure 1.
Synthetic route of the preparation of core 1 and compounds 2–17.

Table 1.
Antibacterial screening and physicochemical parameters for compounds 1–17.
2.2. Antibacterial Screening
The antibacterial profile shows that all compounds except 5 and 6 demonstrated activity against Gram-positive bacteria (methicillin-resistant S. aureus [MRSA] ATCC® 43300, methicillin-susceptible S. aureus [MSSA] ATCC® 29213, and E. faecalis ATCC® 29212). No activity was observed against Gram-negative bacteria (Escherichia coli ATCC® 25922 and P. aeruginosa ATCC® 27853), even at the highest concentrations tested (32 μg/mL, maximum solubility for the compounds). The antibacterial screening results are presented in Table 1. However, preliminary tests showed that adding EDTA to compounds [21] may induce activity against Gram-negative bacteria (unpublished results), through two possible mechanisms: fluidizing the lipid membrane [21] to facilitate passage of the derivatives through the membrane; or enabling the compound to act as a siderophore, in which the compound takes advantage of the high affinity of the iron transport system to facilitate entry into the bacterium [22]. This result suggests that it is possible to achieve activity against Gram-negative bacteria using a strategy that enables penetration of the bacterial membrane.
2.3. Structure-Activity Relationship
Core 1 showed no antibacterial activity, but addition of the thiophenol ring resulted in measurable activity (compound 2). This observation suggests that the aromatic system is essential to activity. Substituents at the 2′-, 3′-, and 4′-positions of the thiophenol ring gave rise to the 2′-, 3′-, and 4′-series, respectively.
The 2′-series was comprised of compounds 3 through 7. Using the activity of compound 2 as a reference, compounds 4 and 7 exhibited the highest antibiotic activity within this series and possessed the bulkiest substituents in the series, the methoxy group and the bromine atom, with MR values of 7.24 and 9.06 cm3/mol, respectively. However, the presence of a methyl group in compound 3 was associated with reduced activity, resulting in MIC values of 32 μg/mL for MRSA and MSSA and >32 μg/mL for E. faecalis. Finally, fluorine and chlorine substituents in the ortho position, which produced compounds 5 and 6, respectively, showed no antibacterial activity. It is interesting to note the steric effect of the substituents at the C-2′ position of the thiophenol ring; apparently, the atomic volume forces the structure to assume a conformation with a semi-perpendicular dihedral angle that may reduce the degrees of freedom of the structure. This observation suggests that bulky groups at C-2′ favor antibacterial activity.
Compounds with meta substitutions, the 3′-series (compounds 8 through 12), showed MIC values of 2 to 32 μg/mL. Within this series, it was not possible to obtain a correlation that explained the results of biological activity obtained according to the LogP, E1/2, or MR molecular descriptors. However, the methyl, methoxy, and fluorine substituents (compounds 8, 9, and 10, respectively) were observed to have the best profiles in Gram-positive bacteria. On the other hand, interestingly, the halogenated compounds (11 and 12) were more potent against resistant than sensitive strains, because these compounds showed MIC values of 2 μg/mL MRSA strains, whereas potency against MSSA strains was lower, at 32 μg/mL.
The 4′-series compounds (13 through 17) showed MIC values ranging from 4 to 16 μg/mL. Compound 16 showed the highest potency. Considering the reported relationships between the quinonic structures, their redox capacity, and their biological activity [24,25], we tackled the study of the 4′-series compounds, considering that the thiophenol substituents could modulate antibacterial activity through control of the descriptor E1/2.
The relationship established by Hammett between the type of substituent in the para position and reactivity is well known [26]. This analysis can be applied to the 4′-series, in that whether the substituent in position 4′ is an electron donor or acceptor will affect the quinonic nucleus through the sulfur atom, modifying the redox potential of the compounds. This effect requires that the aromatic systems, the thiophenolic ring and the quinonic nucleus, are in the same plane to maximize conjugation of the two systems, which can be extrapolated according to the value of the dihedral angle between the two aromatic systems. In order to determine the dihedral angle, compound 16 was crystallized and resolved using X-ray diffraction to produce a three-dimensional image of the structure. In this analysis, the most active compound of the 2′-series (compound 7) was included to determine whether the insertion of a bulky substituent in position 2′ (bromine atom) significantly influences the dihedral angle. Images of the molecular structure showed that the dihedral angle between the aromatic ring and the quinone core was nearly perpendicular, with values of 81.6 and 76.5 degrees for compounds 7 and 16, respectively (Figure 2).
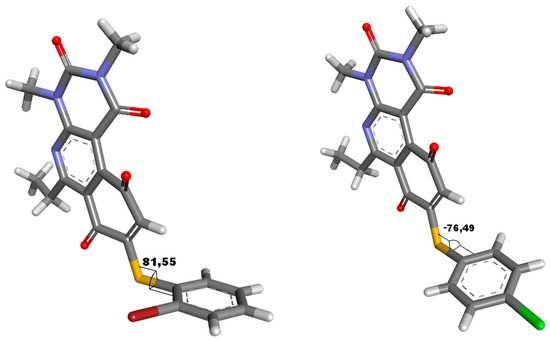
Figure 2.
Dihedral angle for compounds 7 and 16, determinate from structure crystalize resolute by X-ray diffraction.
These results demonstrate than the molecules do not present a dihedral angle sufficient to consider significant conjugation between the aromatic systems; therefore, the electronic effect of the substituent on the half-wave potential must be principally inductive. This observation is concordant with the narrow range of E1/2 values obtained for the compounds (−0.623 to −0.374 mV), suggesting that the inductive effect weakly affected the redox potential of the quinones. Interestingly, the bulky substituent in the ortho or para position did not significantly alter the dihedral angle, because the presence of the bromine atom in position 2′ increased the angle by 5 degrees as compared to molecules with a chloride atom in position 4′. This analysis for the 4′ series shows that the E1/2 parameter does not present a clear correlation with antibacterial activity. With respect to the lipophilicity parameter, no correlation was observed with activity in the 2′ series, since the compound with the lowest (compound 4) and highest (compound 7) logP showed similar activities in the series. For the 3′ series, despite the variation in lipophilicity, a narrow range of MIC values (2 to 8 ug/mL) was observed. Of the MSSA, the most lipophilic compounds of the 3′ series had the lowest activity. For the 4′ series, a linear correlation between logP and MIC was observed, where the compounds with the highest lipophilicity had the highest activities of the series.
In summary, the phenyl ring is essential for activity, the increased size of substituents in the ortho position improve antibacterial activity, and for the meta position, the physicochemical characteristics of the substituents do not show a correlation with antibacterial activity. In the para position, a lipophilic substituent improves antibacterial activity.
2.4. Antibacterial Activity of Compounds 7 and 16 Against Clinical MDR Isolates
To assess the clinical potential of the most promising compounds, we evaluated their antibacterial activity against heterogeneous populations of clinical isolates as well as their toxicity against human and animal cells. Compounds 7 and 16 were selected for this analysis, as these two compounds showed the greatest antibacterial activity.
The MIC50/90 and MBC50/90 against clinical isolates of MRSA and vancomycin-resistant E. faecium (VREF), as well as the geometric mean (MG) of the MIC parameters, were determined for compounds 7 and 16. The MIC50/90 values reflect the effectiveness of antibacterial agents against a heterogeneous bacterial population. Vancomycin (VAN) and gentamicin (GEN) were used as quality controls; if the MIC values for these reference drugs against each bacterial strain were equal to the values reported by the Instituto de Salud Pública de Chile (ISP), the trials were considered valid. The maximum concentration tested was 32 μg/mL, as this value represents the solubility limit of the compounds. The results obtained for compounds 7, 16, and VAN against MRSA and VREF are summarized in Table 2 and Table 3, respectively, and the histograms are presented in Figure 3. For compound 7, the MIC50/90 was 2/2 µg/mL, and the MBC50/90 was 4/4 µg/mL in 32 S. aureus isolates. For compound 16, in contrast, the MIC50/90 was 2/4 µg/mL, and the MBC50/90 was 2/4 µg/mL in 29 S. aureus isolates. For VAN, the MIC50/90 was 1/1 µg/mL. The MBC50/90 was not assessed. For compound 7, the MIC50/90 was 2/4 µg/mL, and the MBC50/90 was 4/4 µg/mL in 44 E. faecium isolates. For compound 16, the MIC50/90 was 4/4 µg/mL and the MBC50/90 was 4/8 µg/mL in 41 E. faecium isolates. For VAN, the MIC50/90 was 256/512 µg/mL, and the MBC50/90 was not assessed.

Table 2.
Summary of antibacterial activity of compounds 7, 16 and vancomycin against MDR S. aureus. MIC and MBC (µg/mL).

Table 3.
Summary of antibacterial activity of compounds 7, 16 and vancomycin against MDR Enterococcus sp. MIC and MBC (µg/mL).
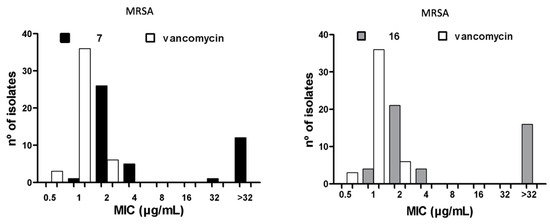
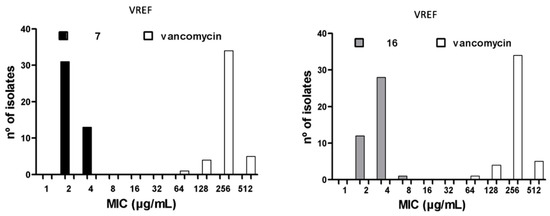
Figure 3.
MIC90 histograms for compounds 7 and 16 against MRSA and VREF vs vancomycin.
2.5. Antibacterial Activity Against Heterogeneous Populations of Clinical Isolates
The bacterial population used to calculate the MIC90 values had a complex susceptibility profile, as most of the isolates tested were resistant to three or more classes of drugs. However, both compounds had MIC90 values near those of VAN against MRSA (MIC90 = 1 μg/mL, p > 0.001). Moreover, the two compounds had significantly lower MIC values than VAN against VREF isolates (64–256 μg/mL, p > 0.001). Compounds 7 and 16 were 128-fold and 64-fold more active than VAN, respectively, against the various clinical isolates of VREF. The MIC90 values for compounds 7 and 16 are similar to those reported for linezolid (2–4 μg/mL) against MRSA [27,28,29,30,31], showing that these molecules may offer potential therapeutic alternatives to VAN for the treatment of skin infections caused by MRSA [32]. Linezolid has been found to have MIC90 values of 2–64 μg/mL against Enterococcus spp. [27,29,33], indicating that linezolid is a therapeutic option for treating urinary tract infections caused by VREF. In addition, compounds 7 and 16 showed activity similar to or greater than VAN and linezolid against MRSA and multidrug-resistant E. faecium isolates. Furthermore, compounds 7 and 16 were found to have MBC values equal to or up to two times greater than the MIC values against all strains tested, including MRSA (32 isolates) and VREF (44 isolates). An MBC/MIC ratio of 2 or lower indicates that a compound has a bactericidal mode of action, according to Craig et al. [34], suggesting that these compounds were successful in killing the clinical isolates of MRSA and VREF.
2.6. Cytotoxicity of Compounds 7 and 16
To test the safety of compounds 7 and 16 in mammalian cells, cytotoxicity studies were performed using the HeLa, HTC-116, SH-SY5Y, and Vero cell lines. A colorimetric assay was performed to estimate the half-maximal inhibitory concentration (IC50) values, which represent the concentration of a drug that is required for 50% inhibition in vitro after 24 h of continuous exposure to the compound. Four serial dilutions (from 0.02 to 20 µg/mL) for each sample were evaluated in triplicate in three reproducible assays, and VAN and GEN were used as reference drugs. The results are shown in Figure 4.

Figure 4.
In vitro toxicity of compounds 7 and 16 (n = 3) over HeLa, HTC-116, SH-SY5Y and Vero cell lines, in the range of 0.02 to 20 µg/mL in triplicated. (+) Positive growth control, RPMI 140 medium. (−) Negative growth control. (−′) Negative growth additional control, 1% DMSO + H2O. Vancomycin and Gentamicin were used as drug reference. ***: p < 0.001 = statistically no significant differences with positive growth control; #: p < 0.001 = statistically no significant differences with vancomycin; †: p < 0.001 = statistically no significant differences with vancomycin.
It was not possible to determine the IC50 values because the viability percentages observed were greater than 75% at all concentrations studied. No significant differences between compounds 7 or 16 (p > 0.001) and the positive control (p > 0.001) or the reference drugs (p > 0.001) were observed.
The toxicity results for mammalian cells, including human (HeLa, HTC-116, and SHSY-5Y) and animal (Vero) cells, showed no significant differences between either of the two compounds and the positive control (p > 0.05), indicating that the compounds did not affect the viability of the cells after 24 h of exposure at any of the concentrations studied (0.002–20 μg/mL). Moreover, there were no significant differences in toxicity between the two novel compounds and the reference antibacterial agents (VAN and GEN) at any of the concentrations tested (p > 0.05). Due to the low solubility of the compounds, it was not possible to determine their IC50 in cell culture. At the maximum concentration tested, 70% cell viability was observed, a percentage that is insufficient for calculation of the IC50.
Finally, there was a wide gap between the antibiotic and toxic concentrations of the compounds. For example, it would be necessary to increase the concentration of compounds 7 and 16, which showed MIC values of 2 and 4 μg/mL against MRSA, respectively, by a factor of 5 to 10 before the viability of mammalian cells (HeLa and Vero) would be affected.
3. Materials and Methods
3.1. Materials
The compounds were synthesized from the commercial precursors 1-(2,5-dihydroxyphenyl)-propan-1-one, 6-amino-1,3-dimethyl-2,4(1H,3H)-pyrimidinedione, 2-bromobenzenethiol, 2-chloro-benzenethiol, 2-fluorobenzenethiol, 2-methoxybenzenethiol, 2-methylbenzenethiol, 3-bromobenzene-thiol, 3-chlorobenzenethiol, 3-fluorobenzenethiol, 3-methoxybenzenethiol, 3-methylbenzenethiol, 4-bromobenzenethiol, 4-chlorobenzenethiol, 4-fluorobenzenethiol, 4-methoxybenzenethiol, and 4-methyl-benzenethiol, purchased from Sigma-Aldrich® (St. Louis, MO, USA), and benzenethiol, from Merck® (Kenilworth, NJ, USA). All solvents were commercially available and of reagent grade and were used without further purification. Melting points (mp) were determined on a SMP3 apparatus (Stuart Scientific, Staffordshire, United Kingdom) and were uncorrected. 1H-NMR spectra (400 MHz) were recorded on an AM-400 instrument (Bruker, Billerica, MA, USA) in deuterochloroform (CDCl3). 13C-NMR spectra were obtained in CDCl3 at 100 MHz. Peak assignment was confirmed by correlation with chemical structures in 2D experiments (HMBC, HSQC) performed by Valderrama et al. [35,36,37,38,39]. The assignments of chemical shifts are expressed in ppm downfield relative to tetramethylsilane (TMS, δ scale), and the coupling constants (J) are reported in Hertz. IR spectra were recorded on a Bruker Vector 22-FT spectrophotometer using KBr discs, and wavenumbers are reported in cm−1. HRMS were obtained on a model MAT 95XP spectrometer (Thermo Finnigan, Barkhausenstr, Germany), Silica gel (70–230 and 230–400 mesh) and TLC aluminum foil 60F254 Merck® were used for preparative column chromatography and analytical TLC, respectively.
3.2. Chemical Synthesis
3.2.1. Procedure for the Synthesis of Compound 1
A suspension of 1-(2,5-dihydroxyphenyl)-propan-1-one 939 mg (5.65 mmol), 6-amino-1, 3-dimethyl-2,4(1H,3H)-pyrimidinedione 1068 mg (6.88 mmol), Ag2O 3794 mg (16.37 mmol), anhydrous MgSO4 1963 mg (16.3 mmol), in dichloromethane (40 mL) was stirred at room temperature for 4 h. The mixture was filtered with Celite® and washed with dichloromethane. The solvent was removed under reduced pressure and the crude of reaction was purified using 65 g of silica gel (230–400 mesh) using a mix of dichloromethane and ethyl acetate = 9:1. The resulting solution was concentrated to dryness under reduced pressure. The product, 6-ethyl-2,4-dimethylpyrimido[4,5-c]isoquinolin-1,3,7,10(2H,4H)-tetraone (1): was obtained as a yellow solid; m.p. 167.6–167.9 °C; IR (KBr), ῡ = 1667 (C=O quinone), 1720 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.11 (d, J = 10.3 Hz, 1H, H-9), 6.81 (d, J = 10.3 Hz, 1H, H-8), 3.76 (s, 3H, 2-NCH3), 3.47 (s, 3H, 4-NCH3), 3.40 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 1.34 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 185.0 (1C, C-10), 183.9 (1C, C-7), 171.2 (1C, C-6), 159.0 (1C, C-4a), 152.9 (1C, C-1), 151.5 (1C, C-3), 146.6 (1C, C-10a), 138.7 (1C, C-8), 138.7 (1C, C-9), 121.2 (1C, C-6a), 105.4 (1C, C-10b), 32.0 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.5 (1C, 6-CH2CH3); HRMS m/z 299.09070 (calcd for C15H13N3O4 [M]+, 299.09061); purified in column chromatography with dichloromethane: ethyl acetate = 9:1; Yield: 84%.
3.2.2. General Procedure for the Synthesis of Compounds 2–17
A solution of 1 (150 mg, 0.4909 mmol) and CeCl3.7H2O (5% mmol respect to 1) in a mix of ethanol: dichloromethane = 1:1 (10 mL), was added dropwise slowly a solution of benzenethiol derivate (0.5 equiv.) in ethanol: dichloromethane = 1:1 (30 mL). The reaction mixture was stirred at room temperature for 16 h. The progress of the reaction was followed by thin-layer chromatography (TLC). The mixture was concentrated and the crude of reaction was purified using 65 g of silica gel (70–230 mesh) and a mix of dichloromethane, light petroleum and ethyl acetate than eluent in determinate proportions. The resulting solution was concentrated to dryness under reduced pressure.
6-Ethyl-8-(phenylthio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (2): orange solid; m.p. 179.4–180.0 °C; IR (KBr), ῡ = 1658, 1676 (C=O quinone), 1720 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.50–7.55 (m, 5H, SC6H5), 6.18 (s, 1H, H-9), 3.76 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.42 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.37 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.8 (1C, C-10), 181.2 (1C, C-7), 171.2 (1C, C-6), 158.8 (1C, C-4a), 157.1 (1C, C-8), 153.1 (1C, C-1), 151.5 (1C, C-3), 147.7 (1C, C-10a), 136.1 (2C, C-2′ and C-6′), 131.1 (1C, C-4′), 130.9 (2C, C-3′ and C-5′), 128.3 (1C, C-9), 127.6 (1C, C-1′), 121.0 (1C, C-6a), 105.8 (1C, C-10b), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.6 (1C, 6-CH2CH3); HRMS m/z 407.09400 (calcd. for C21H17N3O4S [M]+, 407.09398); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 9:8:1; Yield: 67%.
6-Ethyl-8-((2′-methylphenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (3): orange solid; m.p. 206.0–210.9 °C; IR (KBr), ῡ = 1660, 1688 (C=O quinone); 1730 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.49 (d, J = 7.5 Hz, 1H, H-6′), 7.45–7.39 (m, 2H, H-5′ and H-3′), 7.30 (t, J = 6.8 Hz, 1H, H-4′), 6.01 (s, 1H, H-9), 3.75 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.42 (q, J = 6.6 Hz, 2H, 6-CH2CH3), 2.43 (s, 3H, 2′-CH3), 1.38 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.4 (1C, C-10), 181.1 (1C, C-7), 170.9 (1C, C-6), 158.6 (1C, C-4a), 155.6 (1C, C-8), 152.9 (1C, C-1), 151.2 (1C, C-3), 147.6 (1C, C-10a), 143.3 (1C, C-2′), 136.9 (1C, C-6′), 131.9 (1C, C-3′), 131.5 (1C, C-5′), 128.1 (1C, C-4′), 127.6 (1C, C-9), 126.5 (1C, C-1′), 120.9 (1C, C-6a), 105.7 (1C, C-10b), 31.9 (1C, 6-CH2CH3), 30.4 (1C, 2-NCH3), 29.2 (1C, 4-NCH3), 20.7 (1C, CH3-2′), 12.3 (1C, 6-CH2CH3); HRMS m/z 421.10957 (calcd. for C22H19N3O4S [M]+, 421.10963); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 9:10:1; Yield: 72%.
6-Ethyl-8-((2′-methoxyphenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (4): orange solid, m.p. 172.3 (d) °C; IR (KBr), ῡ = 1660, 1688 (C=O quinone); 1727 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.53–7.48 (m, 2H, H-4′ and H-6′), 7.02–7.07 (m, 2H, H-3′ and H-5′), 6.09 (s, 1H, H-9), 3.86 (s, 3H, 2′-CH3O), 3.74 (s, 3H, 2-NCH3), 3.42 (s, 3H, 4-NCH3), 3.41 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.36 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.4 (1C, C-10), 181.2 (1C, C-7), 170.7 (1C, C-6), 160.1 (1C, C-2′), 158.6 (1C, C-4a), 154.8 (1C, C-8), 152.8 (1C, C-1), 151.2 (1C, C-3), 147.4 (1C,10a), 137.6 (1C, C-4′), 133.1 (1C, C-6′), 127.6 (1C, C-9), 122.1 (1C, C-5′), 120.9 (1C, C-6a), 114.6 (1C, C-1′), 112.1 (1C, C-3′), 105.5 (1C, C-10b), 56.2 (1C, 2′-OCH3), 31.8 (1C, 6-CH2CH3), 30.2 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 437.10450 (calcd. for C22H19N3O5S [M]+, 437.10454); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 9:6:1; Yield: 72%.
6-Ethyl-8-((2′-fluorobromophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (5): orange solid; m.p. 218.4 (d) °C; IR (KBr), ῡ = 1660, 1684 (C=O quinone); 1727 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.60–7.54 (m, 2H, H-4′ and H-6′), 7.32–7.26 (m, 2H, H-3′ and H-5′), 6.13 (s, 1H, H-9), 3.76 (s, 3H, 2-NCH3), 3.44 (s, 3H, 4-NCH3), 3.43 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.38 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.6 (1C, C-10), 181.1 (1C, C-7), 171.2 (1C, C-6), 163.1 (1C, d, J = 251.5, C-2′), 158.8 (1C, C-4a), 154.4 (1C, C-8), 153.2 (1C, C-1), 151.5 (1C, C-3), 147.6 (1C, C-10a), 137.9 (1C, C-5′), 133.9 (1C, d, J = 8.1, C-4′), 128.4 (1C, C-9), 126.3 (1C, d, J = 3.9, C-6′), 120.9 (1C, C-6a), 117.6 (1C, d, J = 22.3, C-3′), 114.9 (1C, d, J = 18.6, C-1′), 105.9 (1C, C-10b), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.5 (1C, 6-CH2CH3); HRMS m/z 425.08460 (calcd. for C21H16FN3O4S [M]+, 425.08455); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 9:14:2; Yield: 87%.
6-Ethyl-8-((2′-chlorophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (6): orange solid; m.p. 220.8 (d) °C; IR (KBr), ῡ = 1660, 1678 (C=O quinone); 1720 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.60–7.64 (m, 2H, H-3′ and H-6′); 7.49 (t, J = 7.7 Hz, 1H, H-4′), 7.39 (t, J = 7.5 Hz, 1H, H-5′), 6.05 (s, 1H, H-9), 3.75 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.42 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.37 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.6 (1C, C-10), 181.1 (1C, C-7), 171.2 (1C, C-6), 158.8 (1C, C-4a), 154.2 (1C, C-8), 153.2 (1C, C-1), 151.5 (1C, C-3), 147.6 (1C, C-10a), 140.3 (1C, C-2′), 138.4 (1C, C-6′), 132.9 (1C, C-4′), 131.6 (1C, C-3′), 128.9 (1C, C-5′), 128.3 (1C, C-9), 126.9 (1C, C-1′), 120.9 (1C, C-6a), 105.9 (1C, C-10b), 32.7 (1C, 6-CH2CH3), 30.2 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 12.1 (1C, 6-CH2CH3); HRMS m/z 441.05521 (calcd. for C21H16ClN3O4S [M]+, 441.05500); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 9:14:2; Yield: 82%.
6-Ethyl-8-((2′-bromophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (7): orange solid; m.p. 208.3 (d) °C; IR (KBr), ῡ = 1660; 1688 C=O (quinone); 1730 C=O (uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.79 (d, J = 7.8 Hz, 1H, H-3′), 7.65 (d, J = 7.5 Hz, 1H, H-6′), 7.44 (dt, J = 7.6 Hz, J =1.1 Hz, 1H, H-4′ or H-5′), 7.39 (dt, J = 7.6 Hz, J =1.1 Hz, 1H, H-5′ or H-4′), 6.05 (s, 1H, 9-H), 3.75 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.42 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.39 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.2 (1C, C-10), 180.7 (1C, C-7), 170.9 (1C, C-6), 158.5 (1C, C-4a), 154.1 (1C, C-8), 152.9 (1C, C-1), 151.2 (1C, C-3), 147.3 (1C, C-10a), 138.0 (1C, C-6′), 134.7 (1C, C-3′), 132.5 (1C, C-4′), 130.8 (1C, C-2′), 129.3 (1C, C-5′), 128.9 (1C, C-1′), 127.9 (1C, C-9), 120.7 (1C, C-6a), 105.6 (1C, C-10b), 31.8 (1C, 6-CH2CH3), 30.3 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 485.00455 (calcd. for C21H16BrN3O4S [M]+, 485.00449); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 9:12:1, Yield: 82%.
6-Ethyl-8-((3′-methylphenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (8): orange solid; m.p. 162.0–163.0 °C; IR (KBr), ῡ =1561(C=O quinone), 1661; 1682 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.40–7.36 (m, 1H, H-4′), 7.32–7.30 (m, 3H, H-2′, H-5′ and H-6′), 6.18 (s, 1H, H-9), 3.74 (s, 3H, 2-NCH3), 3.42 (s, 3H, 4-NCH3), 3.41 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 2.40 (s, 3H, 3′-CH3), 1.36 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.5 (1C, C-10), 180.9 (1C, C-7), 170.8 (1C, C-6), 158.5 (1C, C-4a), 157.0 (1C, C-8), 152.8 (1C, C-1), 151.2 (1C, C-3), 147.4 (1C, C-10a), 140.6 (1C, C-3′), 136.2 (1C, C-2′), 132.7 (1C, C-6′), 131.6 (1C, C-5′), 130.3 (1C, C-4′), 128.0 (1C, C-9), 126.9 (1C, C-1′), 120.7 (1C, C-6a), 105.5 (1C, C-10b), 31.8 (1C, 6-CH2CH3), 30.3 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 21.4 (1C, 3′-CH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 421.10960 (calcd. for C22H19N3O4S [M]+, 421.10963); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:3:1; Yield: 47%.
6-Ethyl-8-((3′-methoxyphenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (9): orange solid; m.p. 179.5–180.5 °C; IR (KBr), ῡ = 1560, 1579 (C=O quinone), 1676 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.40 (t, J = 7.7 Hz, 1H, H-5′), 7.10 (d, 1H, H-4′), 7.03 (d, 2H, H-2′ and H-6′), 6.21 (s, 1H, H-9), 3.83 (s, 3H, 3′-OCH3), 3.74 (s, 3H, 2-NCH3), 3.42 (s, 3H, 4-NCH3), 3.40 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.36 (t, J = 7.2 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.4 (1C, C-10), 180.8 (1C, C-7), 170.7 (1C, C-6), 160.8 (1C, C-3′), 158.4 (1C, C-4a), 156.7 (1C, C-8), 152.8 (1C, C-1), 151.1 (1C, C-3), 147.4 (1C, C-10b), 131.3 (1C, C-5′), 128.2 (1C, C-9), 128.0 (1C, C-1′), 127.8 (1C, C-6′), 120.7 (1C, C-2′), 120.6 (1C, C-6a), 116.7 (1C, C-4′), 105.5 (1C, C-10b), 55.6 (1C, 3′-OCH3), 31.8 (1C, 6-CH2CH3), 30.3 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 437.10449 (calcd. for C22H19N3O5S [M]+, 437.10454); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 3:3:1, Yield 66%.
6-Ethyl-8-((3′-fluorophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (10): orange solid; m.p. 170–171 °C; IR (KBr), ῡ = 1563 (C=O quinone); 1667; 1683 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.51 (q, J = 7.5 Hz, 1H, H-5′), 7.35 (d, J = 7.6 Hz, 1H, H-2′), 7.30–7.23 (m, 2H, H-4′ and H-6′), 6.21 (s, 1H, H-9), 3.76 (s, 3H, 2-NCH3), 3.44 (s, 3H, 4-NCH3), 3.42 (q, J = 7.2 Hz, 2H, 6-CH2CH3), 1.37 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.3 (1C, C-10); 180.6 (1C, C-7), 170.9 (1C, C-6), 163.3 (1C, d, J = 251.9, C-3′), 158.4 (1C, C-4a), 155.9 (1C, C-8), 152.9 (1C, C-1), 151.1 (1C, C-3), 147.2 (1C, C-10b), 131.9 (1C, d, J = 8.2, C-5′), 131.6 (1C, d, J = 3.2, C-6′), 129.3 (1C, d, J = 7.6, C-1′), 128.1 (1C, C-9), 122.7 (1C, d, J = 22.1, C-2′), 120.5 (1C, C-6a), 118.16 (1C, d, J = 20.9, C-4′), 105.5 (1C, C-10a), 31.8 (1C, 6-CH2CH3), 30.3 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 425.08457 (calcd. for C21H16FN3O4S [M]+, 425.08455); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:6:1, Yield 71%.
6-Ethyl-8-((3′-chlorophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (11): orange solid; m.p. 156.1–157.1 °C; IR (KBr), ῡ = 1558 (C=O quinone), 1662, 1681 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.54 (s, 1H, H-2′), 7.50 (d, J = 7.1 Hz, 1H, H-4′), 7.47–7.42 (m, 2H, H-6′ and H-5′), 6.19 (s, 1H, H-9), 3.74 (s, 3H, 2-NCH3), 3.42 (s, 3H, 4-NCH3), 3.40 (q, J = 7.4 Hz, 2H, 6-CH2CH3), 1.36 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.3 (1C, C-10), 180.6 (1C, C-7), 170.9 (1C, C-6), 158.4 (1C, C-4a), 155.8 (1C, C-8), 152.9 (1C, C-1), 151.1 (1C, C-3), 147.2 (1C, C-10a), 136.1 (1C, C-3′), 135.5 (1C, C-2′), 133.9 (1C, C-5′), 131.5 (1C, C-4′), 131.1 (1C, C-6′), 129.1 (1C, C-1′), 128.2 (1C, C-9), 120.5 (1C, C-6a), 105.5 (1C, C-10b), 31.8 (1C, 6-CH2CH3), 30.3 (1C, 2-NCH3), 29.1 (1C, 4-NCH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 441.05514 (calcd. for C21H16ClN3O4S [M]+, 441.05500); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:3:1, Yield: 58%.
6-Ethyl-8-((3′-bromophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (12): orange solid; m.p. 138.3–139.3°C; IR (KBr), ῡ = 1559 C=O (quinone); 1668 C=O (uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.69 (s, 1H, H-2′), 7.66 (d, J = 8.0 Hz, 1H, H-6′), 7.48 (d, J = 7.6 Hz, 1H, H-4′), 7.39 (t, J = 7.8 Hz, 1H, H-5′), 6.15 (s, 1H, H-9), 3.75 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.40 (q, J = 7.4 Hz, 2H, 6-CH2CH3), 1.36 (t, J = 7.3 Hz, 3H, 6-CH2CH3). 13C-NMR (CDCl3, 100 MHz) δ 181.3 (1C, C-10), 180.6 (1C, C-7), 170.9 (1C, C-6), 158.4 (1C, C-4a), 155.9 (1C, C-8), 152.9 (1C, C-1), 151.1 (1C, C-3), 147.2 (1C, C-10a), 138.3 (1C, C-2′), 134.4 (1C, C-4′), 134.0 (1C, C-6′), 131.8 (1C, C-5′), 129.5 (1C, C-3′), 128.2 (1C, C-9), 124.0 (1C, C-1′), 120.5 (1C, C-6a), 105.5 (1C, C-10b), 31.8 (1C, 6-CH2CH3), 30.3 (1C, 2-NCH3), 29.2 (1C, 4-NCH3), 12.2 (1C, 6-CH2CH3); HRMS m/z 485.00453 (calcd. for C21H16BrN3O4S [M]+, 485.00449); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:8:1, Yield: 82%.
6-Ethyl-8-((4′-methylphenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (13): orange solid; m.p. 191.0–192.3 °C; IR (KBr), ῡ = 1662; 1687 (C=O quinone); 1726 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.40 (d, J = 8.1 Hz, 2H, H-2′ and H-6′), 7.31 (d, J = 7.9 Hz, 2H, H-3′ and H-5′), 6.17 (s, 1H, H-9), 3.75 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.42 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 2.43 (s, 3H, 4′-CH3), 1.37 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.8 (1C, C-10), 181.3 (1C, C-7), 171.1 (1C, C-6), 158.8 (1C, C-4a), 157.5 (1C, C-8), 153.5 (1C, C-1), 153.1 (1C, C-3), 147.8 (1C, C-10a), 141.6 (1C, C-4′), 136.0 (2C, C-2′ and C-6′), 131.6 (2C, C-3′ and C-5′), 128.2 (1C, C-9), 123.9 (1C, C-1′), 121.0 (1C, C-6a), 105.8 (1C, C-10b), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 21.8 (1C, 4′-CH3), 12.6 (1C, 6-CH2CH3); HRMS m/z 421.10954 (calcd. for C22H19N3O4S [M]+, 421.10963); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:3:1, Yield: 88%.
6-Ethyl-8-((4′-methoxyphenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (14): red solid; m.p. 198.9–201.5 °C; IR (KBr), ῡ = 1662, 1689 (C=O quinone), 1726 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.42 (d, J = 8.8 Hz, 2H, H-2′ and H-6′), 7.01 (d, J = 8.8 Hz, 2H, H-3′ and H-5′), 6.15 (s, 1H, H-9), 3.86 (s, 3H, 4′-OCH3), 3.75 (s, 3H, 2-NCH3), 3.43 (s, 3H, 4-NCH3), 3.40 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 1.36 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.4 (1C, C-10), 181.1 (1C, C-7), 171.1 (1C, C-6), 162.0 (1C, C-4′), 158.8 (1C, C-4a), 157.9 (1C, C-8), 153.1 (1C, C-1), 151.5 (1C, C-3), 147.8 (1C, C-10a), 138.0 (2C, C-2′ and C-6′), 128.2 (1C, C-9), 121.0 (1C, C-6a), 117.6 (1C, C-1′), 116.5 (2C, C-3′ and C-5′), 105.8 (1C, C-10b), 56.0 (1C, 4′-OCH3), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.6 (1C, 6-CH2CH3); HRMS m/z 437.10454 (calcd. for C22H19N3O5S [M]+, 437.10454); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 3:3:1; Yield: 82%.
6-Ethyl-8-((4′-fluorophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (15): orange solid; m.p. 194.9–195.4 °C; IR (KBr), ῡ = 1660; 1675 (C=O quinone); 1720 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.50–7.47 (m, 2H, H-3′ and H-5′), 7.19–7.15 (m, 2H, H-2′ and H-6′), 6.16 (s, 1H, H-9), 3.77 (s, 3H, 2-NCH3), 3.39 (s, 3H, 4-NCH3), 3.37 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 1.38 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.7 (1C, C-10), 181.1 (1C, C-7), 171.2 (1C, C-6), 164.6 (1C, d, J = 251, C-4′), 158.8 (1C, C-4a), 156.9 (1C, C-8), 153.2 (1C, C-1), 151.5 (1C, C-3), 147.6 (1C, C-10a), 138.3 (2C, d, J = 8, C-2′ and C-6′), 128.3 (1C, C-9), 122.9 (1C, C-1′), 120.9 (1C, C-6a), 118.2 (2C, d, J = 22, C-3′ and C-5′), 105.8 (1C, C-10b), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.5 (1C, 6-CH2CH3); HRMS m/z 425.08462 (calcd. for C21H16FN3O4S [M]+, 425.08455); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:6:1, Yield: 61%.
6-Ethyl-8-((4′-chlorophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (16): orange solid; m.p. 196.5–198.3 °C; IR (KBr), ῡ = 1656; 1675 (C=O quinone); 1722 (C=O uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.51–7.46 (m, 4H, H-2′, H-3′, H-5′ and H-6′), 6.17 (s, 1H, H-9), 3.76 (s, 3H, 2-NCH3), 3.44 (s, 3H, 4-NCH3), 3.41 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 1.37 (t, J = 7.3 Hz, 3H, 6-CH2CH3); 13C-NMR (CDCl3, 100 MHz) δ 181.6 (1C, C-10), 180.7 (1C, C-7), 171.2 (1C, C-6), 158.7 (1C, C-4a), 156.4 (1C, C-8), 153.1 (1C, C-1), 151.4 (1C, C-3), 147.6 (1C, C-10a), 137.8 (1C, C-4′), 137.3 (2C, C-2′ and C-6′), 131.1 (2C, C-3′ and C-5′), 128.4 (1C, C-9), 126.0 (1C, C-1′), 120.8 (1C, C-6a), 105.8 (1C, C-10b), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.5 (1C, 6-CH2CH3); HRMS m/z 441.05491 (calcd. for C21H16ClN3O4S [M]+, 441.05500); purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:3:1, Yield: 75%.
6-Ethyl-8-((4′-bromophenyl)thio)-2,4-dimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (17): yellow solid; m.p. 197.9–198.7 °C; IR (KBr), ῡ = 1660; 1677 C=O (quinone); 1722 C=O (uracil) cm−1; 1H-NMR (CDCl3, 400 MHz) δ 7.65 (d, J = 8.4 Hz, 2H, H-3′ and H-5′), 7.40 (d, J = 8.4 Hz, 2H, H-2′ and H-6′), 6.18 (s, 1H, H-9), 3.75 (s, 3H, 2-NCH3), 3.44 (s, 3H, 4-NCH3), 3.41 (q, J = 7.3 Hz, 2H, 6-CH2CH3), 1.36 (t, J = 7.3 Hz, 3H, 6-CH2CH3). 13C-NMR (CDCl3, 100 MHz) δ 181.6 (1C, C-10), 181.0 (1C, C-7), 171.2 (1C, C-6), 158.8 (1C, C-4a), 156.3 (1C, C-8), 153.2 (1C, C-1), 151.4 (1C, C-3), 147.5 (1C, C-10a), 137.6 (2C, C-2′ and C-6′), 134.1 (2C, C-3′ and C-5′), 128.4 (1C, C-9), 126.4 (1C, C-1′), 126.7 (1C, C-4′), 120.8 (1C, C-6a), 105.8 (1C, C-10b), 32.1 (1C, 6-CH2CH3), 30.6 (1C, 2-NCH3), 29.5 (1C, 4-NCH3), 12.5 (1C, 6-CH2CH3). HRMS m/z 485.00438 (calcd. for C21H16BrN3O4S [M]+, 485.00449), purified in column chromatography with dichloromethane: light petroleum: ethyl acetate = 1:8:1, Yield: 68%.
3.3. Crystalography
3.3.1. Preparation of Single Crystals
Single crystals were grown by solvent evaporation at room temperature from the products of synthesis. Crystals suitable for X-ray diffraction studies were obtained from crystallization in saturated solutions: tetrahydropyran for 7 and benzene for 16.
3.3.2. Single Crystal X-ray Diffraction
Measured crystals were prepared under inert conditions and immersed in perfluoropolyether as a protective oil for manipulation. Suitable crystals were mounted on MiTeGen MicromountsTM, and these samples were used for data collection. Data were collected with a D8 Venture diffractometer CuKα, 298 K (Bruker, Karlsruhe, Germany). The data were processed with the APEX3 program [40] and corrected for absorption using SADABS [41]. The structures were resolved using direct methods [42], which revealed the position of all nonhydrogen atoms (Figures S1 and S2). These atoms were refined on F2 by a full-matrix least-squares procedure using anisotropic displacement parameters. All hydrogen atoms were located in difference Fourier maps and were included as fixed contributions riding on attached atoms with isotropic thermal displacement parameters 1.2 (C–H) or 1.5 (methyl) times those of the respective atom. CCDC 1573897 (compound 7) and 1573896 (compound 16) contain the crystallographic data listed in Table S2. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre.
3.4. Evaluation of Antibacterial Activity and Cytotoxicity
3.4.1. Bacterial Strains
As an initial screen for antibacterial activity, the compounds were tested against the following strains: methicillin-resistant Staphylococcus aureus (MRSA) ATCC® 43300, methicillin-susceptible Staphylococcus aureus (MSSA) ATCC® 29213, Enterococcus faecalis ATCC® 29212, Escherichia coli ATCC® 25922, and Pseudomonas aeruginosa ATCC® 27853. The ISP provided the clinical isolates of MRSA [31] and VREF [32] used to calculate the MIC and MBC values of the most potent compounds. MRSA and VREF isolates that were resistant to at least three additional classes of antimicrobial agents were defined as multidrug resistant (MDR) [43]. All isolates were collected in 2014 and were obtained from various Chilean hospitals throughout the country (Table S1, Supplementary Material).
3.4.2. Evaluation of Antibacterial Activity
Minimal Inhibitory Concentration Determination
MIC values were determined using a broth microdilution method, according to recommendations of the Clinical and Laboratory Standards Institute (CLSI) [44]. VAN and GEN were also tested against the strains and the results compared to the MIC ranges reported by the CLSI, as a quality control measure [23]. All compounds tested were dissolved in dimethyl sulfoxide (DMSO), to levels not exceeding 1% per well. MIC50 and MIC90 values were then calculated for the compounds observed to be the most active against the clinical isolates studied were also determined; MIC50 and MIC90 are the concentrations that inhibit 50% and 90% of the tested isolates, respectively. All assays were performed in triplicate and with n = 5.
Minimal bactericidal concentration determination
MBC values were then calculated for the compounds observed to be the most active against clinical isolates, following the recommendations of Pearson et al. [45], using an inoculum of 5 × 105 CFU/mL. All wells with no visible growth observed in the microdilution assay were subcultured, extracting a 10-µL volume. Finally, MBC was determined based on the corresponding subcultured dilution plate in which the growth of microorganisms was less than 0.01% of the initial inoculum of 5 × 105 CFU/mL. Compounds were classified as bactericides if the MBC/MIC ratio was equal to or less than 2 and as bacteriostatic if the ratio was greater than 2, according to Craig et al. [46] and Taylor et al. [47]. All assays were performed in triplicate and with n = 5.
3.4.3. Cell Cultures
The cell lines used were obtained from the ATCC® (Manassas, VA, USA) and included HeLa, human cervix adenocarcinoma (ATCC® CCL-2); HTC-116, human colorectal carcinoma (ATCC® CCL-247); SHSY-5Y, human neuroblastoma (ATCC® CRL-2266); and Vero, monkey kidney fibroblast (ATCC® CCL-81). Cells were grown in RPMI 1640 culture medium supplemented with 10% fetal bovine serum, 100 IU/mL penicillin, and 100 mg/mL streptomycin, in a humidified incubator in air with 5% CO2 at 37 °C.
3.4.4. Evaluation of Cellular Toxicity
Cytotoxicity assays were performed using the MTS reduction method, as described previously [48]. Briefly, cancer cell lines were plated in a flat-bottom 96-well plate at 40,000 cells/mL. The cells were then incubated with test agents and VAN at escalating doses, ranging from 0 to 20 µg/mL, in triplicate in 200 µL of RPMI 1640 supplemented culture medium at 37 °C for 24 h. Five microliters of MTS was added to achieve a final concentration of 0.5 mg/mL and incubated at 37 °C for 2 h. Finally, formazan formation was measured in a multi-well reader at 540 nm (Stat Fax 4200, Awareness Technology, Inc., Palm City, FL, USA). The compounds were dissolved in DMSO, at levels not exceeding 1% per well. All assays were performed in triplicate and with n = 5. As a positive control for growth, wells containing only RPMI 140 medium were used. As a negative control of growth, H2O2 was used. As an additional negative control for growth, H2O + 1% DMSO was used. VAN and GEN were used as drug references.
3.5. Electrochemical Measuring
Cyclic voltammetry: Half-wave potentials (E1/2) were determined using a CHI 650 potentiostat (CH Instruments, Inc., 3700 Tennison Hill Drive Austin, TX, USA). Three-electrode cells with Ag0/AgCl or platinum wires were used as reference and auxiliary electrodes, respectively. The working electrode was prepared by polishing a glassy carbon electrode with 0.3 μm and 0.05 μm alumina and then washing with abundant deionized water. The compounds were dissolved to a concentration of 0.1 M with acetonitrile as a solvent and tetrabutylammonium perchlorate as a support electrolyte.
3.6. Determination of Theoretical Physicochemical Parameters
The lipophilic (LogP), and molar refractivity (MR) parameters were determined for each molecule using ChemDraw Ultra 12.0 (CambridgeSoft, Cambridge, MA, USA, www.cambridgesoft.com).
3.7. Statistical Analysis
The data were analyzed with one-way ANOVA and t-tests, with the criterion for statistical significance set at p < 0.05, using the GraphPad Prism 5.03 program (GraphPad Software, Inc., San Diego, California, USA, www.graphpad.com).
4. Conclusions
We synthesized 17 novel quinone compounds by employing a simple, fast, and economical two-step method with a yield of 47–88% yield. Antibacterial screening showed the compounds to have activity against Gram-positive MRSA, MSSA, and E. faecalis strains but not against Gram-negative E.coli and P. aeruginosa.
Structural analysis showed that the presence of a thiophenolic ring is essential for activity. On the other hand, a study of the LogP, E1/2, and MR parameters showed that the addition of bulky substituents in the ortho position or the addition of lipophilic substituents in the para position improves the antibacterial activity. The results suggest that the antibacterial activity would be a function of more complex parameters that require extensive analysis.
Compounds 7 and 16 were 128- and 64-fold more active against clinical isolates of VREF, respectively, than the drug VAN and did not affect the viability of HeLa, HTC-116, SHSY-5Y, or Vero cells in toxicity assays, demonstrating selectivity for prokaryotic cells. From a drug development point of view, the results of the new scaffold described herein support continuation of preclinical development to guide the synthesis of more potent agents to provide therapeutic options against infectious diseases provoked by Gram-positive MDR strains.
5. Patents
Chilean Patent Application number 201503780, PCT/CL2016/050080, EEUU 16/067,033; EPO 16880235.3; MX/a/2018/008192 titled: “Pyrimidine-Isoquinoline-Quinone Derived Compounds, their Salts, Isomers, Pharmaceutically Acceptable Tautomers; Pharmaceutical Composition; Preparation Procedure; and their Use in the Treatment of Bacterial and Multi-Resistant Bacterial Diseases”.
Supplementary Materials
The following are available online at http://www.mdpi.com/1420-3049/23/7/1776/s1, Table S1. Categorization of isolates by specimen type and multi drug-resistant patterns, Table S2. Crystal data and refinement details for compounds 7 and 16, Figure S1. Molecular structure of 7 showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level. Figure S2. Molecular structure of 16 showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Author Contributions
Conceptualization, J.C.-S. and D.V.-V.; Formal analysis, G.G.R., S.B.D., P.A., F.S., R.V., J.M. and D.V.-V.; Investigation, J.C.-S., J.A.-L., D.C.-L., M.F., E.S.-H., M.K., G.R.G. and D.V.-V.; Methodology, G.G.R., D.C.-L., S.B.D., M.F., F.S., M.K. and J.M.; Resources, G.G.R., P.A. and R.V.; Visualization, J.M.; Writing—original draft, J.C.-S., J.A.-L. and D.V.-V.; Writing—review & editing, J.C.-S., J.A.-L. and D.V.-V.
Funding
This research was funded by FONDECYT grant numbers 11110516 and 79100006.
Acknowledgments
We thanks the FONDECYT, Iniciación en Investigación Grant No. 11110516, Chile; and FONDECYT, Proyecto de Inserción Grant No. 79100006, Chile, and Programa de Estímulo a la Excelencia Institucional PEEI 2017, Universidad de Chile. The authors, J.C.S. and J.A.L. thanks to CONICYT Beca Doctorado Nacional No. 21130643 and 21130628, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014. [Google Scholar]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No eskape! An update from the infectious diseases society of america. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017; p. 7. [Google Scholar]
- CDC-Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/index.html (accessed on 10 July 2018).
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; p. 232. [Google Scholar]
- Werner, G.; Coque, T.M.; Hammerum, A.M.; Hope, R.; Hryniewicz, W.; Johnson, A.; Klare, I.; Kristinsson, K.G.; Leclercq, R.; Lester, C.H.; et al. Emergence and spread of vancomycin resistance among enterococci in europe. Euro Surveill. 2008, 13, 5437–5453. [Google Scholar]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in us hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Moir, D.T.; Opperman, T.J.; Butler, M.M.; Bowlin, T.L. New classes of antibiotics. Curr. Opin. Pharmacol. 2012, 12, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.J.; Vazquez-Rodriguez, S.; Santana, L.; Uriarte, E.; Fuentes-Edfuf, C.; Santos, Y.; Munoz-Crego, A. Looking for new targets: Simple coumarins as antibacterial agents. Med. Chem. 2012, 8, 1140–1145. [Google Scholar] [PubMed]
- Pucci, M.J. Novel genetic techniques and approaches in the microbial genomics era: Identification and/or validation of targets for the discovery of new antibacterial agents. Drugs R D 2007, 8, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Gordaliza, M. Synthetic strategies to terpene quinones/hydroquinones. Mar. Drugs 2012, 10, 358–402. [Google Scholar] [CrossRef] [PubMed]
- Lown, J.W. The mechanism of action of quinone antibiotics. Mol. Cell. Biochem. 1983, 55, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Ray, R.; Hazra, B. Antitubercular and antibacterial activity of quinonoid natural products against multi-drug resistant clinical isolates. Phytother. Res. 2014, 28, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Amani, A.M. Synthesis, characterization and antibacterial and antifungal evaluation of some para-quinone derivatives. Drug Res. 2014, 64, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, V.P.; Assimopoulou, A.N.; Couladouros, E.A.; Hepworth, D.; Nicolaou, K.C. The chemistry and biology of alkannin, shikonin, and related naphthazarin natural products. Angewandte Chem. Int. Ed. 1999, 38, 270–301. [Google Scholar] [CrossRef]
- Kubo, A.; Nakahara, S.; Inaba, K.; Kitahara, Y. Synthesis of renierone, 7-methoxy-1,6-dimethyl-5,8-dihydroisoquinoline-5,8-dione and N-formyl-1,2-dihydrorenierone, antimicrobial metabolites from a marine sponge, reniera sp. Chem. Pharm. Bull. 1986, 34, 4056–4068. [Google Scholar] [CrossRef] [PubMed]
- Swapnaja, K.J.; Yennam, S.; Chavali, M.; Poornachandra, Y.; Kumar, C.G.; Muthusamy, K.; Jayaraman, V.B.; Arumugam, P.; Balasubramanian, S.; Sriram, K.K. Design, synthesis and biological evaluation of diaziridinyl quinone isoxazole hybrids. Eur. J. Med. Chem. 2016, 117, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Subramani, R.; Aalbersberg, W. Three bioactive sesquiterpene quinones from the fijian marine sponge of the genus hippospongia. Nat. Prod. Res. 2013, 27, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Yadav, D.B.; Singh, R.V.; Chaturvedi, A.K.; Shukla, P.K. Synthesis and biological evaluation of novel (l)-alpha-amino acid methyl ester, heteroalkyl, and aryl substituted 1,4-naphthoquinone derivatives as antifungal and antibacterial agents. Bioorg. Med. Chem. Lett. 2005, 15, 5324–5328. [Google Scholar] [CrossRef] [PubMed]
- Tandon, V.K.; Yadav, D.B.; Singh, R.V.; Vaish, M.; Chaturvedi, A.K.; Shukla, P.K. Synthesis and biological evaluation of novel 1,4-naphthoquinone derivatives as antibacterial and antiviral agents. Bioorg. Med. Chem. Lett. 2005, 15, 3463–3466. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992, 56, 395–411. [Google Scholar] [PubMed]
- Neilands, J.B. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.B.; Cockerill, F.R., III; Bradford, P.A.; Eliopoulos, G.M.; Hindler, J.A.; Jenkins, S.G.; Lewis, J.S., II; Limbago, B.; Nicolau, D.P.; Powell, M.; et al. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Clin. Lab. Stand. Inst. 2015, 35, M100-S25. [Google Scholar]
- Verrax, J.; Beck, R.; Dejeans, N.; Glorieux, C.; Sid, B.; Pedrosa, R.C.; Benites, J.; Vasquez, D.; Valderrama, J.A.; Calderon, P.B. Redox-active quinones and ascorbate: An innovative cancer therapy that exploits the vulnerability of cancer cells to oxidative stress. Anti-Cancer Agents Med. Chem. 2011, 11, 213–221. [Google Scholar] [CrossRef]
- Brunmark, A.; Cadenas, E. Redox and addition chemistry of quinoid compounds and its biological implications. Free Radic. Biol. Med. 1989, 7, 435–477. [Google Scholar] [CrossRef]
- Hammett, L.P. Some relations between reaction rates and equilibrium constants. Chem. Rev. 1935, 17, 125–136. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Q.; Zhang, R.; He, W.; Ma, X.; Zhang, J.; Xia, F.; Zhao, F.; Cao, J.; Liu, Y.; et al. In vitro antimicrobial activity of the novel oxazolidinone tedizolid and comparator agents against staphylococcus aureus and linezolid-resistant Gram-positive pathogens: A multicentre study in china. Int. J. Antimicrob. Agents 2014, 44, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Prokocimer, P.; Bien, P.; Deanda, C.; Pillar, C.M.; Bartizal, K. In vitro activity and microbiological efficacy of tedizolid (TR-700) against Gram-positive clinical isolates from a phase 2 study of oral tedizolid phosphate (TR-701) in patients with complicated skin and skin structure infections. Antimicrob. Agents Chemother. 2012, 56, 4608–4613. [Google Scholar] [CrossRef] [PubMed]
- Sahm, D.F.; Deane, J.; Bien, P.A.; Locke, J.B.; Zuill, D.E.; Shaw, K.J.; Bartizal, K.F. Results of the surveillance of tedizolid activity and resistance program: In vitro susceptibility of Gram-positive pathogens collected in 2011 and 2012 from the united states and europe. Diagn. Microbiol. Infect. Dis. 2015, 81, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Schaadt, R.; Sweeney, D.; Shinabarger, D.; Zurenko, G. In vitro activity of TR-700, the active ingredient of the antibacterial prodrug TR-701, a novel oxazolidinone antibacterial agent. Antimicrob. Agents Chemother. 2009, 53, 3236–3239. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K.S.; Goering, R.V. Activity of tedizolid (TR-700) against well-characterized methicillin-resistant staphylococcus aureus strains of diverse epidemiological origins. Antimicrob. Agents Chemother. 2013, 57, 2892–2895. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Baguneid, M.; Bouza, E.; Dryden, M.; Nathwani, D.; Wilcox, M. European perspective and update on the management of complicated skin and soft tissue infections due to methicillin-resistant staphylococcus aureus after more than 10 years of experience with linezolid. Clin. Microbiol. Infect. 2014, 20 (Suppl. 4), 3–18. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Poppe, S.; Schaadt, R.; Brown-Driver, V.; Finn, J.; Pillar, C.M.; Shinabarger, D.; Zurenko, G. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob. Agents Chemother. 2008, 52, 4442–4447. [Google Scholar] [CrossRef] [PubMed]
- WA, C. Post antibiotic effect. In Antibiotics in Laboratory Medicine, 2nd ed.; Victor, L., Ed.; Williams and Wilkins: Baltimore, MD, USA, 1996; pp. 296–329. [Google Scholar]
- Valderrama, J.A.; Gonzalez, M.F.; Colonelli, P.; Vasquez, D. Design and synthesis of angucyclinone 5-aza analogues. Synlett 2006, 2777–2780. [Google Scholar] [CrossRef]
- Valderrama, J.A.; Colonelli, P.; Vasquez, D.; Gonzalez, M.F.; Rodriguez, J.A.; Theoduloz, C. Studies on quinones. Part 44: Novel angucyclinone n-heterocyclic analogues endowed with antitumoral activity. Bioorg. Med. Chem. 2008, 16, 10172–10181. [Google Scholar] [CrossRef] [PubMed]
- Vasquez-Velasquez, D.R. Diseño, Síntesis y Evaluación Antitumoral de Aza-Análogos de Anguciclinonas y Derivados de Aminopirimidoisoquinolinquinonas. Ph.D Thesis, Pontificia Universidad Católica de Chile, Santiago, Chile, 2009. [Google Scholar]
- Vasquez, D.; Rodriguez, J.A.; Theoduloz, C.; Calderon, P.B.; Valderrama, J.A. Studies on quinones. Part 46. Synthesis and in vitro antitumor evaluation of aminopyrimidoisoquinolinequinones. Eur. J. Med. Chem. 2010, 45, 5234–5242. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, J.A.; Ibacache, A.; Rodriguez, J.A.; Theoduloz, C.; Benites, J. Studies on quinones. Part 47. Synthesis of novel phenylaminophenanthridinequinones as potential antitumor agents. Eur. J. Med. Chem. 2011, 46, 3398–3409. [Google Scholar] [CrossRef] [PubMed]
- Francart, T.; van Wieringen, A.; Wouters, J. Apex 3: A multi-purpose test platform for auditory psychophysical experiments. J. Neurosci. Methods 2008, 172, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. SADABS-2012/1. Bruker/Siemens Area Detector Absorption Correction Program; Bruker AXS Inc.: Madison, WI, USA, 2012. [Google Scholar]
- Sheldrick, G. A short history of shelx. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.E.; Sader, H.S.; Flamm, R.K.; Farrell, D.J.; Jones, R.N. Telavancin in vitro activity against a collection of methicillin-resistant staphylococcus aureus isolates, including resistant subsets, from the united states. Antimicrob. Agents Chemother. 2015, 59, 1811–1814. [Google Scholar] [CrossRef] [PubMed]
- Cockerill, F.R.; Wikler, M.A.; Alder, J.; Dudley, M.N.; Eliopoulos, G.M.; Ferraro, M.J.; Hardy, D.J.; Hecht, D.W.; Hindler, J.A.; Patel, J.B.; et al. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition. Clin. Lab. Stand. Inst. 2012, 32, M07-A9. [Google Scholar]
- Pearson, R.D.; Steigbigel, R.T.; Davis, H.T.; Chapman, S.W. Method of reliable determination of minimal lethal antibiotic concentrations. Antimicrob. Agents Chemother. 1980, 18, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Craig, W.A.; Gudmundsson, S. Antibiotics in Laboratory Medicine, 4th ed.; Lorian, V., Ed.; The Williams & Wilkins Co.: Baltimore, MD, USA, 1996; pp. 296–329. [Google Scholar]
- Taylor, P.C.; Schoenknecht, F.D.; Sherris, J.C.; Linner, E.C. Determination of minimum bactericidal concentrations of oxacillin for staphylococcus aureus: Influence and significance of technical factors. Antimicrob. Agents Chemother. 1983, 23, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).